A Semi-Automated Workflow for FAIR Maturity Indicators in the Life Sciences
Abstract
:1. Introduction
2. Materials and Methods
2.1. Use Cases in the Life Sciences
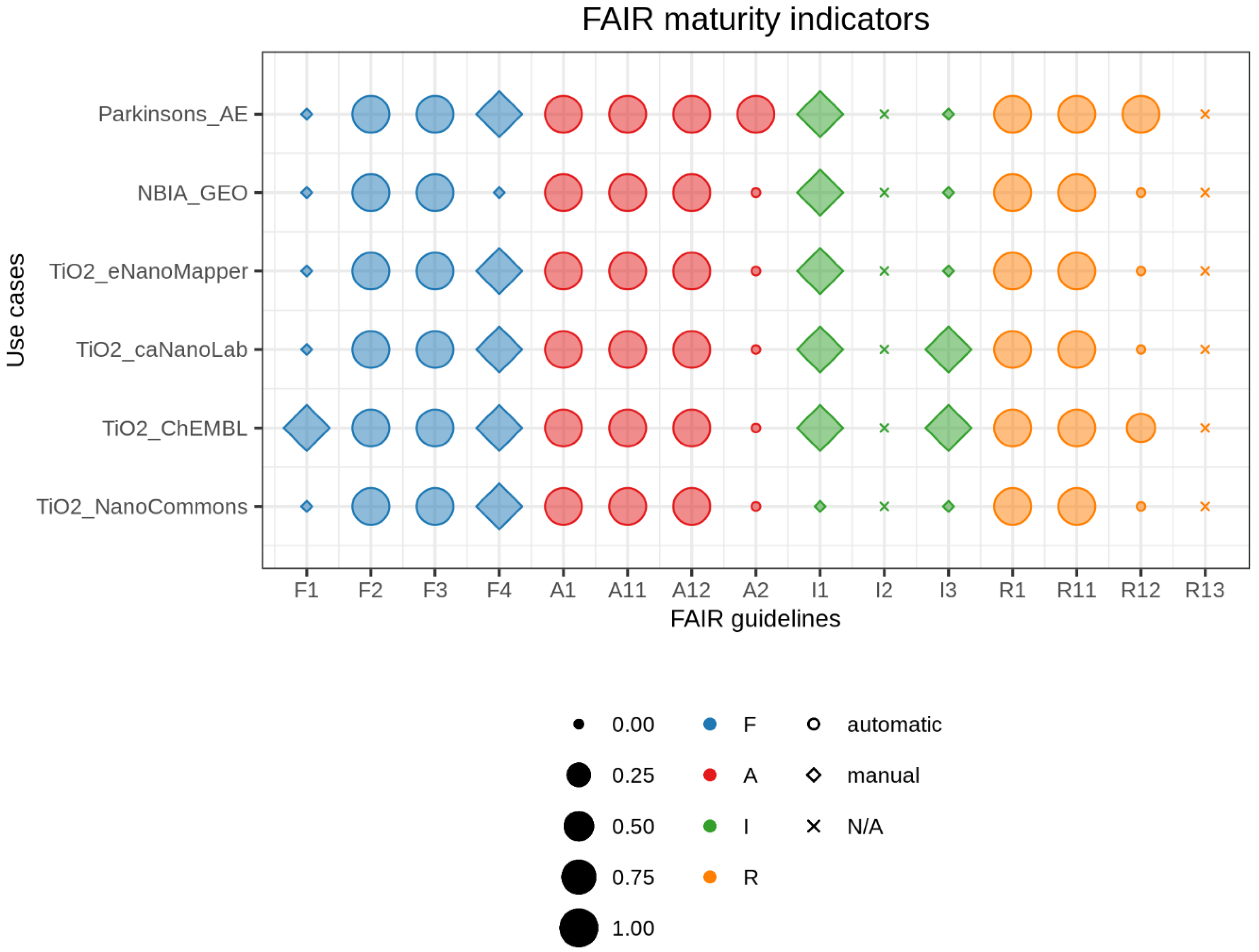
- Parkinsons_AE: What are the differentially expressed genes between normal subjects and subjects with Parkinson’s diseases in the brain frontal lobe? To answer this question, the researcher looked for a dataset in the search engine of ArrayExpress (AE), a repository for microarray gene expression data based at the European Bioinformatics Institute (EBI), United Kingdom [19];
- NBIA_GEO: What is the effect of the WDR45 gene mutation in the brain? In this case, the researcher looked for a dataset in the search engine of Gene Expression Omnibus (GEO), a repository containing gene expression and other functional genomics data hosted at the National Center for Biotechnology Information (NCBI), United States [20].
- TiO2_eNanoMapper, TiO2_caNanoLab, TiO2_ChEMBL and TiO2_NanoCommons: What can these datasets tell us about nanoscale titanium dioxide (TiO2) toxicity? Toxicological studies have shown that TiO2 nanoparticles (NPs) induce oxidative stress in cells resulting in immune response, inflammation, genotoxicity, and cell damage [21]. The researcher looked for a dataset in the search engine of four databases eNanoMapper [22], caNanoLab [23], ChEMBL [24], and NanoCommons [25] for data related to TiO2 induced cytotoxicity, immunotoxicity, genotoxicity, or oxidative stress.
2.2. What is Data and What Is Metadata?
- data: According to the Merriam-Webster online dictionary, data are “information in digital form that can be transmitted or processed” [26];
- metadata: In the Merriam-Webster online dictionary, metadata are defined as “data that provide information about other data” [27];
- (meta)data: We interpreted it as data and/or metadata. We used (meta)data as:
- ∘
- data for the principles R1, R1.1, and R1.2;
- ∘
- metadata for the principles I1 and I3;
- ∘
- data and metadata for the principles F1, F4, and A1.
- data: It is the actual dataset that researchers analyzed to answer their research question. The analysis of the dataset itself is out of the scope of this study;
- metadata: For the following principles, the corresponding metadata are:
- ∘
- F2: Information that allows researchers to find the dataset s/he looks for. It coincides with the keywords used in the search;
- ∘
- F3: Identifier of the dataset in the repository;
- ∘
- I3: Reference to other metadata;
- ∘
- R1: Information about the dataset, other than the search keywords;
- ∘
- R1.1: Data license;
- ∘
- R1.2: Data provenance as publication title, author names, and one author’s email address.
2.3. Calculating FAIR Maturity Indicators
- Data repositories (ArrayExpress, Gene Expression Omnibus, eNanoMapper, caNanoLab, ChEMBL, and NanoCommons): We programmatically queried each repository using the same keywords researchers had used in their manual query when looking for a dataset. From the obtained metadata, we retrieved information to calculate maturity indicators for the principles F2, F3, I1, I3, R1, and R1.2 (see Table 2); The aforementioned principles are assessed directly on the data repository without relying on external sources like registries as in the next point.
- Registry of repositories: We queried re3data.org, a registry containing information about more than 2000 data repositories from various disciplines. We used the retrieved information to compute the maturity indicators for the principles F1, A2, and R1.2; F1 was chosen to be assessed from an external source, because it can be performed in an automated way. re3data.org provides information on whether the data repository provides a persistent unique identifier for its data or not. Obtaining such information from the data source itself is not possible. The same goes for metadata policy (A2) and metadata provenance (R1.2): we need a registry that can be queried programmatically to obtain such information. Otherwise, data repositories can provide this information within unstructured text, and it is not feasible to find it in an automated way.
- Searchable resource: We queried Google Dataset Search, an emerging search engine specific for datasets, to quantify the principle F4, which relates to indexing of the metadata in a searchable resource.
2.4. Visualizing FAIR Maturity Indicators
3. Results
4. Discussion
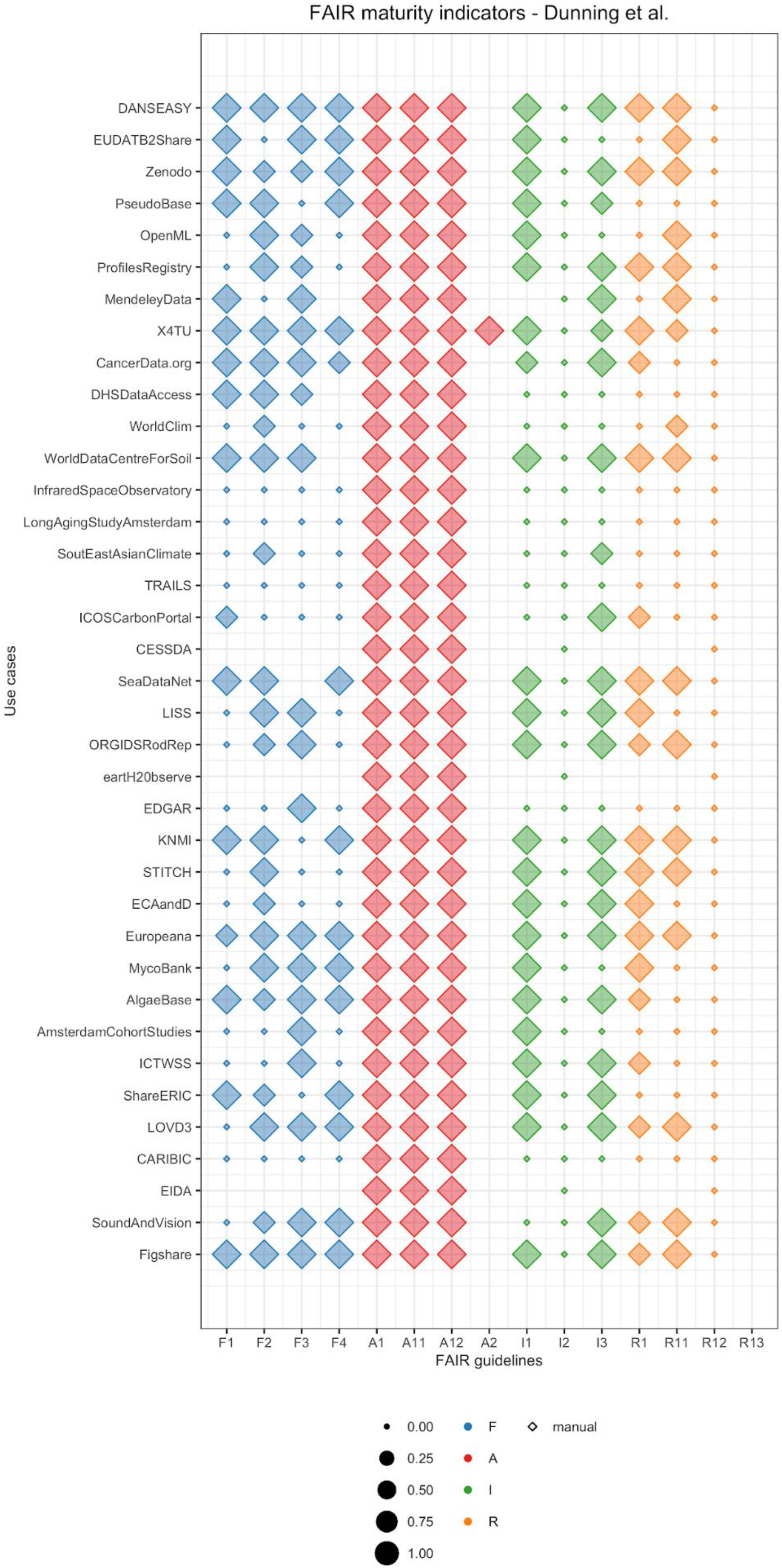
- Findability: The criteria to assess principles F1 (unique identifier), F3 (metadata includes identifier), and F4 ((meta)data are indexed) are similar for all previous studies. In our case, to assess F1 we investigated whether a repository provides a DOI in the registry re3data.org. We chose this registry because it is one of the largest registries of scientific repositories, and it provides an open API. Of course, different communities use different approaches, and FAIRSharing is an important complementary service [12]. For F3, we accepted any dataset identifier provided by the repository as the principle does not explicitly mention restrictions on the characteristics of the identifier. Finally, for F4 we looked for dataset titles in Google Dataset Search. We chose this searchable resource because it could become one of the main search engines specific for data in the future, similar to Google Scholar for publications. However, for Google Dataset Search or the newer DataCite Commons (https://commons.datacite.org/) to recognize datasets, the datasets also need semantic annotation, with, for example, schema.org. This is not tested in the current notebook. Another limitation is that, in contrast to the previous maturity indicators, the implementation of F2 (data are described with rich metadata) has large variations across literature publications. The MIAG recommends to evaluate whether metadata contains “structured” elements, Dunning et al. looked for attributes that favor findability, whereas Weber et al. used metrics of time and space of image acquisition. We followed the criteria suggested by Dunning et al. and looked for the keywords that researchers had used in their manual search to find datasets.
- Accessibility: Similar to the other published approaches, we retrieved our data using the HTTP protocol, which is free, open, and allows for authentication, and thus satisfies all the requirements of the A1 group. Additionally, there is concordance among approaches for the principle A2, which requires that a repository should explicitly provide a policy for data availability. In our implementation, we looked for the policy in re3data.org. However, for integration into research workflows, the mere use of the HTTP is a very narrow definition and choices of protocols on top of HTTP may be needed, e.g., for the authentication.
- Interoperable: Similarly to the MIAG, we assigned a positive score to metadata in a structured file format, such as XML (I1). In contrast, Dunning et al. and Weber et al. suggested that metadata should be in a standardized schema, such as Dublin Core or DataCite, which would increase data interoperability and simplify retrieval. None of the studies assessed I2 (vocabularies are FAIR), because it would require a separate implementation that includes the recursive nature of the FAIR principles. Finally, for I3 all previous studies looked for references to other datasets in metadata. Similarly to accessibility, these metrics are only a first step and not enough to link the various information sources needed to apply workflows for risk governance.
- Reusable: Although the MIAG does not provide any guidelines, the various studies implemented different ways to assess R1 (plurality of relevant attributes). While Weber et al. used the same metrics as for F2, Dunning et al. focused on metadata that provide information on how to reuse a dataset. In our implementation, we assess the presence of metadata attributes other than search keywords. The principles R1.1 (availability of data usage license) and R1.2 (data provenance) had a straight-forward implementation for all approaches. In our approach, we looked for a data license in re3data.org and for authors, author emails, and titles of the corresponding publication in the metadata from the dataset repository. Note that data would ideally be shared before publication and arguably should be shared as independent research output, in which case our implementation of R1.2 would not suffice. Finally, none of the authors evaluated whether metadata follow community standards (R1.3), as community agreements are not formally established yet. It will be clear that here too that these minimal expectations are not a sufficient requirement to ensure research output is practically useful for risk assessment.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability
References
- Curty, R.G.; Crowston, K.; Specht, A.; Grant, B.W.; Dalton, E.D. Attitudes and norms affecting scientists’ data reuse. PLoS ONE 2017, 12, e0189288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, H.H.; Dev, A.; Statham, E.; Bierer, B.E. Credit data generators for data reuse. Nature 2019, 570, 30–32. [Google Scholar] [CrossRef] [PubMed]
- Piwowar, H.A. Who Shares? Who Doesn’t? Factors Associated with Openly Archiving Raw Research Data. PLoS ONE 2011, 6, e18657. [Google Scholar] [CrossRef] [Green Version]
- Wiggins, A.; Young, A.; Kenney, M.A. Exploring visual representations to support data re-use for interdisciplinary science. Proc. Assoc. Inf. Sci. Technol. 2018, 55, 554–563. [Google Scholar] [CrossRef]
- Poole, A.H. How has your science data grown? Digital curation and the human factor: A critical literature review. Arch. Sci. 2015, 15, 101–139. [Google Scholar] [CrossRef]
- Bishop, L.; Kuula-Luumi, A. Revisiting Qualitative Data Reuse: A Decade On. SAGE Open 2017, 7, 215824401668513. [Google Scholar] [CrossRef] [Green Version]
- Vines, T.H.; Albert, A.Y.K.; Andrew, R.L.; Débarre, F.; Bock, D.G.; Franklin, M.T.; Gilbert, K.J.; Moore, J.-S.; Renaut, S.; Rennison, D.J. The Availability of Research Data Declines Rapidly with Article Age. Curr. Biol. 2014, 24, 94–97. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, A.; de Miranda Azevedo, R.; Juty, N.; Batista, D.; Coles, S.; Cornet, R.; Courtot, M.; Crosas, M.; Dumontier, M.; Evelo, C.T.; et al. FAIR Principles: Interpretations and Implementation Considerations. Data Intell. 2020, 2, 10–29. [Google Scholar] [CrossRef]
- Mons, B.; Neylon, C.; Velterop, J.; Dumontier, M.; da Silva Santos, L.O.; Wilkinson, M.D. Cloudy, increasingly FAIR; revisiting the FAIR Data guiding principles for the European Open Science Cloud. Inf. Serv. Use 2017, 37, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Schultes, E.; Strawn, G.; Mons, B. Ready, Set, GO FAIR: Accelerating Convergence to an Internet of FAIR Data and Services. In Proceedings of the Selected Papers of the XX International Conference on Data Analytics and Management in Data Intensive Domains (DAMDID/RCDL 2018), Moscow, Russia, 9–12 October 2018; Volume 2277, pp. 19–23. [Google Scholar]
- The FAIRsharing Community; Sansone, S.-A.; McQuilton, P.; Rocca-Serra, P.; Gonzalez-Beltran, A.; Izzo, M.; Lister, A.L.; Thurston, M. FAIRsharing as a community approach to standards, repositories and policies. Nat. Biotechnol. 2019, 37, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, M.D.; Sansone, S.-A.; Schultes, E.; Doorn, P.; Bonino da Silva Santos, L.O.; Dumontier, M. A design framework and exemplar metrics for FAIRness. Sci. Data 2018, 5, 180118. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, M.D.; Dumontier, M.; Sansone, S.-A.; da Silva Santos, L.O.B.; Prieto, M.; Batista, D.; McQuilton, P.; Kuhn, T.; Rocca-Serra, P.; Crosas, M.; et al. Evaluating FAIR Maturity Through a Scalable, Automated, Community-Governed Framework. Sci. Data 2019, 6, 174. [Google Scholar] [CrossRef] [Green Version]
- Clarke, D.J.B.; Wang, L.; Jones, A.; Wojciechowicz, M.L.; Torre, D.; Jagodnik, K.M.; Jenkins, S.L.; McQuilton, P.; Flamholz, Z.; Silverstein, M.C.; et al. FAIRshake: Toolkit to evaluate the FAIRness of research digital resources. Cell Syst. 2019, 9, 417–421. [Google Scholar] [CrossRef]
- Papadiamantis, A.; Klaessig, F.; Exner, T.; Hofer, S.; Hofstaetter, N.; Himly, M.; Williams, M.A.; Doganis, P.; Hoover, M.D.; Afantitis, A.; et al. Metadata stewardship in nanosafety research: Community-driven organisation of metadata schemas to support FAIR nanoscience data. Nanomaterials 2020, 10, 2033. [Google Scholar] [CrossRef]
- Dunning, A.; De Smaele, M.; Böhmer, J. Are the FAIR Data Principles fair? Int. J. Digit. Curation 2017, 12, 177–195. [Google Scholar] [CrossRef]
- Weber, T.; Kranzlmuller, D. How FAIR Can you Get? Image Retrieval as a Use Case to Calculate FAIR Metrics. In Proceedings of the IEEE 14th International Conference on e-Science (e-Science), Amsterdam, The Netherlands, 12–16 August 2018; pp. 114–124. [Google Scholar]
- Brazma, A. ArrayExpress—A public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003, 31, 68–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Skocaj, M.; Filipic, M.; Petkovic, J.; Novak, S. Titanium dioxide in our everyday life; is it safe? Radiol. Oncol. 2011, 45. [Google Scholar] [CrossRef]
- Jeliazkova, N.; Chomenidis, C.; Doganis, P.; Fadeel, B.; Grafström, R.; Hardy, B.; Hastings, J.; Hegi, M.; Jeliazkov, V.; Kochev, N.; et al. The eNanoMapper database for nanomaterial safety information. Beilstein J. Nanotechnol. 2015, 6, 1609–1634. [Google Scholar] [CrossRef] [Green Version]
- Gaheen, S.; Hinkal, G.W.; Morris, S.A.; Lijowski, M.; Heiskanen, M.; Klemm, J.D. caNanoLab: Data sharing to expedite the use of nanotechnology in biomedicine. Comput. Sci. Discov. 2013, 6, 014010. [Google Scholar] [CrossRef] [Green Version]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [Green Version]
- Afantitis, A.; Lynch, I. Driving the Nanoinformatics Wave; Zenodo: Geneva, Switzerland, 2019. [Google Scholar] [CrossRef]
- Data|Definition of Data by Merriam-Webster. Available online: https://www.merriam-webster.com/dictionary/data (accessed on 19 October 2020).
- Metadata|Definition of Metadata by Merriam-Webster. Available online: https://www.merriam-webster.com/dictionary/metadata (accessed on 19 October 2020).
- This Repository Contains the Results of the FAIR Metrics Group: FAIRMetrics/Metrics. 2019. Available online: https://github.com/FAIRMetrics/Metrics (accessed on 19 October 2020).
- Wickham, H. ggplot2; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Hegi, M.; Hardy, B.; Willighagen, E. Requirements Analysis and System Design; Zenodo: Geneva, Switzerland, 2015. [Google Scholar] [CrossRef]
- Hendren, C.O.; Powers, C.M.; Hoover, M.D.; Harper, S.L. The Nanomaterial Data Curation Initiative: A collaborative approach to assessing, evaluating, and advancing the state of the field. Beilstein J. Nanotechnol. 2015, 6, 1752–1762. [Google Scholar] [CrossRef] [PubMed]
- Lobaskin, V.; Puzyn, T.; Verheyen, G. EU US Roadmap Nanoinformatics 2030; Zenodo: Geneva, Switzerland, 2018. [Google Scholar] [CrossRef]
- Marchese Robinson, R.L.; Lynch, I.; Peijnenburg, W.; Rumble, J.; Klaessig, F.; Marquardt, C.; Rauscher, H.; Puzyn, T.; Purian, R.; Åberg, C.; et al. How should the completeness and quality of curated nanomaterial data be evaluated? Nanoscale 2016, 8, 9919–9943. [Google Scholar] [CrossRef]
- Brazma, A.; Hingamp, P.; Quackenbush, J.; Sherlock, G.; Spellman, P.; Stoeckert, C.; Aach, J.; Ansorge, W.; Ball, C.A.; Causton, H.C.; et al. Minimum information about a microarray experiment (MIAME)—Toward standards for microarray data. Nat. Genet. 2001, 29, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Rivera, S.C.; Moher, D.; Calvert, M.J.; Denniston, A.K. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: The CONSORT-AI Extension. BMJ 2020, 370, m3164. [Google Scholar] [CrossRef]
- Faria, M.; Björnmalm, M.; Thurecht, K.J.; Kent, S.J.; Parton, R.G.; Kavallaris, M.; Johnston, A.P.R.; Gooding, J.J.; Corrie, S.R.; Boyd, B.J.; et al. Minimum information reporting in bio–nano experimental literature. Nat. Nanotechnol. 2018, 13, 777–785. [Google Scholar] [CrossRef]
- Comandella, D.; Gottardo, S.; Rio-Echevarria, I.M.; Rauscher, H. Quality of physicochemical data on nanomaterials: An assessment of data completeness and variability. Nanoscale 2020, 12, 4695–4708. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Cruz, M.L.; Hernández-Moreno, D.; Catalán, J.; Cross, R.K.; Stockmann-Juvala, H.; Cabellos, J.; Lopes, V.R.; Matzke, M.; Ferraz, N.; Izquierdo, J.J.; et al. Quality evaluation of human and environmental toxicity studies performed with nanomaterials—The GUIDEnano approach. Environ. Sci. Nano 2018, 5, 381–397. [Google Scholar] [CrossRef]
- Isigonis, P.; Afantitis, A.; Antunes, D.; Bartonova, A.; Beitollahi, A.; Bohmer, N.; Bouman, E.; Chaudhry, Q.; Cimpan, M.R.; Cimpan, E.; et al. Risk Governance of Emerging Technologies Demonstrated in Terms of its Applicability to Nanomaterials. Small 2020, 16, 2003303. [Google Scholar] [CrossRef]
- EFSA Scientific Committee; Hardy, A.; Benford, D.; Halldorsson, T.; Jeger, M.J.; Knutsen, H.K.; More, S.; Naegeli, H.; Noteborn, H.; Ockleford, C.; et al. Guidance on the use of the weight of evidence approach in scientific assessments. EFSA J. Eur. Food Saf. Auth. 2017, 15, e04971. [Google Scholar] [CrossRef] [Green Version]
- EFSA Scientific Committee; Hardy, A.; Benford, D.; Halldorsson, T.; Jeger, M.J.; Knutsen, H.K.; More, S.; Naegeli, H.; Noteborn, H.; Ockleford, C.; et al. Guidance on the assessment of the biological relevance of data in scientific assessments. EFSA J. Eur. Food Saf. Auth. 2017, 15, e04970. [Google Scholar] [CrossRef]
| Authors | Questionnaire/Platform | Manual Assessment | Automatic Assessment | Data/Code Repository | ||
|---|---|---|---|---|---|---|
| - | - | - | Code/Language | Metadata Format | Protocol/Library | |
| FAIRness evaluators | ||||||
| Wilkinsons et al. [13] | - | ✓ | - | - | - | GitHub |
| Australian Research Data Commons | FAIR self-assessment tool | ✓ | - | - | - | - |
| Commonwealth Scientific and Industrial Research Organization | 5 star data rating tool | ✓ | - | - | - | - |
| Data Archiving and Networked Services | FAIR enough? and FAIR data assessment tool | ✓ | - | - | - | - |
| GOFAIR consortium | FAIR ImplementationMatrix | ✓ | - | - | - | Open Science Framework |
| EUDAT2020 | How FAIR are your data? | ✓ | - | - | - | Zenodo |
| Wilkinsons et al. [14] | FAIR evaluation services | - | Ruby on Rails | JSON, Microformat, JSON-LD, RDFa | nanopublications | GitHub |
| Clark et al. [15] | FAIRshake | - | Django and Python | RDF | Extruct | GitHub |
| Studies assessing FAIRness of repositories | ||||||
| Dunning et al. [17] | - | ✓ | - | - | - | Institutional repository |
| Weber et al. [18] | - | - | Python | DataCite | OAI-PMH | GitLab |
| Our approach | - | ✓ (partially) | Jupyter notebook with Python | XML, JSON | request | GitHub |
| FAIR Principles [8] | Guidelines by the Maturity Indicator Authoring Group [28] | Dunning et al. [17] | Weber et al. [18] | Our Approach |
|---|---|---|---|---|
| F1: (meta)data are assigned a globally unique and persistent identifier | The GUID matches a scheme that is globally unique and persistent in FAIRsharing | Persistent identifier is DOI or similar | Pass (embedded in DataCite) | “doi” icon is enabled in www.re3data.org (data and metadata) (M) |
| F2: data are described with rich metadata (defined by R1 below) | Metadata contains “structured” elements (micrograph, JSON) or linked data (JSON-LD, RDFa) | Title, creator, date, contributors, keywords, temporal and spatial coverage | Qgeo, Qchrono | Search keywords are in dataset metadata (A) |
| F3: metadata clearly and explicitly include the identifier of the data they describe | Metadata contains both its own GUID and the data GUID | DOI of data is in metadata | Pass (embedded in DataCite) | Dataset metadata contains dataset ID (A) |
| F4: (meta)data are registered or indexed in a searchable resource | The digital resource can be found using web-based search engines | Dataset title found in google.com or duckduckgo.com | Pass | Dataset title found in Google Dataset Search (data and metadata) (M) |
| A.1 (meta)data are retrievable by their identifier using a standardized communications protocol | N/A | HTTP request returns 200 | Qret | HTTP request returns 200 (data and metadata) (A) |
| A1.1 the protocol is open, free, and universally implementable | The resolution protocol is universally implementable with an open protocol | Accomplished if protocol is HTTP | Q ret | Accomplished if protocol is HTTP (A) |
| A1.2 the protocol allows for an authentication and authorization procedure, where necessary | The resolution protocol supports authentication and authorization for access to restricted content | Accomplished if protocol is HTTP | Qret | Accomplished if protocol is HTTP (A) |
| A2. metadata are accessible, even when the data are no longer available | There is a policy for metadata | Repository has a clear policy statement | N/A | “data availability policy” is filled in registry metadata (A) |
| I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation | If hash-style metadata (e.g., JSON) or Linked Data are found, pass | Metadata is structured (e.g., Dublin Core) | Pass (embedded in DataCite) | Dataset metadata is structured (e.g., xml) (metadata) (M) |
| I2. (meta)data use vocabularies that follow FAIR principles | (meta)data use vocabularies that are, themselves, FAIR | N/A | N/A | N/A |
| I3. (meta)data include qualified references to other (meta)data | Metadata contain links that are not from the same source (domain/host) | Links to publications and terms definitions | N/A | Dataset metadata include reference to other dataset IDs (metadata) (M) |
| R1. meta(data) are richly described with a plurality of accurate and relevant attributes | N/A | Metadata provide information on how to reuse a dataset | Qgeo, Qchrono | Dataset metadata contain more information than search keywords (F2) (data) (A) |
| R1.1. (meta)data are released with a clear and accessible data usage license | Metadata contain a pointer to the data license | Metadata license is present | Qlic | “datalicensename” and “datalicenseurl” are filled in registry metadata (data) (A) |
| R1.2. (meta)data are associated with detailed provenance | N/A | Documentation on how data was created | N/A | “authors”, “email” and “title” are filled in dataset metadata (data) (A) |
| R1.3. (meta)data meet domain-relevant community standards | N/A | N/A | N/A | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ammar, A.; Bonaretti, S.; Winckers, L.; Quik, J.; Bakker, M.; Maier, D.; Lynch, I.; van Rijn, J.; Willighagen, E. A Semi-Automated Workflow for FAIR Maturity Indicators in the Life Sciences. Nanomaterials 2020, 10, 2068. https://doi.org/10.3390/nano10102068
Ammar A, Bonaretti S, Winckers L, Quik J, Bakker M, Maier D, Lynch I, van Rijn J, Willighagen E. A Semi-Automated Workflow for FAIR Maturity Indicators in the Life Sciences. Nanomaterials. 2020; 10(10):2068. https://doi.org/10.3390/nano10102068
Chicago/Turabian StyleAmmar, Ammar, Serena Bonaretti, Laurent Winckers, Joris Quik, Martine Bakker, Dieter Maier, Iseult Lynch, Jeaphianne van Rijn, and Egon Willighagen. 2020. "A Semi-Automated Workflow for FAIR Maturity Indicators in the Life Sciences" Nanomaterials 10, no. 10: 2068. https://doi.org/10.3390/nano10102068
APA StyleAmmar, A., Bonaretti, S., Winckers, L., Quik, J., Bakker, M., Maier, D., Lynch, I., van Rijn, J., & Willighagen, E. (2020). A Semi-Automated Workflow for FAIR Maturity Indicators in the Life Sciences. Nanomaterials, 10(10), 2068. https://doi.org/10.3390/nano10102068






