Abstract
Marine cyanobacteria are a source of bioactive natural compounds, with a wide range of biotechnological applications. However, information on sponge-associated cyanobacteria are relatively scarce to date. In this paper, we carried out the morphological and molecular characterization of eight cyanobacterial strains, previously isolated from the Mediterranean sponge Petrosia ficiformis, and evaluated their biological activities on epithelial- and neuron-like cultured cells of human and murine origin. The new analysis allowed maintaining the assignment of three strains (Cyanobium sp., Leptolyngbya ectocarpi, and Synechococcus sp.), while two strains previously identified as Synechococcus sp. and Leptolyngbya sp. were assigned to Pseudanabaena spp. One strain, i.e., ITAC104, and the ITAC101 strain corresponding to Halomicronema metazoicum, shared extremely high sequence identity, practically representing two clones of the same species. Finally, for only one strain, i.e., ITAC105, assignment to a specific genus was not possible. Concerning bioactivity analyses, incubation of cyanobacterial aqueous cell supernatants induced variable responses in cultured cells, depending on cell type, with some of them showing toxic activity on human epithelial-like cells and no toxic effects on human and rat neuron-like cells. Future investigations will allow to better define the bioactive properties of these cyanobacteria strains and to understand if they can be useful for (a) therapeutic purpose(s).
1. Introduction
Cyanobacteria are sources of bioactive compounds with manifold biotechnological applications [1,2]. The ability of cyanobacteria to produce molecules with antibacterial, antiviral and antifungal properties [3,4,5], as well as to exert anti-proliferative or toxic activities on cancer cells results of particular interest to date. Marine cyanobacterial biomolecules can act on cancer cells affecting, e.g., cytoskeleton and enzymes that can modulate cell death and apoptosis [6,7,8,9]. Over 400 biomolecules produced by marine cyanobacterial strains have recently been reported [10].
The high discovery rate (>95%) of novel compounds from cyanobacteria is largely due to the unexplored nature of this group of microalgae. Thus, collecting cyanobacterial strains from unexplored localities [11] and/or new substrata (e.g., water and invertebrates), besides hard and soft substrates, is strategic to discover new species and new bioactive compounds [12,13,14].
Among the substrata suitable to isolate new cyanobacterial strains, sponges seem to be very good candidates. To defend themselves from predators or spatial competitors, sponges produce bioactive molecules [15], some of which have high biomedical potential [16]. For some time, it had been unclear if the sponges or the microbial community living in association with the sponges are responsible for the metabolites’ production, but some early results demonstrated that the antimicrobial activity showed by the Mediterranean species Petrosia ficiformis was due to the epibiotic heterotrophic bacterial community isolated from the sponge [17]. Other studies evidenced that, besides heterotrophic bacteria, the most commonly observed symbionts of marine sponge are cyanobacteria [18,19,20], which in some cases, are the most conspicuous components of the associated microbial community [21]. Their role in association with sponge is wide because cyanobacteria through photosynthesis can provide up to 80% of the host’s energy requirements [22]. They also assist in the removal of nitrogenous wastes from the sponge host [23] and should provide biochemical defense throughout the production of bioactive metabolites ([24,25,26,27] and references therein).
Our previous research on cyanobacteria living in association with P. ficiformis allowed us to isolate eight strains of cyanobacteria, and to verify their different ability to produce molecules with biological activity. Indeed, the examined cyanobacteria aqueous cell supernatant should be responsible for the lysis of human erythrocytes, mortality of brine shrimp (Artemia salina) nauplii and inefficiency of the sea urchin (Paracentrotus lividus) gametes and embryo [28]. Among the tested strains, the filamentous forms proved to be the most toxic.
In this paper, by combining morphological and molecular studies, we specifically report on the identification of such cyanobacterial strains isolated from the sponge P. ficiformis. We have also investigated the isolated strains for their differential and cell type-specific effects on different cultured cells of human (epithelial-like (cervical adenocarcinoma) and neuron-like (neuroblastoma)) and murine (neuron-like (neuroblastoma/glioblastoma)) origin, as preliminary screening for the possible massive production of bioactive molecules.
2. Materials and Methods
2.1. Cyanobacterial Characterization and Biomass Production
Cyanobacterial strains have been previously isolated from the Mediterranean marine sponge Petrosia ficiformis, as described in Pagliara and Caroppo [28]. Monospecific cultures were obtained after repeated self-isolation on agar [29]. Cyanobacterial morphological characterization was based on available literature [30,31,32]. Moreover, cell average dimensions for the cyanobacterial strains were determined by measuring 25 cells.
Eight monospecific cyanobacterial strains were cultivated on a large scale for biological activity screening. Biomass was produced by cultivating for two months the isolated strains in two liters of MN medium enriched with B12 vitamin (5 μg/L) [33]. The cultures were incubated at 26.0 ± 1.0 °C under white fluorescent light, at a photosynthetic photon flux abundance of 20 μmol photon m2/s [34], and an illumination cycle of 12D:12N.
2.2. Genomic DNA Extraction
For each strain, aliquots of cultures were concentrated by centrifugation, then the supernatant was eliminated and the sediment volume was exposed to 1:1 (v/v) buffer for cell wall lysis, i.e., TEN (10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA and 10 mmol/L NaCl) containing 50 μg/mL lysozyme; 20 μg/mL proteinase K. The solution was incubated at 4 °C in agitation for 1 h, then SDS was added at 1% (w/v) (final concentration), and the solution was incubated again at 4 °C in agitation for additional 2 h. Subsequently, a first extraction step was performed by adding phenol:chloroform:isoamyl alcohol (25:24:1) and centrifuging at 3000× g, 20 min, 4 °C. Then, repeating the same conditions, the supernatant was subject to a second extraction step, and the second supernatant (aqueous solution containing nucleic acids) was collected. Then, 40 μg/mL RNAse A were added to avoid RNA contamination, and incubated at 37 °C for 30 min. The third extraction was performed by adding phenol:chloroform:isoamyl alcohol (25:24:1) and centrifuging at 6000× g, 15 min, 4 °C. The third supernatant was collected and added with 0.1 volumes NaOAc (3 mol/L) pH 7.0 and 2 volumes EtOH absolute. The mixture was left overnight at room temperature for genomic DNA precipitation. The day after, the mixture was centrifuged at 13,000× g, 30 min, 4 °C; the genomic DNA pellet was washed with EtOH absolute and finally resuspended in TE buffer. DNA visualization was performed by gel electrophoresis with 0.5% (w/v) agarose in standard TAE; DNA quantitation and purity were assessed by using a NanoDrop™ (ND-1000, Thermo-Fisher Scientific, Waltham, MA, USA) spectrophotometer.
2.3. Amplification of the 16S rRNA Gene
Amplification assays were performed on 100 ng genomic DNA per sample type. Amplifications were performed by adapting the Platinum Taq™ DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) standard protocol, according to the manufacturer’s instructions. The PCR and primer design strategy were based on approach adapted from Bruno and coll. [35] In brief, the amplification of the 16S23S rRNA operons from the investigated strains was performed utilizing the primer 1 (5′-AGAGTTTGATCCTGGCTCAG-3′; nucleotides 8–27 of the 16S rRNA gene in Synechocystis sp. strain PCC 6301), and primer 18m (5′-TCTGTGTGCCTAGGTATCC-3′; nucleotides 26–45 of the 23S rRNA gene in Synechocystis sp. strain PCC 6301). PCR amplicons (size ~2000 bp) were purified on an agarose gel with a Qiaquick gel extraction kit (Qiagen, Hilden, Germany), and used as templates for amplifying the 16S rRNA gene. The partitions of 16S rRNA genes were amplified using the following primers: CYA359F (forward primer; 5′-GGGGAATYTTCCGCAATGGG-3′), and the universal primer C up16S-CR (5′-ACGGGCGGTGTGTAC-3′), corresponding to Escherichia coli positions 1406 to 1392. PCR amplicons (size ~1.100 bp) were purified from gel, as described above. Purified PCR products were commercially sequenced independently on both strands, using primers previously adopted for amplification.
The 16S rRNA nucleotide sequences were submitted (October 2019) to GenBank. Both sequences and GenBank accession numbers (Acc. Nos.) are reported in Supplementary Table S1.
2.4. Molecular Analysis
All our new sequences were blasted (BLASTn) and the closest relative(s) for each sequence were included in the phylogenetic trees. For the molecular analyses, we selected sequences (for details see Supplementary Table S1) belonging to non-heterocystous taxa, to examine the phylogenetic position of our strains. Multiple sequence alignments were conducted using the Clustal Omega program [36,37]. Phylogenetic trees were constructed in MEGA7 [38] using the neighbor-joining (NJ) method, on a Jukes and Cantor distance matrix model. The robustness of the inferred phylogenies was determined by a bootstrap analysis based on 1000 resamplings of data.
Phylogenetic relationships among coccoid and filamentous cyanobacteria strains were examined separately, strictly referring to the species and sequence selection reported by Konstantinou and coll. [14,20,39] and refined by Gkelis and coll. [13]
2.5. Preparation of Cyanobacterial Aqueous Cell Supernatants
Cyanobacteria cells were harvested after two months growth, by either centrifugation or filtration through a 20 µm pore net (filamentous forms), freeze-dried and stored at −20 °C. Freeze-dried cyanobacterial material was lyophilized, then suspended in distilled water (15 mg dry weight/mL), and maintained for 1 h in the dark at room temperature. Then, suspensions were sonicated with an ultra-sound probe (Sonifer sonicator Model 250/240, Brain Ultrasonic Corporation, Danbury, CT, USA), 5 times for 50 s on ice. Sonicated samples were checked under the microscope to ensure cell breakage. Samples were left on ice for 1 h in the dark before centrifugation at 20,000× g for 30 min. The supernatants were recovered and stored at −20 °C until use.
2.6. Cell Culture Conditions and Treatments
HeLa (CLS Cat# 300194/p772_HeLa, RRID:CVCL_0030), SH-SY5Y (ATCC Cat# CRL-2266, RRID:CVCL_0019) and B-104 (ICLC Cat# ATL99008, RRID:CVCL_0154) cells were grown at 37 °C, 5% CO2, in Dulbecco’s modified Eagle’s medium (D-MEM; Sigma Aldrich Italy), supplemented with 10% (v/v) fetal bovine serum, 2 mM glutamine and antibiotics (penicillin-streptomycin 5000 U/mL–5 mg/mL Sigma-Aldrich, St. Louis, MO, USA). Culture propagation occurred every third day post-seeding (70–80% confluence), and all the experiments were conducted between passage 3 and 10 of propagation. Aqueous cell supernatants were diluted 1:100 (final concentration 150 μg/mL) in the routine culture medium, which was administered to cells at 80% confluence; effects were evaluated at 6 h (h) post-administration, by viability/proliferation and morphology assays.
2.7. Cell Morphology Analysis by Hematoxylin-Eosin Staining
Firstly, 1 × 105 cells were seeded on UV-sterilized cover glasses (size 22 mm × 22 mm) in six-well plates. The day after, cells were treated as described above then washed twice with Dulbecco’s phosphate buffer saline (D-PBS) and fixed for 1 h at room temperature with 4% (w/v) paraformaldehyde (PFA) in D-PBS. After fixation, cells on coverslips were washed trice with D-PBS and stained according to standard hematoxylin-eosin staining protocol before mounting the coverslips with Eukitt® mounting medium (Sigma-Aldrich, St. Louis, MO, USA). Bright field image acquisition was performed with an Eclipse 50i microscopy system (Nikon, Tokyo, Japan).
2.8. MTT Assay for Evaluation of Cell Viability
The standard MTT assay was used to evaluate the effects of the aqueous cell supernatants from cyanobacteria on mitochondrial activity and cell viability. Cells were seeded in 96-well plates (1.5 × 104 cells per well) and incubated for 24 h (37 °C, 5% CO2). After incubation, the medium was removed and replaced with routine culture medium containing cyanobacterial aqueous cell supernatants (at the concentration of 150 μg/mL). After 6 h treatments, MTT solution (5 mg/mL in sterile filtered PBS, pH 7.4) was added to each well, to reach the final concentration of 0.5 mg/mL, and plates were incubated at 37 °C for an additional 3 h. The dark-blue formazan crystals produced by MTT metabolization were then solubilized by lysing cells with 200 μL 2-propanol/HCl 4N per well, and absorbance values of the solutions were measured at λ = 550 nm with a microplate reader. Absorbance arbitrary units were normalized and mean values ± SEM are reported as percent (%) with respect to control mean values.
2.9. Apoptosis Assay
Cell lines were treated with cyanobacteria aqueous cell supernatants (150 μg/mL) for 6 h. After incubation, cell death was detected by TdT-mediated dUTP terminal nick-end labeling kit (In Situ Cell Death Detection Kit, Roche Applied Science, Penzberg, Germany). Nuclei were counterstained with 1 μg/mL DAPI for 3 min. Slides were examined by a fluorescence microscope (Eclipse 50i; Nikon) and images were acquired with a digital camera. Apoptotic data are reported as apoptosis percentage, obtained by determining the number of apoptotic cells vs. the total number of cells. For each sample, a minimum of 3 counts involving a minimum of 100–200 cells/count were scored. Apoptotic data are presented as the mean ± SD for three independently performed experiments.
2.10. In Vitro Proliferation Assays
Here, 5-Bromo-2′-deoxyuridine (BrdU) incorporation assay was performed by using an appropriate kit (5-Bromo-2′-deoxy-uridine Labeling and Detection Kit I; Roche Applied Science, Germany). Briefly, cells were seeded in 24-well culture plates at a density of 5 × 105, grown to 50% sub-confluence. After 6 h of cyanobacteria treatment, a medium containing BrdU (10 mM) was added to each well, and incubated for 3 h at 37 °C and 5% CO2. After washing the cells 3 times with the washing buffer, a solution containing fluorescein isothiocyanate-labeled anti-BrdU antibody was added. Staining was visualized using a fluorescent microscope (Eclipse 50i; Nikon). Cell proliferation was determined by counting the number of total and BrdU-positive cells in eight alternative areas, to determine the percentage of BrdU-positive cells.
2.11. Statistical Analysis
In the MTT viability assays, data are represented as mean values ± s.e.m. of three independent experiments with n = 8 biological replicates. Student’s t-test was used to analyze the data; differences were indicated as statistically significant with p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***).
3. Results
3.1. Cyanobacterial Strains
Cyanobacterial strains isolated from the marine sponge P. ficiformis were successfully cultured and characterized, both morphologically and molecularly. The morphological characteristics of the strains are shown in Figure 1 and summarized in Table 1.
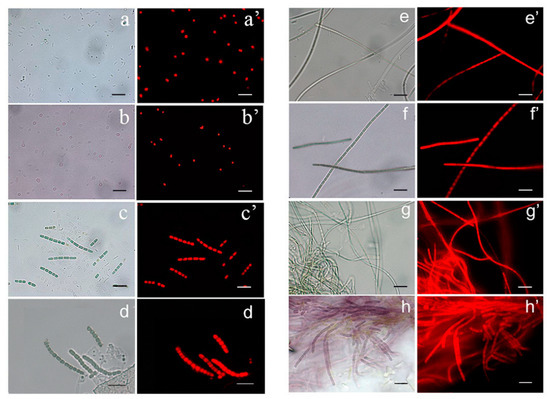
Figure 1.
Light micrographs of the cyanobacterial strains and respective micrographs highlighting the red fluorescence of their photosynthetic pigments. (a,a’) strain Cyanobium sp. ITAC108; (b,b’) Synechococcus sp. ITAC107; (c,c’) Pseudanabaena sp. ITAC106; (d,d’) Pseudanabaena sp. ITAC102; (e,e’) Leptolyngbya ectocarpi ITAC103; (f,f’) undetermined Oscillatoriales ITAC105; (g,g’) Halomicronema cf. metazoicum ITAC104; (h,h’) Halomicronema metazoicum ITAC101. Scale bars = 10 µm.

Table 1.
Morphological features of the cyanobacterial strains isolated from Petrosia ficiformis and observed with light microscopy.
The strain ITAC108 (Figure 1a,a’) were green solitary cells almost spherical or oval (1.6 ± 0.3 µm wide and 2.0 ± 0.5 µm long), with no mucilaginous envelopes. ITAC108 exhibited a 99.70% sequence identity to Cyanobium sp. LEGE 06012 (GenBank Acc. No. KU951687.1), and were included in the phylogenetic tree, among other highly similar Cyanobium sequences from the NCBI database (Figure 2A and for details see Supplementary Figure S1 and Supplementary Table S1).
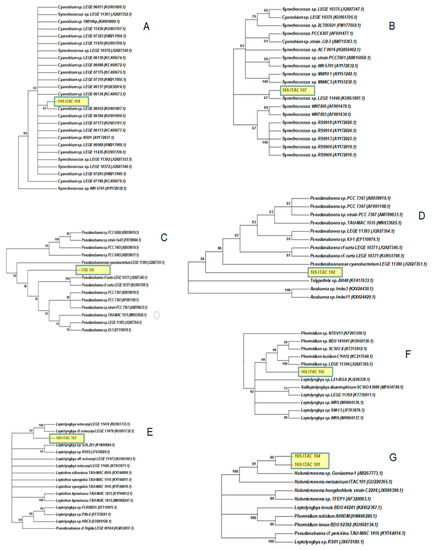
Figure 2.
Phylogenetic relationships of the studied cyanobacteria strains based on 16S with other cyanobacteria taxa: (A) ITAC108; (B) ITAC107; (C) ITAC106; (D) ITAC102; (E) ITAC103; (F) ITAC105; (G) ITAC104 and ITAC101. The trees were constructed in MEGA7 using the Neighbor-Joining (NJ) method on a Jukes and Cantor distance matrix model. The robustness of the inferred phylogenies was determined by bootstrap analysis based on 1000 resamplings of data.
The isolate ITAC107 (Figure 1b,b’) were solitary red cells almost long oval or cylindrical (0.8 ± 0.2 µm wide and 1.5 ± 0.2 µm long), with no mucilaginous envelopes. This strain is responsible for the color of the sponge. Molecular analyses (Figure 2B and for details see Supplementary Figure S2 and Supplementary Table S1) confirmed that the strain is a Synechococcus sp, because of a 96.17% identity with the sequence of Synechococcus sp. LEGE 11466 (GenBank Acc. No. KU951807.1) and many other highly similar Synechococcus spp. available from the NCBI database.
The phenotypic features of ITAC106 (Figure 1c,c’) and ITAC102 (Figure 1d,d’) strains and the molecular analysis (for details, see Supplementary Figures S3 and S4 and Supplementary Table S1) fit the Pseudanabaena morphotype. These two strains presented cylindrical or barrel cells with fine mucilaginous envelopes. ITAC106 presents almost straight or arcuate trichomes consisting of 3–8 units, with conspicuous constrictions at cross-walls. Cells were always longer than wide (1.2 ± 0.2 µm wide and 3.1 ± 0.6 µm long) and not differentiated in the apical region. Cultures of the strain and ITAC102 showed freely entangled filaments with numerous cylindrical cells (1.3 ± 0.3 µm wide and 2.8 ± 0.5 µm long). Cell constrictions were evident. The identity of the two strains was of 93.73% for ITAC106 (Figure 2C) and of 94.35% for ITAC102 (Figure 2D) with Pseudanabaena sp. LEGE 11383 (GenBank Acc. No. JQ927354.1), respectively (with an observed 95.02% percent identity of ITAC106 vs. ITAC102). Based on these findings, we considered these as two different Pseud anabaena spp.
The isolate ITAC103 (Figure 1e,e’) showed flexuous filaments densely and irregularly entangled with colorless mucilaginous sheaths. Cells were cylindrical and longer than wide (0.7 ± 0.2 µm wide and 1.8 ± 0.3 µm long). Apical cells rounded or conical-rounded, lengthened, and acute were observed. Molecular analyses (Figure 2E, and for details see Supplementary Figure S5 and Supplementary Table S1) confirmed that this strain is close to Leptolyngbya ectocarpi LEGE 11474 (GenBank Acc. No. KU951733.1) and to Leptolyngbya cf. ectocarpi LEGE 14779 (GenBank Acc. No. KU951732.1) (in both cases, percent identity: 98.25%).
Filaments of ITAC105 appeared variously curved and entangled (Figure 1f,f’) with thin and colorless sheaths. Trichomes were distinctly constricted at the cross-walls. Cells were longer than wide (0.9 ± 0.3 µm wide and 4.0 ± 0.8 µm long), and pointed, narrowed or rounded apical cells were present. Molecular analyses (Figure 2F, and for details, see Supplementary Figure S6 and Supplementary Table S1) indicated that this strain falls in a Phormidium spp. cluster, with the most similar sequence available from GenBank being that of Phormidium sp. SCSIO X (GenBank Acc. No. KT315912.1) (percent identity: 97.41%). However, morphological features were not typical of Phormidium genus and the ITAC105 strain was considered as an undetermined Oscillatoriales.
Filaments of ITAC104 were entangled with colorless, firm, distinct and very thin (Figure 1g,g’). Trichomes had slight constriction at the cross-walls. Cells were 0.8 ± 0.3 µm in diameter and 3.1 ± 0.5 µm long. The morphology of these isolates fits with that of the strain ITAC101 [40] and molecular analyses (Figure 2G, and for details see Supplementary Figure S7 and Supplementary Table S1) evidenced that the strain shows the highest identity (percentage identity: 99.35%) to Halomicronema metazoicum (GenBank Acc. No. GU220365.1) (that is the ITAC101 strain).
Last, the ITAC101 strain, which had previously been assigned to the species H. metazoicum [40], is shown (Figure 1h,h’).
3.2. Effects of the Extracts of Cyanobacteria Strains on Cell Line Viability
The cytotoxic effect of the eight strains was assessed by MTT assay, a direct indicator of the metabolic activity of the cells and an indirect indicator of cell viability [41,42,43]. As shown in Figure 3, in the human epithelial-like (cervical adenocarcinoma) HeLa cells after 6 h exposure to cyanobacterial aqueous cell supernatants, the mitochondrial activity was reduced by aqueous cell supernatants from each of the strains, according to the following pattern: ITAC106 ≤ ITAC108 < ITAC102 < ITAC104 ≤ ITAC101 < ITAC107 ≈ ITAC103 < ITAC105 << control (CTRL). Specifically, the lowest mitochondrial activity was induced by ITAC106, which exerted the most toxic effects (62.1 ± 1.9% vs. 100% of the untreated control), while ITAC105 induced a non-significant reduction, showing the lowest toxicity (82.8 ± 4.7%). In parallel, in the human neuron-like SH-SY5Y cells (neuroblastoma), after 6 h treatment the mitochondrial activity was found to be both lower and higher than the control cells, showing the following pattern: ITAC106 << ITAC108 < CTRL ≈ ITAC102 < ITAC103 < ITAC104 ≤ ITAC105 << ITAC101 << ITAC107. In particular, ITAC106 strain induced the significant reduction of the mitochondrial activity (83.5 ± 3.5% vs. 100% control), while no significant changes occurred with the ITAC108, ITAC107, ITAC102, ITAC103, ITAC104 and ITAC105 strain aqueous cell supernatants. Interestingly, the ITAC101 and ITAC107 aqueous cell supernatant induced a slight but significant increase of mitochondrial activity (118.7 ± 8.7% and 125.6 ± 5.9% vs. 100% control, respectively) (Figure 3). Again, in the rat neuron-like B104 cells treatment with the cyanobacterial aqueous cell supernatants induced both increase and decrease of the mitochondrial activity with respect to control cells, with the following pattern: ITAC106 << ITAC108 < CTRL <<ITAC102 << ITAC103 << ITAC107 << ITAC104 ≈ ITAC101 << ITAC105. ITAC103, ITAC107, ITAC104, ITAC101 and ITAC107. Strains were found to induce significant increase of mitochondrial activity, ranging from 132 ± 12% to 166.1 ± 21.6%. No significant differences with respect to control were detected for ITAC102 and ITAC108. ITAC106 was the only strain aqueous cell supernatant that reduced the mitochondrial activity (80.5 ± 6.6%) of B104 cells, even though without statistical significance. Taken together, these results indicate ITAC106 as the most effective strain in terms of MTT-detected cytotoxicity on HeLa, SH-SY5Y and B104 cells.
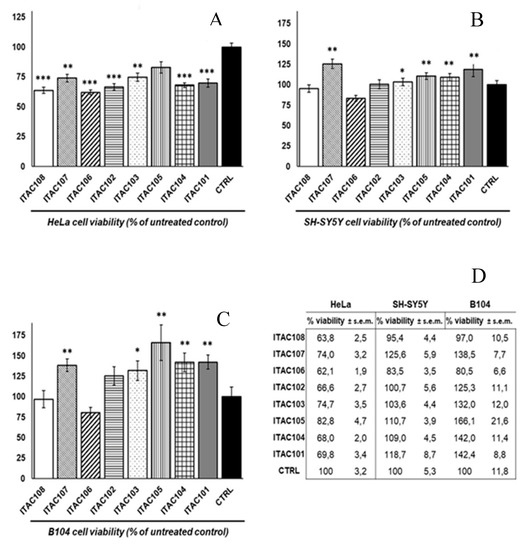
Figure 3.
Effects of cyanobacterial aqueous cell supernatants on the viability of cultured cells evaluated by MTT assays on the human HeLa (A), SH-SY5Y cells (B) and the rat B104 cells (C), exposed for 6 h to cyanobacterial aqueous cell supernatants. In the graphs and the numerical table (D), data are represented as percentage (%) mean values ± s.e.m., with respect to the untreated control cells (CTRL, 100%). Data derive from three independent experiments, each with n = 8 biological replicates. Statistical analysis: Student’s t-test; differences vs. CTRL are indicated as statistically significant with p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***).
3.3. Effects of the Aqueous Cell Supernatants of Cyanobacteria Strains on Cell Line Morphology
Treated cells observed after eosin/hematoxylin staining showed some morphological changes generally consisting in alterations of the cytoplasm organization. When aqueous cell supernatants were employed, cells with apoptotic-like appearance as rounded shape and cytoplasmic blebs were observed (for details on cells treated with aqueous cell supernatants from ITAC106 and ITAC108, see Figure 4). On the contrary, apoptotic-like cells were rarely found in controls. The incidence of apoptosis was evaluated by counting the number of cells with a typical apoptotic aspect in five optic fields (at least 100 cells were counted).
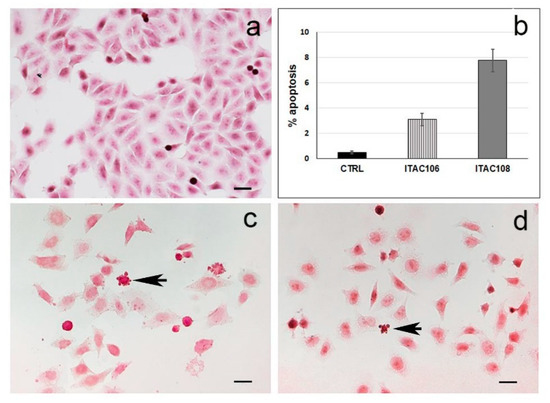
Figure 4.
HeLa cells treated with cyanobacteria aqueous cell supernatants. (a) control; (b) percentage (%) of apoptotic cells after treatment with ITAC106 and ITAC108 cyanobacteria aqueous cell supernatants; (c,d) HeLa cells with apoptotic morphology after treatment with Pseudanabaena sp. ITAC106 and Cyanobium sp. ITAC108, respectively. Arrows indicate cells with apoptotic-like morphology (i.e., blebs). Scale bar = 20 μm.
Notably, the increase in apoptosis was evident for HeLa cells challenged with ITAC108 (8% of apoptotic cells); a lower increase than the previous was recorded with ITAC106, but also in this case, the increase was significant (p < 0.01) compared to the control (see Figure 4b). The presence of apoptotic-like cells was not significant (p > 0.01) after treatment with the other cyanobacteria aqueous cell supernatants (data not shown).
No morphological differences were evidenced after SH-SY5Y cell line treatment with the eight cyanobacterial strains aqueous cell supernatants.
Microscopic observations of B104 cells after treatment with cyanobacteria revealed different responses of cells to the different strains. In particular, ITAC106 induced, in some cases, apoptotic morphology with round shape and cytoplasmic blebs formation. On the contrary, after treatment with the other cyanobacteria strains, B104 cells consistently showed dividing cells (see Figure 5), with the metaphase plate often clearly visible. Since B104 cells after treatment with cyanobacterial aqueous cell supernatants showed an increase of their mitochondrial activity, we performed a BrdU assay to verify whether an increase in cell proliferation occurred. BrdU test did not show an increase in the proliferation rate after cell treatment with cyanobacterial aqueous cell supernatants. In fact, in the control sample, a percentage of 69% of cells positive to BrdU was counted, while after 6 h cyanobacteria treatment, this value was never reached. Remarkably, a significant decrease (p < 0.01) in proliferation was assessed after ITAC101 (28.6 ± 1.1%) and ITAC108 treatment (27.6 ± 3.6%).
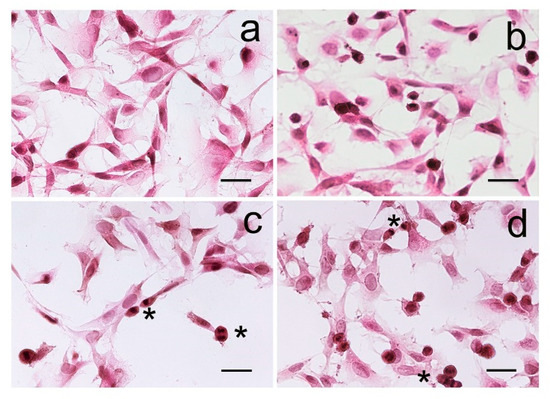
Figure 5.
B104 cells treated with cyanobacteria aqueous cell supernatants. (a) control; (b) B104 cells after treatment with Halomicronema cf. metazoicum ITAC104; (c) B104 cells after treatment with Synechococcus sp. ITAC107; (d) B104 cells after treatment with the undetermined Oscillatoriales ITAC105. Asterisks indicate dividing cells. Scale bar = 20 μm.
4. Discussion
Cyanobacteria are photosynthetic microorganisms that can live in symbiosis with a variety of eukaryotic hosts, and, to date, the association with more than 300 sponge species has been reported [20,27,44,45]. Cyanobacteria strains living in association with sponges can easily be isolated and cultured [20,28], which allows production on a large-scale of secondary metabolites from cultured strains and/or screening for cyanobacterial extracts without the need of culturing the sponge tissue itself [46]. However, due to the well-known phenotypic plasticity of cyanobacteria [47]; often the isolated species are not easily identifiable by using only morphological criteria, and to date, the molecular method (e.g., 16S rRNA gene sequencing) is considered a basic procedure for implementing taxonomic classification [48,49,50]. Such molecular techniques not only provide taxonomic information, but, for the last decades, they have provided new data about sponge-cyanobacteria associations, which is the base for many biotechnological applications [1]. A polyphasic approach, which includes the combined use of microscopic, cytomorphological, ecophysiological, biochemical and molecular methods, therefore results in a more effective characterization of the cyanobacterial strains [40,47,50,51]. In this study, we carried out molecular analyses, to review the identification of cyanobacterial species living in symbiosis with the Mediterranean sponge P. ficiformis, and to investigate their properties for potential applications in pharmaceutics. Harboring a rich community of symbiotic bacteria [52], this sponge is a treasure chest of biodiversity. In fact, although Usher et al. [18] asserted that P. ficiformis contains only one cyanobacterium, i.e., a Synechococcus species, more recently, Pagliara and Caroppo [28] reported the isolation from this sponge of eight cyanobacterial strains, of which the classification was based on the specificity of the morphological traits and the phylogenetic analysis of the 16S rRNA gene. Kostantinou and coll. [20] have extended the list of cyanobacteria species associated to P. ficiformis with a Leptolyngbya species (Leptolyngbya sp. TAU-MAC 0915).
Our re-classification of the cultured strains described in Pagliara and Caroppo [28] takes advantage of the increasing number of cyanobacterial sequences yearly entered in the databases. The new analysis in this paper allows maintaining the assignment of three strains (ref. ITAC108, ITAC107 and ITAC103 to Cyanobium sp., Synechococcus sp. and Leptolyngbya ectocarpi, respectively), while the other strains result in being differently classified. In particular, the previous analysis assigned the ITAC106 and ITAC102 strains to Synechococcus and Leptolyngbya genera, respectively, but the present BLASTn-based analyses assign them more properly to the Pseudanabaena genus. The identification of these two Pseudanabaena species confirm the association between this genus and marine sponge as already described to occur on marine sponges, such as Axinella damicornis [20] or Clathria prolifera [53].
Moreover, the sequence analysis of ITAC105 strain succeeded in assigning the strain to a not well-defined genus (undetermined Oscillatoriales), by considering the molecular as well as the morphological approach, while previously ITAC105 was as to belong to the Leptolyngbya genus for its phenotypic features. Further analyses (polyphasic approach) are necessary to conclusively re-assign the taxonomy of this microorganism. Finally, of interest is the case of ITAC104 strain, since its morphology and that of ITAC101, a strain already classified as H. metazoicum [40], just after their isolation seemed different; but, over time, the morphology and color of the two colonies did change, converging to highly similar macroscopic forms. Such morphological observations, matched with the very high sequence identity at the DNA level, led us to consider the two strains as the same; however, this classification needs to be confirmed by electron microscopy. Due to its bioactivities, the Halomicronema genus has been receiving particular attention in recent times [54,55]. Notably, H. metazoicum has been isolated in a free-living state from Posidonia oceanica leaves and, as it affects the vitality and the life cycle of other organisms, authors suppose that it can play important roles in the ecology of benthic and planktonic communities [54].
The identification of new cultivable cyanobacteria has significant implications for their capacity to produce new bioactive molecules and make them available as new drugs, pharmaceuticals, nutraceuticals, etc. Many cyanobacteria produce bioactive compounds that can affect enzymatic activities, interfere with signaling pathways, and cause apoptosis events, also leading to mortality as an endpoint [5,9]. Moreover, it is important to point out that a high degree of novel biodiversity among natural product producing strains revealed in some studies, was previously not recognized due to the lack of adequate classification systems. In some cases, the data reveal a large amount of new biodiversity which, however, is not reflected in a more dispersed taxonomic distribution of natural products [56]. In this respect, the interest in revising already identified strains also derives from the need of better understanding previous observations on their bioactivity. In a preliminary investigation, Pagliara and Caroppo [28] observed for our strain extracts (i) a cytolytic effect on human erythrocytes; (ii) toxic activity against A. salina nauplii; and (iii) negative influence on the sea urchin embryo development. In the present study, the ability of the different cyanobacteria to cause biological effects extends to in vitro cellular systems, namely on mammalian human and rodent cell lines.
Extracts from the cyanobacteria of marine, freshwater, or terrestrial origin, are commonly tested for their cytotoxicity by MTT test [57,58,59,60]. Notably, as in previous similar cytotoxicity screenings, the interaction between cyanobacteria extracts and cell cultures may induce rather different responses depending on cyanobacterial extract and the cell type tested [4,61,62]. Indeed, in this study, we observed that the cyanobacterial effect was cell type-specific, inducing a general toxicity on the human epithelial-like HeLa cells, with up to 40% of mitochondrial activity reduction. However, no toxic effect was recorded by any of the strains tested on the other human cell line, i.e., the neuron-like SH-SY5Y neuroblastoma cells. On these cells a general—even though faint—increase in mitochondrial activity was found (with the highest response observed with ITAC107 and ITAC101 strain aqueous cell supernatants). It is worth noticing that, when the strain aqueous cell supernatants were administered to the rat neuron-like B104 cells (neuroblastoma-derived cells), an evident increase of mitochondrial activity was registered for almost all strain aqueous cell supernatants. Interestingly, the most toxic effect was found for the Pseudanabaena sp. ITAC106 strain, which is actually the only strain with a toxic impact on each of the three cell lines tested. This cyanobacterial strain results of particular interest, considering that it was found very effective also on sea urchin development [28], inducing a pronounced inhibitory effect on mitosis. Taken together, these results suggest that this Pseudanabaena sp. ITAC106 strain has a general aggressive potential against mammalian cultured cells, a question that should be further investigated. Our findings on this strain are in line with observations on the toxic effects of extracts obtained from other Pseudanabaena species on Cladocerans [63], or on mice when administered intraperitoneally [64]. On the other side, further studies are required for the undetermined Oscillatoriales ITAC105. Indeed, even if it induced a non-significant effect on the cell lines here used, it definitely affected sea urchin gametes and larval development [28].
5. Conclusions
In summary, our results confirm and extend the concept that P. ficiformis harbors both filamentous and coccoid cyanobacteria species. Some of the isolated strains still need to be defined at the species level. In this respect, a polyphasic approach, sequence analyses involving genes other than 16S, and the comparison of 16S sequences to novel sequences in databanks are all required to achieve the taxonomic assignment of the isolated strains from this sponge. Due to their rather controllable effects on human and rodent cell lines, our strains are promising for future biotechnological, chemical and/or pharmacological applications. In this respect, emphasis should be given to ITAC106 and ITAC108 strains for their toxicological properties, although other work is needed to chemically and pharmacologically characterize their complete bioactivity. Last, but not least, since these strains are not found to grow naturally in large densities, the implementation of protocols for massive growth under laboratory conditions is necessary for biomass production and future biotechnological applications.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-1312/8/9/638/s1, Figures S1–S7: Molecular phylogenetic analysis by Neighbor-Joining method, Table S1: 16S rRNA nucleotide sequences.
Author Contributions
Conceptualization, P.P. and C.C.; methodology, P.P. and C.C.; software, T.V.; validation, P.P., C.C., A.B. and T.V.; investigation, A.B., P.P.; writing—original draft preparation, P.P. and C.C.; writing—review and editing, P.P. and C.C.; visualization, P.P., C.C. and T.V.; supervision, P.P., C.C. and T.V.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abed, R.M.; Dobretsov, S.; Sudesh, K. Applications of cyanobacteria in biotechnology. J. Appl. Microbiol. 2009, 106, 1–12. [Google Scholar] [CrossRef]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- Nunnery, J.K.; Mevers, E.; Gerwick, W.H. Biologically active secondary metabolites from marine cyanobacteria. Curr. Opin. Biotech. 2010, 21, 787–793. [Google Scholar] [CrossRef]
- Costa, M.; Garcia, M.; Costa Rodrigues, J.; Costa, M.S.; Ribeiro, M.J.; Fernandes, M.H.; Barros, P.; Barreiro, A.; Vasconcelos, V.; Martins, R. Exploring bioactive properties of marine cyanobacteria isolated from the Portuguese coast: High potential as a source of anticancer compounds. Mar. Drugs 2013, 12, 98–114. [Google Scholar] [CrossRef]
- Huang, I.S.; Zimba, P.V. Cyanobacterial bioactive metabolites—A review of their chemistry and biology. Harmful Algae 2019, 83, 42–94. [Google Scholar] [CrossRef]
- Williams, P.G. Panning for chemical gold: Marine bacteria as a source of new therapeutics. Trends Biotechnol. 2009, 27, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T. Filamentous tropical marine cyanobacteria: A rich source of natural products for anticancer drug discovery. J. Appl. Phycol. 2010, 22, 659–676. [Google Scholar] [CrossRef]
- Regueiras, A.; Pereira, S.; Costa, M.S.; Vasconcelos, V. Differential toxicity of cyanobacteria isolated from marine sponges towards echinoderms and crustaceans. Toxins 2018, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Demay, J.; Bernard, C.; Reinhardt, A.; Marie, B. Natural products from cyanobacteria: Focus on beneficial activities. Mar. Drugs 2019, 17, 320. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Zhang, J.; He, S.; Yan, X. New peptides isolated from marine cyanobacteria, an overview over the past decade. Mar. Drugs 2017, 15, 132. [Google Scholar] [CrossRef]
- Uzair, B.; Tabassum, S.; Rasheed, M.; Rehman, S.F. Exploring marine cyanobacteria for lead compounds of pharmaceutical importance. Sci. World J. 2012, 2012, 179782. [Google Scholar] [CrossRef] [PubMed]
- Caroppo, C.; Francavilla, M.; Pagliara, P. Mediterranean cyanobacterial biodiversity and bioactivity. In Microbes: An Innovative Approach; Goyal, P., Chauhan, A., Kaushik, P., Eds.; Plant SBW Publishers: New Delhi, India, 2014; pp. 18–32. [Google Scholar]
- Gkelis, S.; Panou, M.; Konstantinou, D.; Apostolidis, P.; Kasampali, A.; Papadimitriou, S.; Kati, D.; Di Lorenzo, G.M.; Ioakeim, S.; Zervou, S.K.; et al. Diversity, cyanotoxin production, and bioactivities of cyanobacteria isolated from freshwaters of Greece. Toxins (Basel) 2019, 11, 436. [Google Scholar] [CrossRef] [PubMed]
- Konstantinou, D.; Mavrogonatou, E.; Zervou, S.K.; Giannogonas, P.; Gkelis, S. Bioprospecting sponge-associated marine cyanobacteria to produce bioactive compounds. Toxins (Basel) 2020, 12, 73. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, J.R.; Steindler, L.; Henkel, T.P.; Beer, S.; Ilan, M. Chemical warfare on coral reefs: Sponge metabolites differentially affect coral symbiosis in situ. Limnol. Oceanogr. 2007, 52, 907–911. [Google Scholar] [CrossRef]
- Munro, M.H.G.; Blunt, J.W.; Dumdei, E.J.; Hickfort, S.J.H.; Lill, R.E.; Li, S.; Battershill, C.N.; Duckworth, A.R. The discovery and development of marine compounds with pharmaceutical potential. J. Biotechnol. 1999, 70, 15–25. [Google Scholar] [CrossRef]
- Chelossi, E.; Milanese, M.; Milano, A.; Pronzato, R.; Riccardi, G. Characterisation and antimicrobial activity of epibiotic bacteria from Petrosia ficiformis (Porifera, Demospongiae). J. Exp. Mar. Biol. Ecol. 2004, 309, 21–33. [Google Scholar] [CrossRef]
- Usher, K.M.; Fromont, J.; Sutton, D.C.; Toze, S. The biogeography and phylogeny of unicellular cyanobacterial symbionts in selected sponges from Australia and the Mediterranean. Microb. Ecol. 2004, 48, 167–177. [Google Scholar] [CrossRef]
- Regueiras, A.; Alex, A.; Pereira, S.; Costa, M.S.; Antunes, A.; Vasconcelos, V. Cyanobacterial diversity in the marine sponge Hymeniacidon perlevis from a temperate region (Portuguese coast, northeast Atlantic). Aquat. Microb. Ecol. 2017, 79, 259–272. [Google Scholar] [CrossRef]
- Konstantinou, D.; Gerovasileiou, V.; Voultsiadou, E.; Gkelis, S. Sponges-cyanobacteria associations: Global diversity overview and new data from the eastern mediterranean. PLoS ONE 2018, 13, e0195001. [Google Scholar] [CrossRef]
- Pfannkuchen, M.; Schlesinger, S.; Fels, A.; Brümmer, F. Microscopical techniques reveal the in situ microbial association inside Aplysina aerophoba, Nardo 1886 (Porifera, Demospongiae, Verongida) almost exclusively consists of cyanobacteria. J. Exp. Mar. Biol. Ecol. 2010, 390, 169–178. [Google Scholar] [CrossRef]
- Cheshire, A.C.; Wilkinson, C.R.; Seddon, S.; Westphalen, G. Bathymetric and seasonal changes in photosynthesis and respiration of the phototrophic sponge Phyllospongia lamellosa in comparison with respiration by the heterotrophic sponge Ianthella basta on Davies Reef, Great Barrier Reef. Mar. Freshw. Res. 1997, 48, 589–599. [Google Scholar] [CrossRef]
- Diaz, M.C.; Ward, B.B. Sponge-mediated nitrification in tropical benthic communities. MEPS 1997, 156, 97–107. [Google Scholar] [CrossRef]
- Unson, M.D.; Holland, N.D.; Faulkner, D.J. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol. 1994, 119, 1–11. [Google Scholar] [CrossRef]
- König, G.M.; Wright, A.D.; Stiche, O.; Angerhofer, C.K.; Pezzuto, J.M. Biological activities of selected marine natural products. Planta Med. 1994, 60, 532–537. [Google Scholar] [CrossRef]
- Usher, K.M. The ecology and phylogeny of cyanobacterial symbionts in sponges. Mar. Ecol. 2008, 29, 178–192. [Google Scholar] [CrossRef]
- Thacker, R.; Freeman, C.J. Sponge-microbe symbioses: Recent advances and new directions. Adv. Mar. Biol. 2012, 62, 57–111. [Google Scholar] [CrossRef]
- Pagliara, P.; Caroppo, C. Cytotoxic and antimitotic activities in aqueous extracts of eight cyanobacterial strains isolated from the marine sponge Petrosia ficiformis. Toxicon 2011, 57, 889–896. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobateria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar]
- Kana, T.M.; Glibert, P.M.G. Effect of irradiances up to 2000 mE m−2 s−1 on marine Synechococcus WH7803-I. Growth, pigmentation and cell composition. Deep Sea Res. 1987, 34, 479–495. [Google Scholar] [CrossRef]
- Castenholz, R.W. Culturing methods for cyanobacteria. In Cyanobacteria, Methods in Enzymology; Packer, L., Glazer, A.N., Eds.; Academic Press Inc.: New York, NY, USA, 1988; Volume 167, pp. 68–93. [Google Scholar]
- Boone, D.R.; Castenholz, R.W.; Garrity, G.M. Bergey’s m Anual of Systematic Bacteriology, 2nd ed.; The Archaea and the Deeply Branching and Phototrophic Bacteria; Springer: New York, NY, USA, 2001; Volume 1, pp. 1–722. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota, Part 1: Chroococcales. In Süsswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Spektrum Akademischer Verlag: Heidelberg, Germany, 2008; pp. 1–548. [Google Scholar]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota, Part 2: Oscillatoriales. In Süsswasserflora von Mitteleuropa, Bd19/2; Büdel, B., Gärtner, G., Krienitz, L., Schagerl, M., Eds.; Spektrum Akademischer Verlag: Heidelberg, Germany, 2008; pp. 1–759. [Google Scholar]
- Bruno, L.; Billi, D.; Bellezza, S.; Albertano, P. Cytomorphological and genetic characterization of troglobitic Leptolyngbya strains isolated from Roman hypogea. Appl. Environ. Microbiol. 2009, 75, 608–617. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014, 1079, 105–116. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Konstantinou, D.; Voultsiadou, E.; Panteris, E.; Zervou, S.K.; Hiskia, A.; Gkelis, S. Leptothoe, a new genus of marine cyanobacteria (Synechococcales) and three new species associated with sponges from the Aegean Sea. J. Phycol. 2019, 55, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Caroppo, C.; Albertano, P.; Bruno, L.; Montinari, M.; Rizzi, M.; Vigliotta, G.; Pagliara, P. Identification and characterization of a new Halomicronema species (Cyanobacteria) isolated from the Mediterranean marine sponge Petrosia ficiformis (Porifera). Fottea Olomouc 2012, 12, 315–326. [Google Scholar] [CrossRef]
- Stoddart, M.J. Cell viability assays: Introduction. Methods Mol. Biol. 2011, 740, 1–6. [Google Scholar] [CrossRef]
- Sumantran, V.N. Cellular chemosensitivity assays: An overview. Methods Mol. Biol. 2011, 731, 219–236. [Google Scholar] [CrossRef]
- Adan, A.; Kiraz, Y.; Baran, Y. Cell proliferation and cytotoxicity assays. Curr. Pharm. Biotechnol. 2016, 17, 1213–1221. [Google Scholar] [CrossRef]
- Adams, D.G.; Bergman, B.; Nierzwicki-Bauer, S.A.; Rai, A.N.; Schüssler, A. Cyanobacterial-plant symbioses. In The Prokaryotes: A Handbook on the Biology of Bacteria, 3rd ed.; Symbiotic Associations, Biotechnology, Applied Microbiology; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 1, pp. 331–363. [Google Scholar]
- Steinert, G.; Taylor, M.W.; Deines, P.; Simister, R.L.; De Voogd, N.J.; Hoggard, M.; Schupp, P.J. In four shallow and mesophotic tropical reef sponges from Guam the microbial community largely depends on host identity. PeerJ 2016, 4, e1936. [Google Scholar] [CrossRef]
- Lafi, F.F.; Garson, M.J.; Fuerst, J.A. Culturable bacterial symbionts isolated from two distinct sponge species (Pseudoceratina clavata and Rhabdastrella globostellata) from the Great Barrier Reef display similar phylogenetic diversity. Microb. Ecol. 2005, 50, 213–220. [Google Scholar] [CrossRef]
- Komárek, J. A polyphasic approach for the taxonomy of cyanobacteria: Principles and applications. Eur. J. Phycol. 2016, 51, 346–353. [Google Scholar] [CrossRef]
- Komárek, J. The modern classification of Cyanoprokaryotes (Cyanobacteria). Oceanol. Hydrobiol. Stud. 2005, 34, 5–17. [Google Scholar]
- Komárek, J. Recent changes (2008) in cyanobacteria taxonomy based on a combination of molecular background with phenotype and ecological consequences (genus and species concept). Hydrobiologia 2010, 639, 245–259. [Google Scholar] [CrossRef]
- Caroppo, C. Ecology and biodiversity of picoplanktonic cyanobacteria in coastal and brackish environments. Biodivers. Conserv. 2015, 24, 949–971. [Google Scholar] [CrossRef]
- Jung, P.; Briegel-Williams, L.; Schermer, M.; Büdel, B. Strong in combination: Polyphasic approach enhances arguments for cold-assigned cyanobacterial endemism. Microbiol. Open 2018, 8, e00729. [Google Scholar] [CrossRef]
- Schmitt, S.; Hentschel, U.; Taylor, M.W. Deep sequencing reveals diversity and community structure of complex microbiota in five Mediterranean sponges. Hydrobiologia 2012, 687, 341–351. [Google Scholar] [CrossRef]
- Isaacs, L.T.; Kan, J.; Nguyen, L.; Videau, P.; Anderson, M.A.; Wright, T.L.; Hill, R.T. Comparison of the bacterial communities of wild and captive sponge Clathria prolifera from the Chesapeake Bay. Mar. Biotechnol. 2009, 11, 758–770. [Google Scholar] [CrossRef]
- Ruocco, N.; Mutalipassi, M.; Pollio, A.; Costantini, S.; Costantini, M.; Zupo, V. First evidence of Halomicronema metazoicum (Cyanobacteria) free-living on Posidonia oceanica leaves. PLoS ONE 2018, 13, e0204954. [Google Scholar] [CrossRef]
- Zupo, V.; Mutalipassi, M.; Ruocco, N.; Glaviano, F.; Pollio, A.; Langellotti, A.L.; Romano, G.; Costantini, M. Distribution of toxigenic Halomicronema spp. in adjacent environments on the Island of Ischia: Comparison of strains from thermal waters and free living in Posidonia oceanica meadows. Toxins 2019, 11, 99. [Google Scholar] [CrossRef]
- Engene, N.; Gunasekera, S.P.; Gerwick, W.H.; Paul, V.J. Phylogenetic inferences reveal a large extent of novel biodiversity in chemically rich tropical marine cyanobacteria. Appl. Environ. Microbiol. 2013, 79, 1882–1888. [Google Scholar] [CrossRef]
- Niccolai, A.; Bigagli, E.; Biondi, N.; Rodolfi, L.; Cinci, L.; Luceri, C.; Tredici, M.R. In vitro toxicity of microalgal and cyanobacterial strains of interest as food source. J. Appl. Phycol. 2017, 29, 199–209. [Google Scholar] [CrossRef]
- Hrouzek, P.; Kapuścik, A.; Vacek, J.; Voráčová, K.; Paichlová, J.; Kosina, P.; Voloshkod, L.; Ventura, S.; Kopecký, J. Cytotoxicity evaluation of large cyanobacterial strain set using selected human and murine in vitro cell models. Ecotoxicol. Environ. Saf. 2016, 124, 177–185. [Google Scholar] [CrossRef]
- Freitas, S.; Martins, R.; Campos, A.; Azevedo, J.; Osório, H.; Costa, M.; Barros, P.; Vasconcelos, V.; Urbatzka, R. Insights into the potential of picoplanktonic marine cyanobacteria strains for cancer therapies—Cytotoxic mechanisms against the RKO colon cancer cell line. Toxicon 2016, 119, 140–151. [Google Scholar] [CrossRef] [PubMed]
- El Semary, N.; Fouda, M. Anticancer activity of Cyanothece sp. strain extracts from Egypt: First record. Asian Pac. J. Trop. Biomed. 2015, 5, 992–995. [Google Scholar] [CrossRef]
- Surakka, A.; Sihvonen, L.M.; Lehtimaki, J.M.; Wahlsten, M.; Vuorela, P.; Sivonen, K. Benthic cyanobacteria from the Baltic Sea contain cytotoxic Anabaena, Nodularia, and Nostoc strains and an apoptosis-inducing Phormidium strain. Environ. Toxicol. 2005, 20, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Mian, P.; Heilmann, J.; Burgi, H.R.; Sticher, O. Biological screening of terrestrial and freshwater cyanobacteria for antimicrobial activity, brine shrimp lethality, and cytotoxicity. Pharm. Biol. 2003, 41, 243–247. [Google Scholar] [CrossRef]
- Olvera-Ramírez, R.; Centeno-Ramos, C.; Martínez-Jerónimo, F. Toxic effects of Pseudanabaena tenuis (Cyanobacteria) on the cladocerans Daphnia magna and Ceriodaphnia dubia. Hidrobiológica 2010, 20, 203–221. [Google Scholar]
- Rangel, M.; Martins, J.C.G.; Nunes Garcia, A.; Conserva, G.A.A.; Costa-Neves, A.; Leite Sant’Anna, C.; De Retz Carvalho, L. Analysis of the toxicity and histopathology induced by the oral administration of Pseudanabaena galeata and Geitlerinema splendidum (Cyanobacteria) extracts to mice. Mar. Drugs 2014, 12, 508–524. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).