Molecular Characterization, Evolution and Expression Analysis of TNFSF14 and Three TNFSF Receptors in Spotted Gar Lepisosteus oculatus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Fish
2.2. Gene Cloning
2.3. Sequence Analysis
2.4. Tissue Expression Analysis
2.5. Expression Analysis during Infection of Klebsiella Pneumoniae
2.6. Statistical Analysis of Data
3. Results
3.1. Sequence Analysis
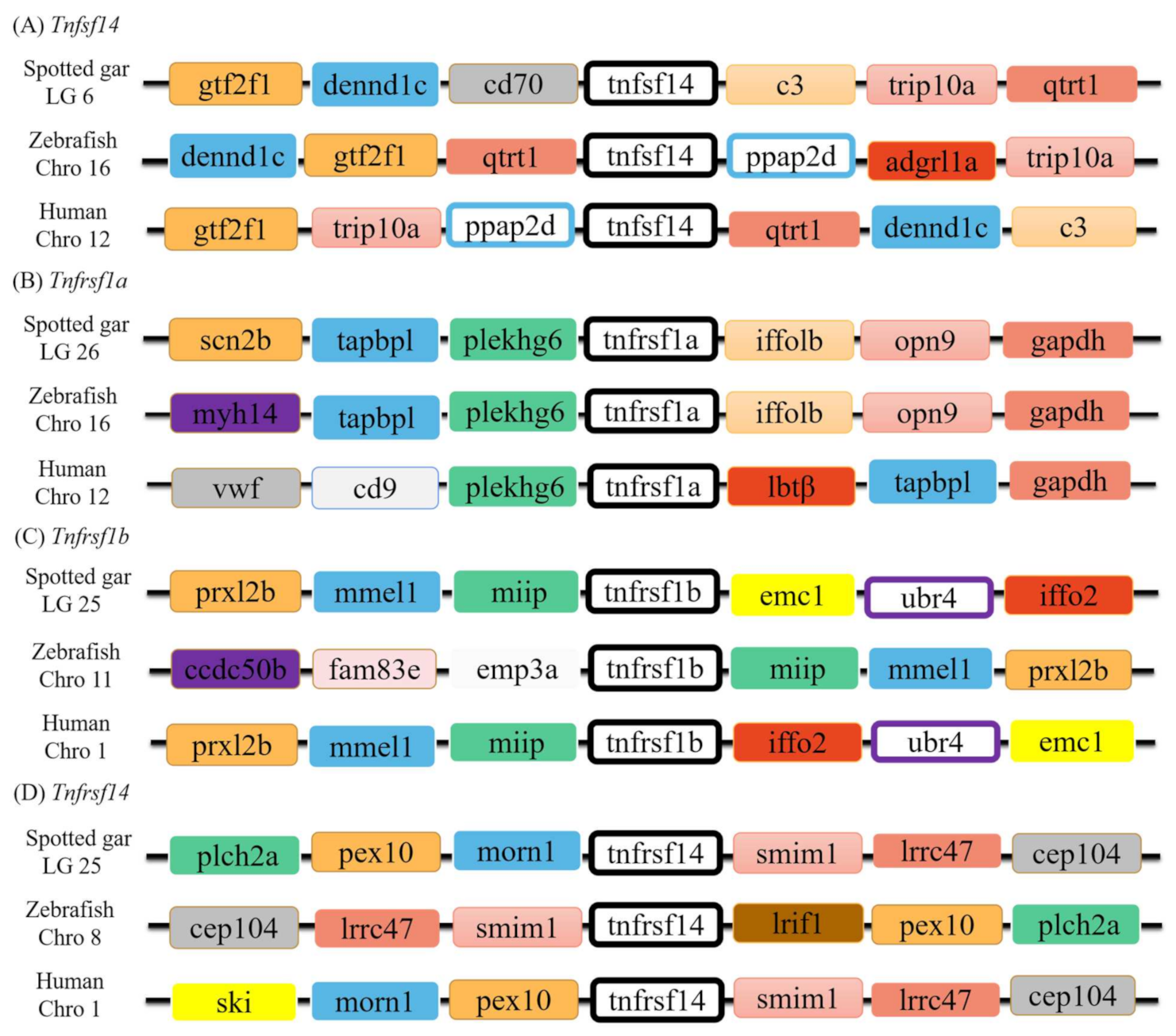
3.2. Genomic Structure and Synteny
3.3. Phylogenetic Analysis
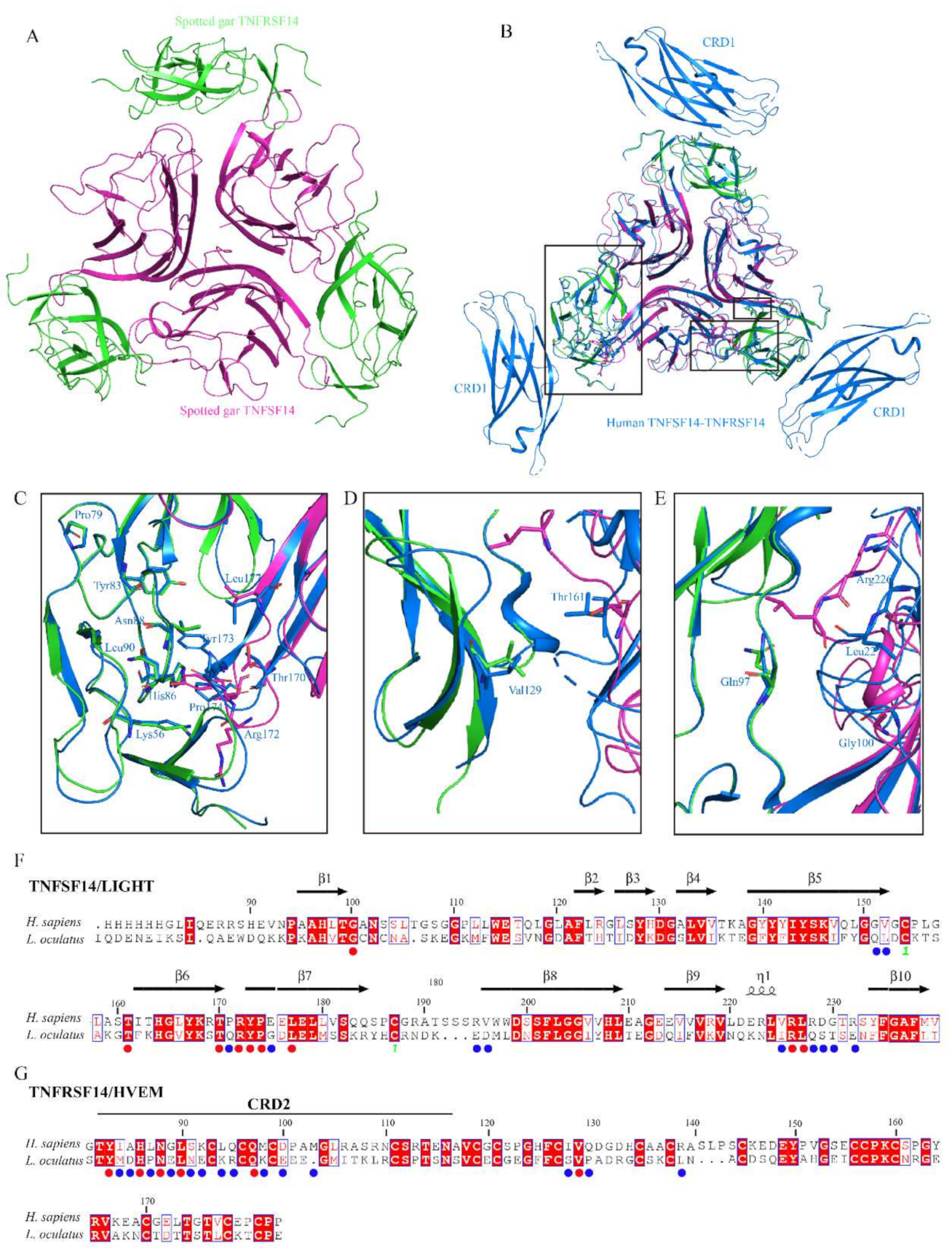
3.4. Protein Structure Modeling
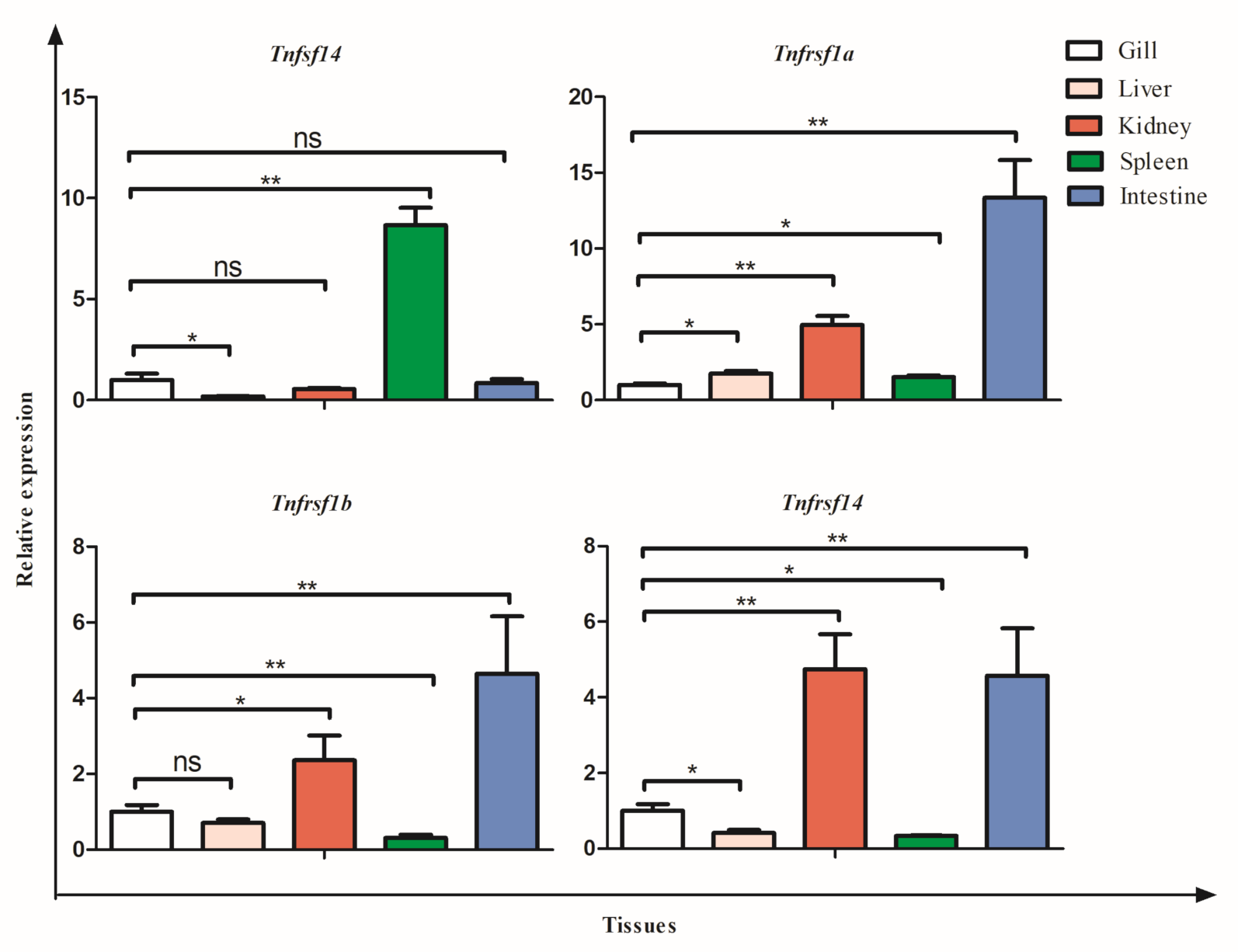
3.5. Tissue Expression
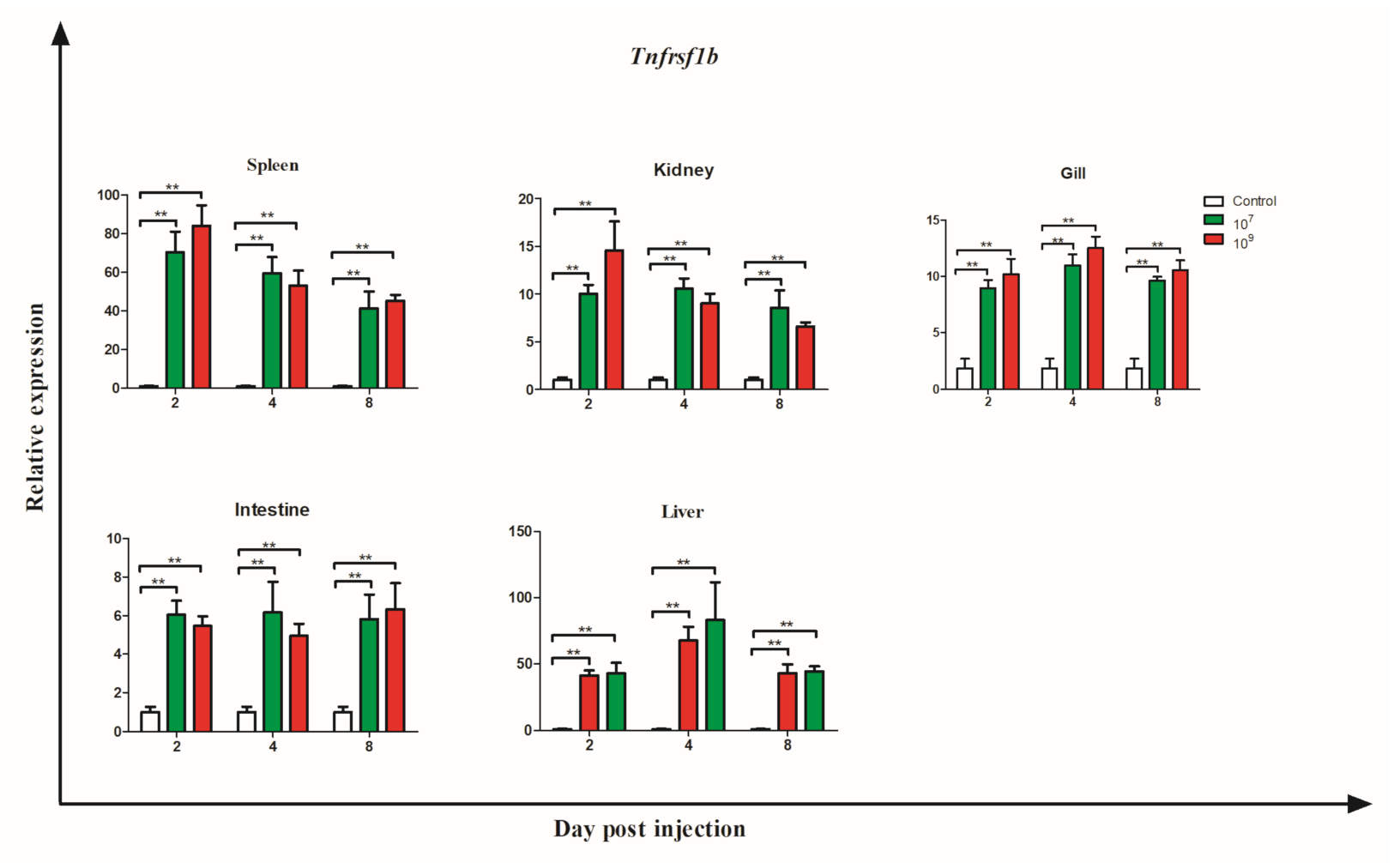
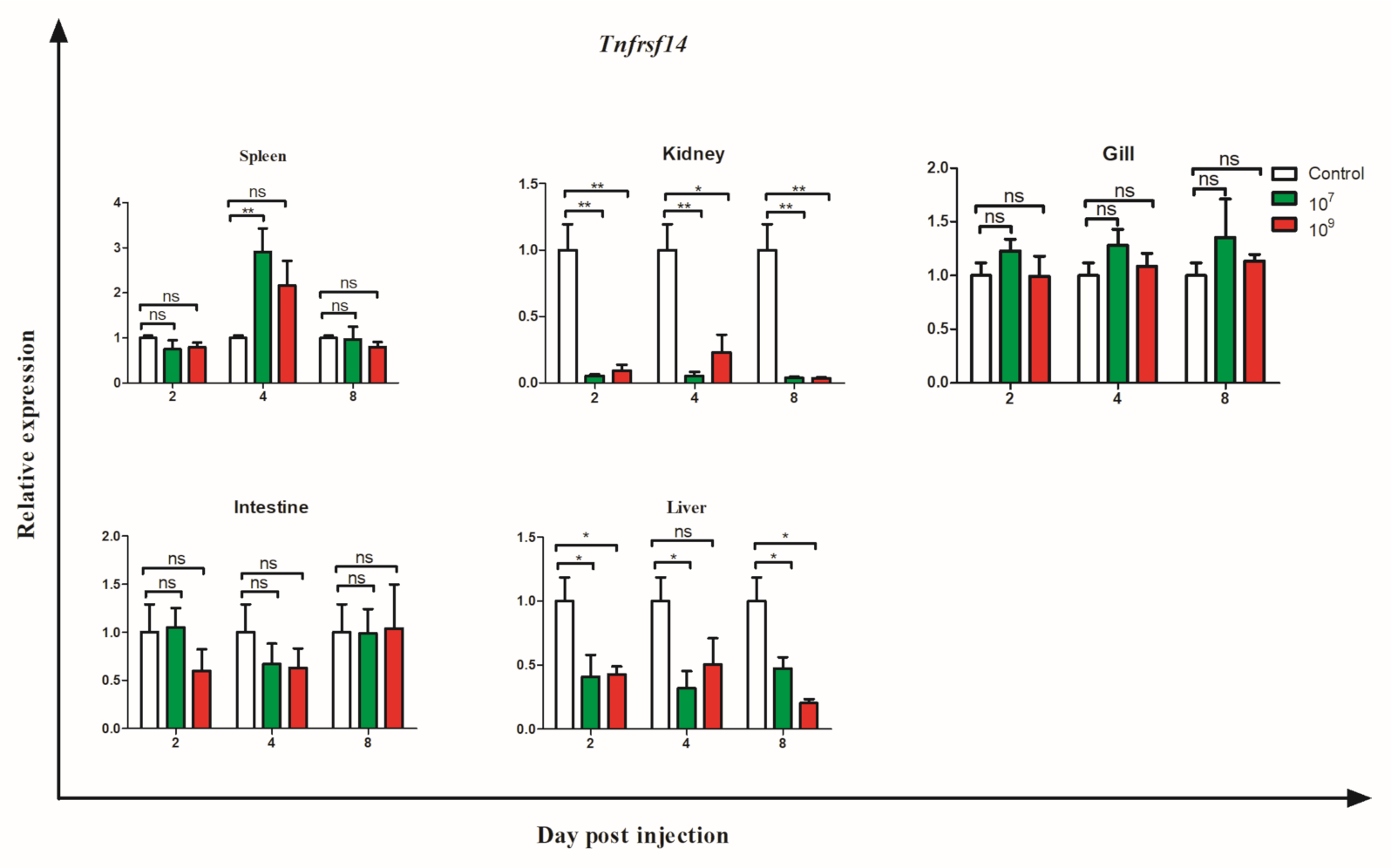
3.6. Gene Expression during Bacterial Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Sun, Y.; Liu, C.; Huang, M.; Huang, J.; Liu, C.; Zhang, J.; Postlethwait, J.-H.; Wang, H. The molecular evolution of circadian clock genes in spotted gar (Lepisosteus oculatus). Genes 2019, 10, 622. [Google Scholar] [CrossRef] [Green Version]
- Sukeena, J.M.; Galicia, C.A.; Wilson, J.D.; McGinn, T.; Boughman, J.W.; Robison, B.D.; Postlethwait, J.H.; Braasch, I.; Stenkamp, D.L.; Fuerst, P.G. Characterization and evolution of the spotted gar retina. J. Exp. Zool. B Mol. Dev. Evol. 2016, 326, 403–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wcisel, D.J.; Ota, T.; Litman, G.W.; Yoder, J.A. Spotted Gar and the Evolution of Innate Immune Receptors. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 666–684. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.; Lage, C.R.; Yoder, J.A.; Postlethwait, J.H.; Kim, C.H. Evolutionary divergence of the vertebrate TNFAIP8 gene family: Applying the spotted gar orthology bridge to understand ohnolog loss in teleosts. PLoS ONE 2017, 12, e0179517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Wang, X.; Li, C.; Feng, S.; Zhang, A.; Yang, K.; Zhou, H. Identification and functional characterization of grass carp (Ctenopharyngodon idella) tumor necrosis factor receptor 2 and its soluble form with potentiality for targeting inflammation. Fish Shellfish Immunol. 2019, 86, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, J.; Li, Q.; Pang, Y. Characterization of lamprey (Lampetra japonica) tnfr10-like gene: A potential granulocyte marker molecule and its immune functions. Mol. Immunol. 2020, 124, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Tafalla, C.; Granja, A.G. Novel insights on the regulation of B cell functionality by members of the tumor necrosis factor superfamily in jawed fish. Front. Immunol. 2018, 9, 1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodmer, J.L.; Schneider, P.; Tschopp, J. The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 2002, 27, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Pang, Y.; Li, Q. Molecular evolution of the tnfr gene family and expression profiles in response to pathogens in lamprey (Lethenteron reissneri). Fish Shellfish Immunol. 2020, 96, 336–349. [Google Scholar] [CrossRef]
- Hong, S.; Wang, T.Y.; Secombes, C.J.; Wang, T. Different origins of paralogues of salmonid TNR1 and TNFR2: Characterisation and expression analysis of four TNF receptor genes in rainbow trout Oncorhynchus mykiss. Dev. Comp. Immunol. 2019, 99, 103403. [Google Scholar] [CrossRef] [PubMed]
- Pozzolini, M.; Scarfì, S.; Ghignone, S.; Mussino, F.; Vezzulli, L.; Cerrano, C.; Giovine, M. Molecular characterization and expression analysis of the first Porifera tumor necrosis factor superfamily member and of its putative receptor in the marine sponge Chondrosia reniformis. Dev. Comp. Immunol. 2016, 57, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, T.; Zou, J. Fish TNF and TNF receptors. Sci. China Life Sci. 2021, 64, 196–220. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Kang, Y.J.; Koh, E.M.; Ahn, K.S.; Cha, H.S.; Lee, W.H. LIGHT is involved in the pathogenesis of rheumatoid arthritis by inducing the expression of pro-inflammatory cytokines and MMP-9 in macrophages. Immunology 2005, 114, 272–279. [Google Scholar] [CrossRef]
- Tian, A.Y.; Yang, H.J.; Zhu, S.C.; Zhang, Y.S.; Jiang, Z.A.; Song, J.Y.; Liu, H.Z.; Zhang, S.Q. Isolation and characterization of LIGHT (TNFSF14) gene homologue in zebrafish (Danio rerio). Int. Immunopharmacol. 2012, 14, 629–634. [Google Scholar] [CrossRef]
- Li, C.; Shen, Y.; Liang, D.; Yin, F.; Ai, H.; Sun, B.; Lin, S.; Zhang, S. Cloning, expression, and characterization of TNFSF14 (LIGHT) gene in mefugu, Takifugu obscurus. Mol. Cell Biochem. 2013, 379, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Suetake, H.; Odaka, T.; Miyadai, T. Original ligand for LTβR is LIGHT: Insight into evolution of the LT/LTβR system. J. Immunol. 2018, 201, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Quistad, S.D.; Stotland, A.; Barott, K.L.; Smurthwaite, C.A.; Hilton, B.J.; Grasis, J.A.; Wolkowicz, R.; Rohwer, F.L. Evolution of TNF-induced apoptosis reveals 550 My of functional conservation. Proc. Natl. Acad. Sci. USA 2014, 111, 9567–9572. [Google Scholar] [CrossRef] [Green Version]
- Balkwill, F. Tumour necrosis factor and cancer. Nat. Rev. Cancer 2009, 9, 361–371. [Google Scholar] [CrossRef]
- Eimon, P.M.; Kratz, E.; Varfolomeev, E.; Hymowitz, S.G.; Stern, H.; Zha, J.; Ashkenazi, A. Delineation of the cell-extrinsic apoptosis pathway in the zebrafish. Cell Death Differ. 2006, 13, 1619–1630. [Google Scholar] [CrossRef] [Green Version]
- Grayfer, L.; Belosevic, M. Molecular characterization of tumor necrosis factor receptors 1 and 2 of the goldfish (Carassius aurutus L.). Mol. Immunol. 2009, 46, 2190–2199. [Google Scholar] [CrossRef]
- Candel, S.; de Oliveira, S.; López-Muñoz, A.; García-Moreno, D.; Espín-Palazón, R.; Tyrkalska, S.D.; Cayuela, M.L.; Renshaw, S.A.; Corbalán-Vélez, R.; Vidal-Abarca, I.; et al. Tnfa signaling through tnfr2 protects skin against oxidative stress-induced inflammation. PLoS Biol. 2014, 12, e1001855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Z.W.; Kong, L.L.; Zhao, F.; Tan, A.P.; Deng, Y.T.; Jiang, L. Two types of TNF-α and their receptors in snakehead (Channa argus): Functions in antibacterial innate immunity. Fish Shellfish Immunol. 2020, 104, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Espín, R.; Roca, F.J.; Candel, S.; Sepulcre, M.P.; González-Rosa, J.M.; Alcaraz-Pérez, F.; Meseguer, J.; Cayuela, M.L.; Mercader, N.; Mulero, V. TNF receptors regulate vascular homeostasis in zebrafish through a caspase-8, caspase-2 and P53 apoptotic program that bypasses caspase-3. Dis Models Mech. 2013, 6, 383–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen-Chi, M.; Laplace-Builhé, B.; Travnickova, J.; Luz-Crawford, P.; Tejedor, G.; Lutfalla, G.; Kissa, K.; Jorgensen, C.; Djouad, F. TNF signaling and macrophages govern fin regeneration in zebrafish larvae. Cell Death Dis. 2017, 8, e2979. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.X.; Yang, Y.B.; Dang, H.F.; Xiao, H.H.; Huang, W.J.; Jia, Z.; Zhao, X.; Chen, K.Y.; Ji, N.; Guo, J.H.; et al. Molecular identification of Klebsiella pneumoniae and expression of immune genes in infected spotted gar Lepisosteus oculatus. Fish Shellfish Immunol. 2021, 119, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Jiang, X.; Gao, J.; Li, X.; Xu, J.; Wang, J.; Gao, Q.; Zou, J. Functional characterisation of interleukin 34 in grass carp Ctenopharyngodon idella. Fish Shellfish Immunol. 2019, 92, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Qin, Y.; Liu, D.; Wang, B.; Jia, Z.; Wang, J.; Gao, Q.; Zou, J.; Pang, Y. The evolution and functional characterization of CXC chemokines and receptors in lamprey. Dev. Comp. Immunol. 2021, 116, 103905. [Google Scholar] [CrossRef]
- Shrestha, R.; Garrett-Thomson, S.C.; Liu, W.; Almo, S.C.; Fiser, A. Redesigning HVEM interface for selective binding to LIGHT, BTLA, and CD160. Structure 2020, 28, 1197–1205.e2. [Google Scholar] [CrossRef]
- Palanisamy, R.; Kumaresan, V.; Harikrishnan, R.; Arasu, M.V.; Al-Dhabi, N.A.; Arockiaraj, J. Functional roles and gene regulation of tumor necrosis factor receptor 1 in freshwater striped murrel. Mol. Immunol. 2015, 66, 240–252. [Google Scholar] [CrossRef]
- Park, C.I.; Kurobe, T.; Hirono, I.; Aoki, T. Cloning and characterization of cDNAs for two distinct tumor necrosis factor receptor superfamily genes from Japanese flounder Paralichthys olivaceus. Dev. Comp. Immunol. 2003, 27, 365–375. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Brand, D.D.; Zheng, S.G. Role of TNF-TNF receptor 2 signal in regulatory T cells and its therapeutic implications. Front. Immunol. 2018, 9, 784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Zhang, R.; Ma, T.; Qiu, X.; Wang, X.; Zhang, A.; Zhou, H. Identification and functional characterization of tumor necrosis factor receptor 1 (TNFR1) of grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2016, 58, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Brisse, S.; Fevre, C.; Passet, V.; Issenhuth-Jeanjean, S.; Tournebize, R.; Diancourt, L.; Grimont, P. Virulent clones of Klebsiella pneumoniae: Identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS ONE 2009, 4, e4982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Acharya, S.; Behera, B.K.; Paria, P.; Bhowmick, S.; Parida, P.K.; Das, B.K. Isolation, identification and characterization of Klebsiella pneumoniae from infected farmed Indian Major Carp Labeo rohita (Hamilton 1822) in West Bengal, India. Aquaculture 2018, 482, 111–116. [Google Scholar] [CrossRef]
- Gopi, M.; Thankappanpillai, T.; Kumar, A.; Prakash, S. Opportunistic pathogen Klebsiella pneumoniae isolated from Maldive’s clown fish Amphiprion nigripes with hemorrhages at Agatti Island, Lakshadweep archipelago. Int. J. Fish. Aquat. Stud. 2016, 4, 464–467. [Google Scholar]
- Hu, Y.H.; Zhou, X.; Jiang, X.X.; Zhang, G.R.; Shi, Z.C.; Ji, W.; Ma, X.F.; Luo, Z.L.; Wei, K.J. Molecular characterization, expression analysis and function identification of Pf_TNF-α and its two receptors Pf_TNFR1 and Pf_TNFR2 in yellow catfish (Pelteobagrus fulvidraco). Int. J. Biol. Macromol. 2021, 185, 176–193. [Google Scholar] [CrossRef]







| Primer Name | Primer Sequences (5′-3′) | Application |
|---|---|---|
| tnfsf14-F1 | GGTCATCAAGACTGAAGGC | 3′-RACE |
| tnfsf14-F2 | GAGACCTGGAGCTCATGAGC | |
| tnfrsf1a-F1 | AGCACAAACAGCCTGAGCTG | 3′-RACE |
| tnfrsf1a-F2 | GTATAACATCAGCCTGGAG | |
| tnfrsf1b-F1 | GCACTACCACAACAACTGC | 3′-RACE |
| tnfrsf1b-F2 | GTAGCTGTCATCGACAAGAC | |
| tnfrsf14-F1 | ATGTGTGAGGCATGTGATTC | 3′-RACE |
| tnfrsf14-F2 | GCTGTCCTAAGTGCAATCGTG | |
| tnfsf14-R1 | GCCTTCAGTCTTGATGACC | 5′-RACE |
| tnfsf14-R2 | CCATCCACCACAAAGACTG | |
| tnfrsf1a-R1 | CTCCAGGCTGATGTTATAC | 5′-RACE |
| tnfrsf1a-R2 | CACAGCTGGATCATGTAGTAC | |
| tnfrsf1b-R1 | GTCTTGTCGATGACAGCTAC | 5′-RACE |
| tnfrsf1b-R2 | GCAGTTGTTGTGGTAGTGC | |
| tnfrsf14-R1 | GTCCTTCTGAACATACAGTG | 5′-RACE |
| tnfrsf14-R2 | CACGATTGCACTTAGGACAGC | |
| tnfsf14-F3 | CTCTCACGAAGGTCTTCCTG | Verify the full length |
| tnfsf14-R3 | GAGACATAGAGCTGAATGTAG | |
| tnfrsf1a-F3 | CACGTTGGAGACCTCGATG | Verify the full length |
| tnfrsf1a-R3 | GATGTGCTTATGCTCATGAC | |
| tnfrsf1b-F3 | CGAGTTGAGAGGGCGTGCTG | Verify the full length |
| tnfrsf1b-R3 | CTGGTACTGCCCTTGCATG | |
| tnfrsf14-F3 | GAATTCCAGGGTCCACGTGGG | Verify the full length |
| tnfrsf14-R3 | TTCAAAGTATTCTTGATGTAAATGTTGC | |
| tnfsf14-qF | CAGCATCAATGAAAACAGTCC | qRT-PCR |
| tnfsf14-qR | CAGATTCCCAGAACATCTTTCC | |
| tnfrsf1a-qF | AGCACAAACAGCCTGAGCTG | qRT-PCR |
| tnfrsf1a-qR | CACAGCTGGATCATGTAGTAC | |
| tnfrsf1b-qF | CACACTGACTGTGCCTCACAC | qRT-PCR |
| tnfrsf1b-qR | GTCTTGTCGATGACAGCTAC | |
| tnfrsf14-qF | GGGTGTTGCATTTTGGCTGAG | qRT-PCR |
| tnfrsf14-qR | GCATCTTTTGCACTCATTCAG | |
| ef-1α-qF | CAAGGATATCCCGTCGTGGCA | qRT-PCR |
| ef-1α-qR | AATACGCCAAGAGGAGG | |
| UPM-Long | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | |
| UPM-Short | CTAATACGACTCACTATAGGGC | |
| NUP | AAGCAGTGGTATCAACGCAGAGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Jia, Z.; Dang, H.; Xiao, H.; Huang, W.; Liu, Q.; Chen, K.; Zhang, L.; Zou, J.; Wang, J. Molecular Characterization, Evolution and Expression Analysis of TNFSF14 and Three TNFSF Receptors in Spotted Gar Lepisosteus oculatus. J. Mar. Sci. Eng. 2022, 10, 1035. https://doi.org/10.3390/jmse10081035
Wu Y, Jia Z, Dang H, Xiao H, Huang W, Liu Q, Chen K, Zhang L, Zou J, Wang J. Molecular Characterization, Evolution and Expression Analysis of TNFSF14 and Three TNFSF Receptors in Spotted Gar Lepisosteus oculatus. Journal of Marine Science and Engineering. 2022; 10(8):1035. https://doi.org/10.3390/jmse10081035
Chicago/Turabian StyleWu, Yaxin, Zhao Jia, Huifeng Dang, Hehe Xiao, Wenji Huang, Qin Liu, Kangyong Chen, Lei Zhang, Jun Zou, and Junya Wang. 2022. "Molecular Characterization, Evolution and Expression Analysis of TNFSF14 and Three TNFSF Receptors in Spotted Gar Lepisosteus oculatus" Journal of Marine Science and Engineering 10, no. 8: 1035. https://doi.org/10.3390/jmse10081035
APA StyleWu, Y., Jia, Z., Dang, H., Xiao, H., Huang, W., Liu, Q., Chen, K., Zhang, L., Zou, J., & Wang, J. (2022). Molecular Characterization, Evolution and Expression Analysis of TNFSF14 and Three TNFSF Receptors in Spotted Gar Lepisosteus oculatus. Journal of Marine Science and Engineering, 10(8), 1035. https://doi.org/10.3390/jmse10081035






