Prediction of Wheat Stripe Rust Occurrence with Time Series Sentinel-2 Images
Abstract
:1. Introduction
2. Materials and Methods
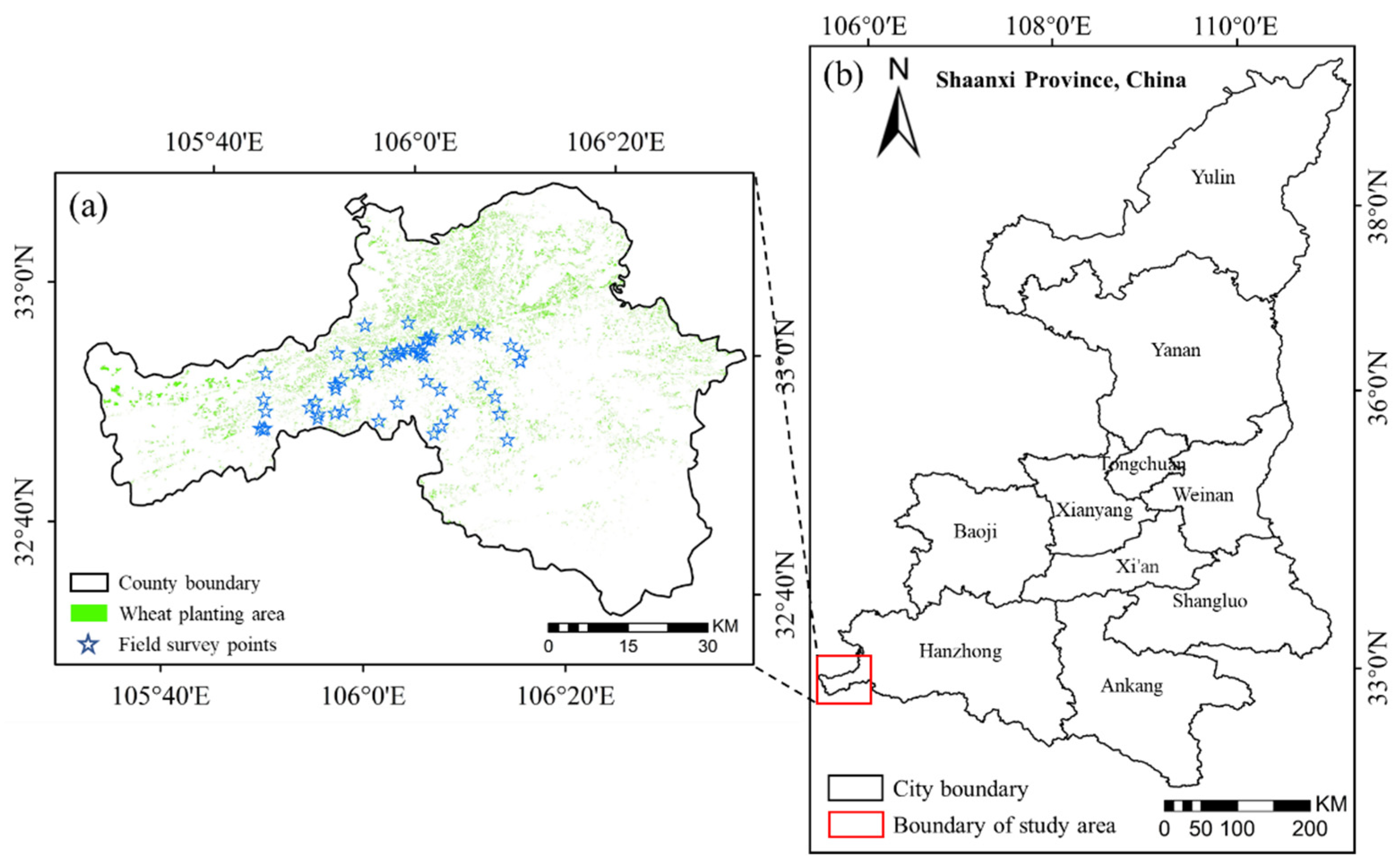
2.1. Study Site
2.2. Data Acquisition and Preprocessing
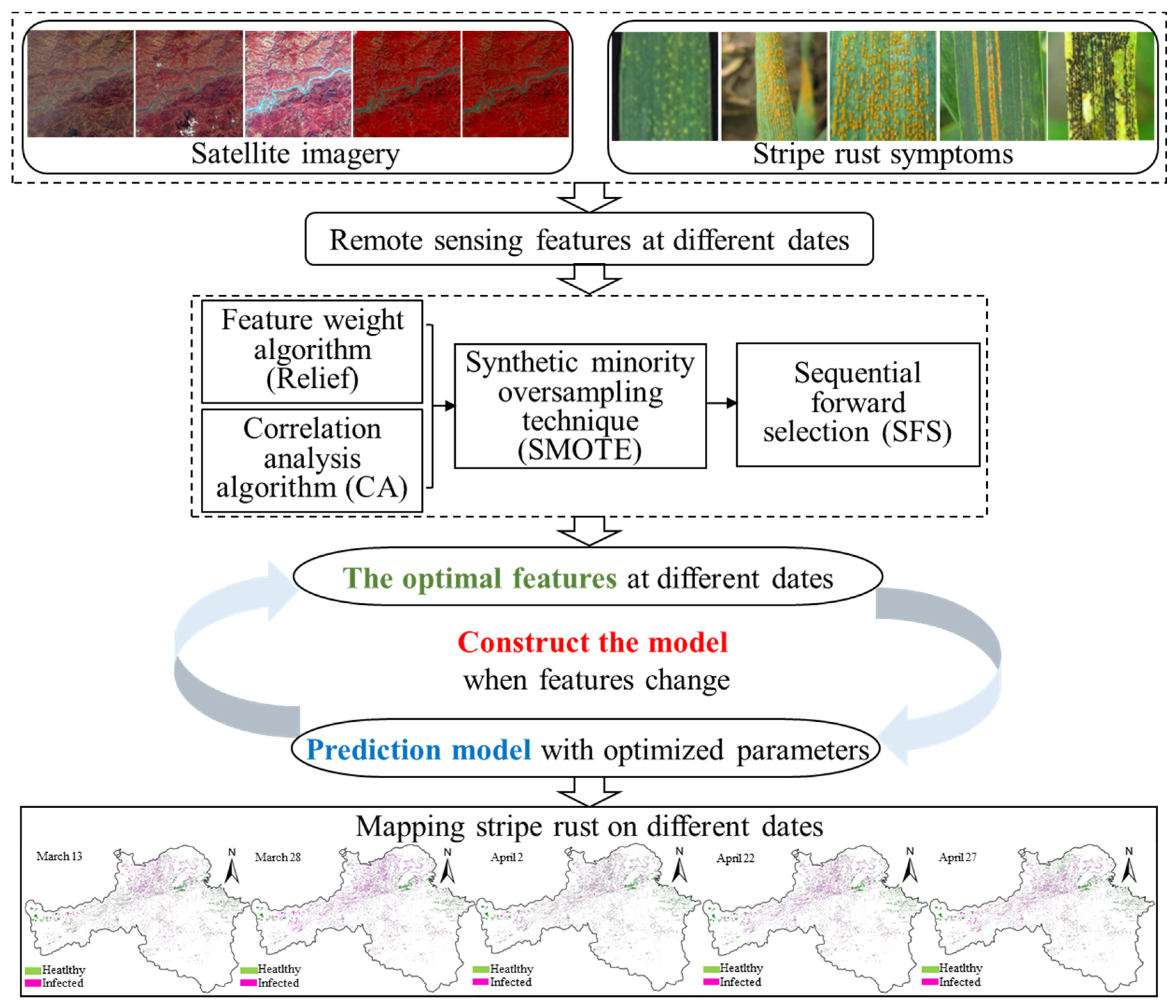
2.3. Prediction of Wheat Stripe Rust by Using Time Series Remote Sensing Images
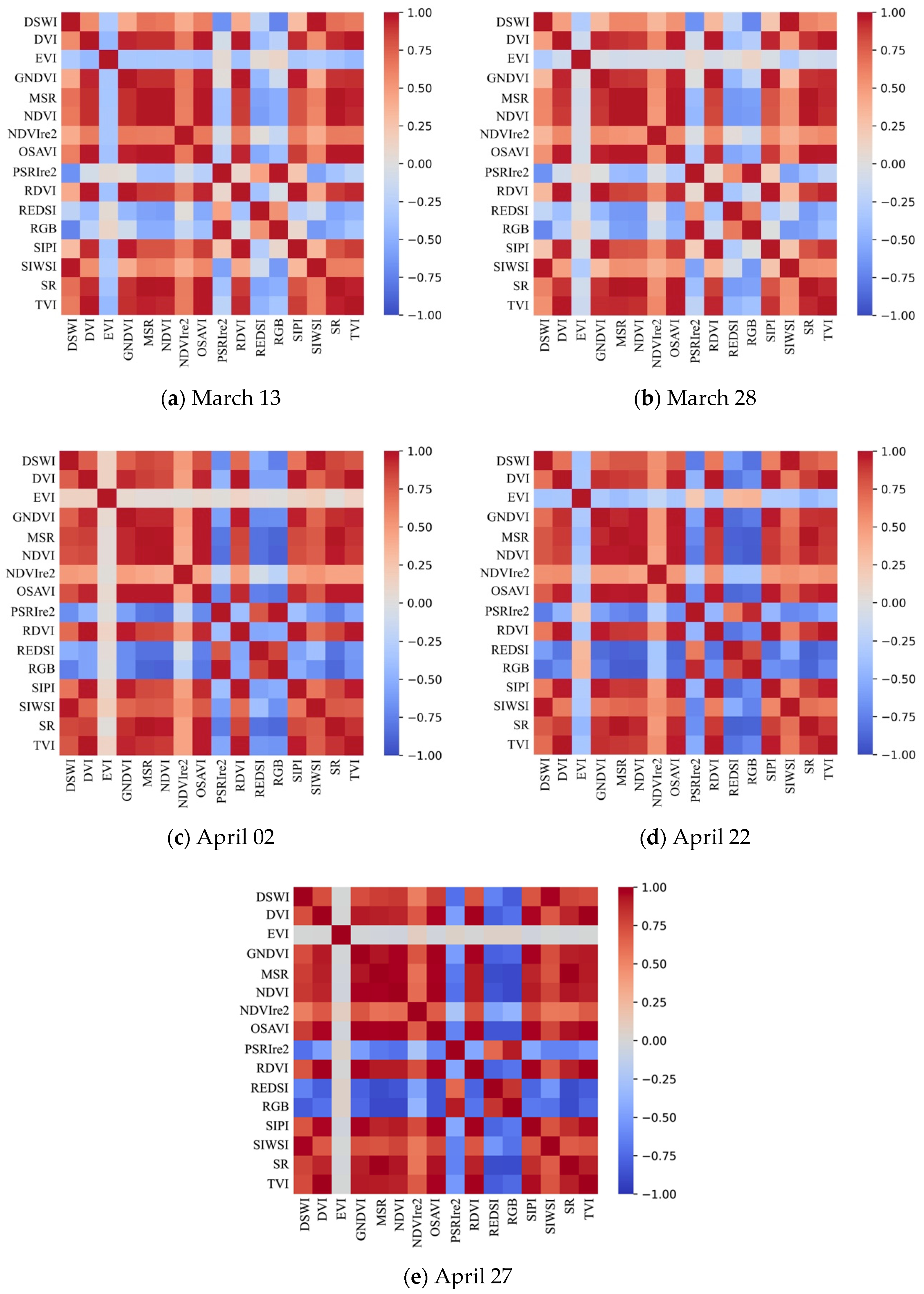
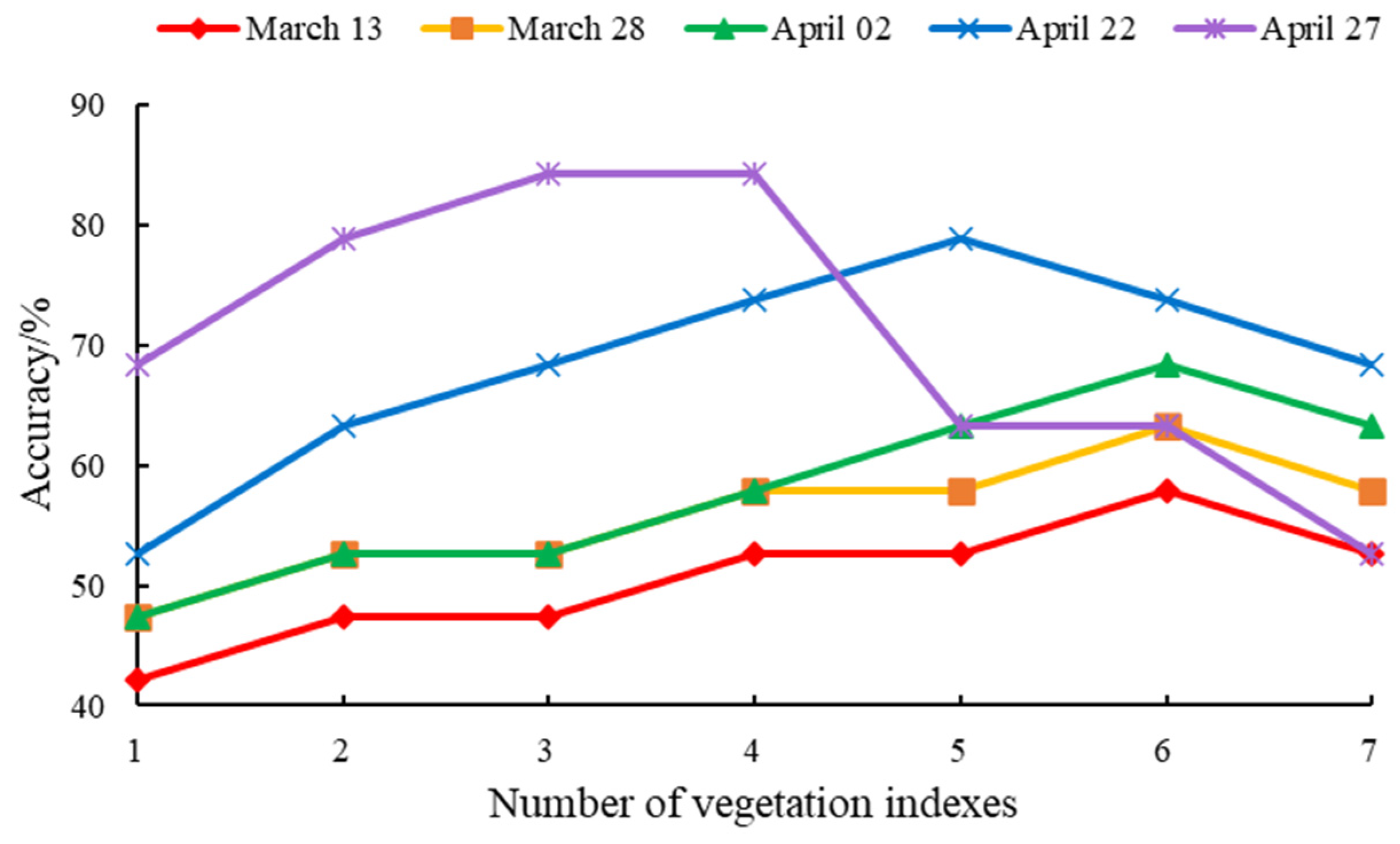
2.3.1. Selection of Optimal Vegetation Indice Combinations
| Vegetation Indices Definition | Formula | Correlation |
|---|---|---|
| Normalized difference Vegetation index, NDVI [27] | LAI, biomass | |
| Normalized difference vegetation Index red-edge2, NDVIre2 [47] | LAI, biomass | |
| Green normalized difference Vegetation index, GNDVI [48] | Pigment concentration | |
| Ratio of red and green, RGB [49] | Leaf pigment content | |
| Modified simple ratio index, MSR [50] | Vegetation status | |
| Simple ratio index, SR [51] | Photosynthetic area | |
| Enhanced vegetation index, EVI [52] | Sensitive to canopy | |
| Renormalized difference Vegetation index, RDVI [53] | Vegetation coverage | |
| Structural independent Pigment index, SIPI [54] | Pigment content | |
| Difference vegetation index, DVI [55] | Vegetation coverage | |
| Optimized soil-adjusted Vegetation index, OSAVI [56] | Minimize brightness-related soil effects | |
| Plant senescence reflectance Index, PSRI2 [31] | Pigment content, vegetation health | |
| Red-edge disease stress Index, REDSI [20] | Sensitive to stripe rust | |
| Triangular vegetation index, TVI [57] | Vegetation status | |
| Shortwave infrared water Stress index, SIWSI [29] | Water status | |
| Disease water stress index, DSWI [58] | Water status |
2.3.2. Establishment and Performance Test of the Wheat Stripe Rust Prediction Model
| Name | Formula | References |
|---|---|---|
| Overall accuracy, OA | [70] | |
| Producer accuracy, PA | [70] | |
| User accuracy, UA | [70] | |
| Kappa coefficient | [71] |
3. Results
3.1. Time Series Remote Sensing Stripe Rust Feature Extraction
3.2. Establishment and Verification of the Prediction Model of Wheat Stripe Rust
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wellings, C.R. Global status of stripe rust: A review of historical and current threats. Euphytica 2011, 179, 129–141. [Google Scholar] [CrossRef]
- Ali, S.; Rodriguez-Algaba, J.; Thach, T.; Sorensen, C.K.; Hansen, J.G.; Lassen, P.; Nazari, K.; Hodson, D.P.; Justesen, A.F.; Hovmoller, M.S. Yellow rust epidemics worldwide were caused by pathogen races from divergent genetic lineages. Front. Plant Sci. 2017, 8, 1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Pathol. 2005, 27, 314–337. [Google Scholar] [CrossRef]
- Chen, X. Pathogens which threaten food security: Puccinia striiformis, the wheat stripe rust pathogen. Food Secur. 2020, 12, 239–251. [Google Scholar] [CrossRef]
- Khanfri, S.; Boulif, M.; Lahlali, R. Yellow rust (Puccinia striiformis): A serious threat to wheat production worldwide. Not. Sci. Biol. 2018, 10, 410. [Google Scholar] [CrossRef] [Green Version]
- Boshoff, W.; Visser, B.; Lewis, C.; Adams, T.M.; Pretorius, Z.A. First report of Puccinia striiformis f. sp. tritici, causing stripe rust of wheat, in Zimbabwe. Plant Dis. 2019, 104, 290. [Google Scholar] [CrossRef]
- Boshoff, W.; Pretorius, Z.A.; Niekerk, B.V. Fungicide efficacy and the impact of stripe rust on spring and winter wheat in South Africa. S. Afr. J. Plant Soil 2003, 20, 11–17. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Line, R.F. Stripe rust of wheat and barley in North America: A retrospective historical review. Annu. Rev. Phytopathol. 2002, 40, 75–118. [Google Scholar] [CrossRef]
- Rapilly, F. Yellow rust epidemiology. Annu. Rev. Phytopathol. 1979, 17, 59–73. [Google Scholar] [CrossRef]
- Birr, T.; Verreet, J.A.; Klink, H. Prediction of deoxynivalenol and zearalenone in winter wheat grain in a maize-free crop rotation based on cultivar susceptibility and meteorological factors. J. Plant Dis. Prot. 2019, 126, 13–27. [Google Scholar] [CrossRef]
- Jarroudi, M.; Kouadio, L.; Bock, C.H.; Jarroudi, M.E.; Junk, J.; Pasquali, M.; Maraite, H.; Delfosse, P. A threshold-based weather model for predicting stripe rust infection in winter wheat. Plant Dis. 2016, 101, 693–703. [Google Scholar] [CrossRef] [Green Version]
- Jarroudi, M.; Lahlali, R.; Kouadio, L.; Denis, A.; Tychon, B. Weather-based predictive modeling of wheat stripe rust infection in Morocco. Agronomy 2020, 10, 280. [Google Scholar] [CrossRef] [Green Version]
- Allen-Sader, C.; Thurston, W.; Meyer, M.; Nure, E.; Bacha, N.; Alemayehu, Y.; Stutt, R.O.; Safka, D.; Craig, A.P.; Derso, E. An early warning system to predict and mitigate wheat rust diseases in Ethiopia. Environ. Res. Lett. 2019, 14, 115004. [Google Scholar] [CrossRef]
- Meyer, M.; Cox, J.; Hitchings, M.; Burgin, L.; Hort, M.; Hodson, D.; Gilligan, C. Quantifying airborne dispersal routes of pathogens over continents to safeguard global wheat supply. Nat. Plants 2017, 3, 780–786. [Google Scholar] [CrossRef]
- Yuan, L.; Zhang, J.; Shi, Y.; Nie, C.; Wei, L.; Wang, J. Damage mapping of powdery mildew in winter wheat with high-resolution satellite image. Remote Sens. 2014, 6, 3611–3623. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Huang, W.; Jing, Y.; Dong, Y.; Huang, L. Remote sensing monitoring of wheat powdery mildew based on AdaBoost model combining mRMR algorithm. Trans. Chin. Soc. Agric. Eng. 2017, 33, 162–169. [Google Scholar]
- Yuan, L.; Pu, R.; Zhang, J.; Wang, J.; Yang, H. Using high spatial resolution satellite imagery for mapping powdery mildew at a regional scale. Precis. Agric. 2016, 17, 332–348. [Google Scholar] [CrossRef]
- Zhang, J.; Yuan, L.; Nie, C.; Wei, L.; Yang, G. Forecasting of powdery mildew disease with multi-sources of remote sensing information. In Proceedings of the 2014 The Third International Conference on Agro-Geoinformatics, Beijing, China, 11–14 August 2014; pp. 1–5. [Google Scholar]
- Zheng, Q.; Huang, W.; Cui, X.; Yue, S.; Liu, L. New spectral index for detecting wheat yellow rust using Sentinel-2 multispectral imagery. Sensors 2018, 18, 868. [Google Scholar] [CrossRef] [Green Version]
- Dutta, S.; Singh, S.K.; Khullar, M. A case study on forewarning of yellow rust affected areas on wheat crop using satellite data. J. Indian Soc. Remote Sens. 2014, 42, 335–342. [Google Scholar] [CrossRef]
- Du, X.; Li, Q.; Shang, J.; Liu, J.; Qian, B.; Jing, Q.; Dong, T.; Fan, D.; Wang, H.; Zhao, L. Detecting advanced stages of winter wheat yellow rust and aphid infection using RapidEye data in North China Plain. GISci. Remote Sens. 2019, 56, 1093–1113. [Google Scholar] [CrossRef]
- Pryzant, R.; Ermon, S.; Lobell, D. In Monitoring Ethiopian wheat fungus with satellite imagery and deep feature learning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 39–47. [Google Scholar]
- Dong, Y.; Xu, F.; Liu, L.; Du, X.; Ren, B.; Guo, A.; Geng, Y.; Ruan, C.; Ye, H.; Huang, W. Automatic system for crop pest and disease dynamic monitoring and early forecasting. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 4410–4418. [Google Scholar] [CrossRef]
- Sørensen, C. Infection Biology and Aggressiveness of Puccinia striiformis on Resistant and Susceptible Wheat. Ph.D. Thesis, University of Aarhus, Research Center Flakkebjerg, Aarhus, Denmark, 2012. [Google Scholar]
- Zhang, J.; Huang, Y.; Pu, R.; Gonzalez-Moreno, P.; Yuan, L.; Wu, K.; Huang, W. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165, 104943. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.W.; Schell, J.A.; Deering, D.W.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; NASA/GSFC Type III. Final Report; Texas A & M University, Remote Sensing Center: Greenbelt, MD, USA, 1974. [Google Scholar]
- Salarux, C.; Kaewplang, S. Estimation of algal bloom biomass using UAV-Based remote sensing with NDVI and GRVI. Mahasarakham Int. J. Eng. Technol. 2020, 6, 1–6. [Google Scholar]
- Fensholt, R.; Sandholt, I. Derivation of a shortwave infrared water stress index from MODIS near-and shortwave infrared data in a semiarid environment. Remote Sens. Environ. 2003, 87, 111–121. [Google Scholar] [CrossRef]
- Olsen, J.L.; Ceccato, P.; Proud, S.R.; Fensholt, R.; Grippa, M.; Mougin, E.; Ardö, J.; Sandholt, I. Relation between seasonally detrended shortwave infrared reflectance data and land surface moisture in Semi-Arid Sahel. Remote Sens. 2013, 5, 2898–2927. [Google Scholar] [CrossRef] [Green Version]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Kang, Z.; Ma, Z.; Xu, S.; Jin, S.; Jiang, Y. Integrated management of wheat stripe rust caused by Puccinia striiformis f. sp. tritici in China. Sci. Agric. Sin. 2013, 46, 4254–4262. [Google Scholar]
- Xie, F.Z.; Feng, X.J.; Wen, Y.D.; Wang, B.T.; Zhi, X.M.; Kang, Z.S. Preliminary study on over-summer of Puccinia striiformis f. sp. tritici in Shaanxi Province during 2004–2011. J. Triticeae Crop. 2012, 32, 774–778. [Google Scholar]
- Zheng, Q.; Ye, H.; Huang, W.; Dong, Y.; Jiang, H.; Wang, C.; Li, D.; Wang, L.; Chen, S. Integrating spectral information and meteorological data to monitor wheat yellow rust at a regional scale: A case study. Remote Sens. 2021, 13, 278. [Google Scholar] [CrossRef]
- Dengke, L.; Zhao, W.; Feizhou, X. Occurrence regularity and meteorological influencing factors of wheat stripe rust in Shaanxi province. J. Catastrophol. 2019, 34, 59–65. [Google Scholar]
- Korhonen, L.; Packalen, P.; Rautiainen, M. Comparison of Sentinel-2 and Landsat 8 in the estimation of boreal forest canopy cover and leaf area index. Remote Sens. Environ. 2017, 195, 259–274. [Google Scholar] [CrossRef]
- Li, Z.; Yang, P.; Zhou, Q.; Wang, Y.; Wu, W.; Zhang, L.; Zhang, X. Research on spatiotemporal pattern of crop phenological characteristics and cropping system in North China based on NDVI time series data. Acta Ecol. Sin. 2009, 29, 6216–6226. [Google Scholar]
- Wang, L.; Liu, J.; Yang, F.; Fu, C.; Teng, F.; Gao, J. Early recognition of winter wheat area based on GF-1 satellite. Trans. Chin. Soc. Agric. Eng. 2015, 31, 194–201. [Google Scholar]
- Huang, L.; Ruan, C.; Huang, W.; Shi, Y.; Peng, D.; Ding, W. Wheat powdery mildew monitoring based on GF-1 remote sensing image and relief-mRMR-GASVM model. Trans. Chin. Soc. Agric. Eng. 2018, 34, 167–175. [Google Scholar]
- Zhang, L.X.; Wang, J.X.; Zhao, Y.N.; Yang, Z.H. Combination feature selection based on relief. J. Fudan Univ. Nat. Sci. Ed. 2004, 43, 893–898. [Google Scholar]
- Ma, H.; Huang, W.; Jing, Y.; Pignatti, S.; Laneve, G.; Dong, Y.; Ye, H.; Liu, L.; Guo, A.; Jiang, J. Identification of Fusarium Head Blight in winter wheat ears using continuous wavelet analysis. Sensors 2020, 20, 20. [Google Scholar] [CrossRef] [Green Version]
- Ververidis, D.; Kotropoulos, C. Sequential forward feature selection with low computational cost. In Proceedings of the 13th European Signal Processing Conference, Antalya, Turkey, 4–8 September 2005; pp. 1–4. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Liu, X.Y.; Wu, J.; Zhou, Z.H. Exploratory undersampling for class-imbalance learning. IEEE Trans. Syst. Man Cybern. Part B Cybern. 2008, 39, 539–550. [Google Scholar]
- Ma, H.; Huang, W.; Jing, Y.; Yang, C.; Han, L.; Dong, Y.; Ye, H.; Shi, Y.; Zheng, Q.; Liu, L. Integrating growth and environmental parameters to discriminate powdery mildew and aphid of winter wheat using bi-temporal Landsat-8 imagery. Remote Sens. 2019, 11, 846. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Luo, Z.; Huang, J.; Feng, Y.; Liu, Z. A novel ensemble method for imbalanced data learning: Bagging of extrapolation-SMOTE SVM. Comput. Intell. Neurosci. 2017, 2017, 1–11. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Quantitative estimation of chlorophyll-a using reflectance spectra: Experiments with autumn chestnut and maple leaves. J. Photochem. Photobiol. B Biol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Gamon, J.; Surfus, J. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Chen, J.M. Evaluation of vegetation indices and a modified simple ratio for boreal applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Peñuelas, J.; Filella, I. Visible and near-infrared reflectance techniques for diagnosing plant physiological status. Trends Plant Sci. 1998, 3, 151–156. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Roujean, J.-L.; Breon, F.-M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Peñuelas, J.; Inoue, Y. Reflectance indices indicative of changes in water and pigment contents of peanut and wheat leaves. Photosynthetica 1999, 36, 355–360. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Galvao, L.S.; Formaggio, A.R.; Tisot, D.A. Discrimination of sugarcane varieties in Southeastern Brazil with EO-1 Hyperion data. Remote Sens. Environ. 2005, 94, 523–534. [Google Scholar] [CrossRef]
- Amari, S.I.; Wu, S. Improving support vector machine classifiers by modifying kernel functions. Neural Netw. 1999, 12, 783–789. [Google Scholar] [CrossRef]
- Pal, M.; Foody, G.M. Feature selection for classification of hyperspectral data by SVM. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2297–2307. [Google Scholar] [CrossRef] [Green Version]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.C.; Dehne, H.W.; Plümer, L. Early detection and classification of plant diseases with support vector machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Thanh Noi, P.; Kappas, M. Comparison of random forest, k-nearest neighbor, and support vector machine classifiers for land cover classification using Sentinel-2 imagery. Sensors 2018, 18, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.W.; Chang, C.C.; Lin, C.J. A Practical Guide to Support Vector Classification; National Taiwan University: Taipei, Taiwan, 2003. [Google Scholar]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-validation. Encycl. Database Syst. 2009, 5, 532–538. [Google Scholar]
- Congalton, R.G.; Mead, R.A. A quantitative method to test for consistency and correctness in photointerpretation. Photogramm. Eng. Remote Sens. 1983, 49, 69–74. [Google Scholar]
- Ma, H.; Jing, Y.; Huang, W.; Shi, Y.; Dong, Y.; Zhang, J.; Liu, L. Integrating early growth information to monitor winter wheat powdery mildew using multi-temporal Landsat-8 imagery. Sensors 2018, 18, 3290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Wang, X.C.; Shi, F.; Yu, L.; Li, Y. Cases Analysis of MATLAB Neural Network; Beijing University of Aeronautics and Astronautics: Beijing, China, 2013; pp. 59–62. [Google Scholar]
- Story, M.; Congalton, R.G. Accuracy assessment: A user’s perspective. Photogramm. Eng. Remote Sens. 1986, 52, 397–399. [Google Scholar]
- Rosenfield, G.H.; Fitzpatrick-Lins, K. A coefficient of agreement as a measure of thematic classification accuracy. Photogramm. Eng. Remote Sens. 1986, 52, 223–227. [Google Scholar]
- Devadas, R.; Lamb, D.; Simpfendorfer, S.; Backhouse, D. Evaluating ten spectral vegetation indices for identifying rust infection in individual wheat leaves. Precis. Agric. 2009, 10, 459–470. [Google Scholar] [CrossRef]
- Nilsson, H. Remote sensing and image analysis in plant pathology. Annu. Rev. Phytopathol. 1995, 33, 489–528. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lamb, D.W.; Niu, Z.; Zhang, Y.; Liu, L.; Wang, J. Identification of yellow rust in wheat using in-situ spectral reflectance measurements and airborne hyperspectral imaging. Precis. Agric. 2007, 8, 187–197. [Google Scholar] [CrossRef]
- Lizarazo, I. SVM-based segmentation and classification of remotely sensed data. Int. J. Remote Sens. 2008, 29, 7277–7283. [Google Scholar] [CrossRef]
- Xiao, Y.; Dong, Y.; Huang, W.; Liu, L.; Ma, H.; Ye, H.; Wang, K. Dynamic remote sensing prediction for wheat fusarium head blight by combining host and habitat conditions. Remote Sens. 2020, 12, 3046. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, W.; González-Moreno, P.; Luke, B.; Dong, Y.; Zheng, Q.; Ma, H.; Liu, L. Wavelet-based rust spectral feature set (WRSFs): A novel spectral feature set based on continuous wavelet transformation for tracking progressive host–pathogen interaction of yellow rust on wheat. Remote Sens. 2018, 10, 525. [Google Scholar] [CrossRef] [Green Version]
- Coakley, S.M.; Line, R.F.; McDaniel, L.R. Predicting stripe rust severity on winter wheat using an improved method for analyzing meteorological and rust data. Phytopathology 1988, 78, 543–550. [Google Scholar] [CrossRef]
- Guo, X.; Wang, M.T.; Zhang, G.Z. Prediction model of meteorological grade of wheat stripe rust in winter-reproductive area, Sichuan Basin, China. J. Appl. Ecol. 2017, 28, 3994–4000. [Google Scholar]
- Rodríguez-Moreno, V.M.; Jiménez-Lagunes, A.; Estrada-Avalos, J.; Mauricio-Ruvalcaba, J.E.; Padilla-Ramírez, J.S. Weather-data-based model: An approach for forecasting leaf and stripe rust on winter wheat. Meteorol. Appl. 2020, 27, e1896. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Liu, Y.; Chen, H.; Zhang, C.C. Estimation model of winter wheat disease based on meteorological factors and spectral information. Food Prod. Process. Nutr. 2020, 2, 1–7. [Google Scholar] [CrossRef]

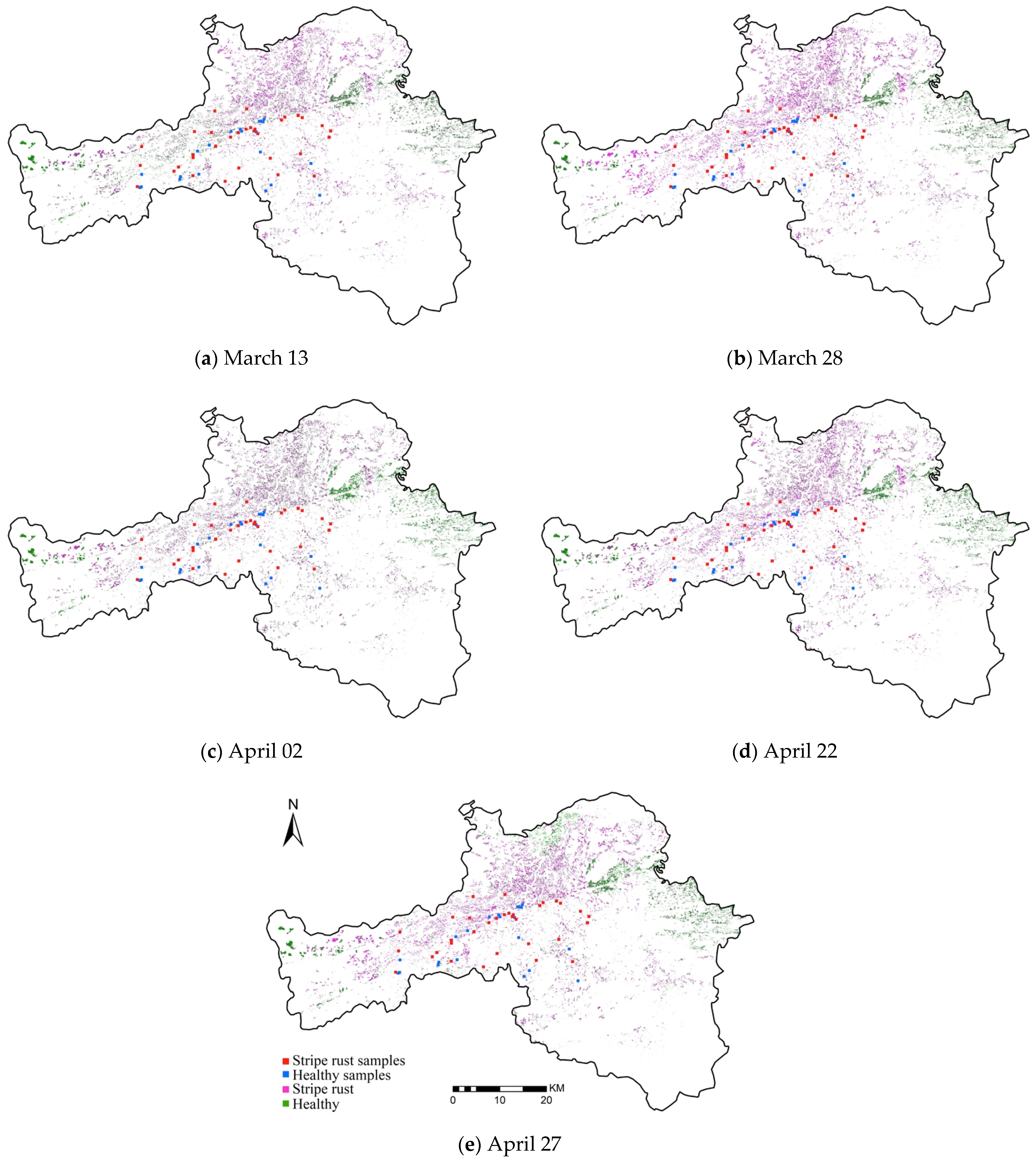
| Date | Vegetation Indices Combination |
|---|---|
| March 13 | NDVI, RGB, NDVIre2, RDVI, REDSI, DSWI, EVI |
| March 28 | NDVI, RGB, NDVIre2, RDVI, REDSI, DSWI, EVI |
| April 02 | PSRIre2, NDVI, NDVIre2, TVI, REDSI, DSWI, EVI |
| April 22 | PSRIre2, NDVI, NDVIre2, TVI, REDSI, DSWI, EVI |
| April 27 | PSRIre2, NDVI, NDVIre2, TVI, REDSI, DSWI, EVI |
| Type | Original Training Samples | Balanced Training Samples after Using SMOTE | ||
|---|---|---|---|---|
| Number | Ratio | Number | Ratio | |
| Healthy | 15 | 38.5% | 24 | 50.0% |
| Diseased | 24 | 61.5% | 24 | 50.0% |
| Sum | 39 | 100% | 48 | 100% |
| Date | Vegetation Indice Combinations |
|---|---|
| March 13 | NDVI, REDSI, DSWI, NDVIre2, RDVI, RGB |
| March 28 | NDVI, REDSI, DSWI, NDVIre2, RDVI, RGB |
| April 02 | NDVI, REDSI, DSWI, NDVIre2, TVI, PSRIre2 |
| April 22 | NDVI, REDSI, DSWI, TVI, PSRIre2 |
| April 27 | NDVI, REDSI, PSRIre2 |
| Model | Date | Parameter | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | γ | ||||||||||
| SVM | March 13 | 90.5097 | 0.0221 | ||||||||
| March 28 | 90.5097 | 0.3535 | |||||||||
| April 02 | 2.8248 | 0.0441 | |||||||||
| April 22 | 45.2548 | 0.011 | |||||||||
| April 27 | 1.0 | 0.0625 | |||||||||
| Model | Date | Parameter | |||||||||
| k | |||||||||||
| KNN | March 13 | 5 | |||||||||
| March 28 | 5 | ||||||||||
| April 02 | 5 | ||||||||||
| April 22 | 3 | ||||||||||
| April 27 | 3 | ||||||||||
| Model | Date | The number of layers | |||||||||
| Input | Hidden | Output | |||||||||
| BPNN | March 13 | 6 | 7 | 2 | |||||||
| March 28 | 6 | 7 | 2 | ||||||||
| April 02 | 6 | 7 | 2 | ||||||||
| April 22 | 5 | 6 | 2 | ||||||||
| April 27 | 3 | 4 | 2 | ||||||||
| Date | Method | Healthy | Infected | Sum | UA | OA | Kappa | |
|---|---|---|---|---|---|---|---|---|
| March 13 | SVM | Healthy | 13 | 11 | 24 | 54.2% | 65.5% | 0.280 |
| Infected | 9 | 25 | 34 | 73.5% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 59.1% | 69.4% | ||||||
| BPNN | Healthy | 12 | 12 | 24 | 50.0% | 62.1% | 0.208 | |
| Infected | 10 | 24 | 34 | 70.1% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 54.5% | 66.7% | ||||||
| KNN | Healthy | 11 | 13 | 24 | 45.8% | 58.6% | 0.136 | |
| Infected | 11 | 23 | 34 | 67.6% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 50.0% | 63.9% | ||||||
| March 28 | SVM | Healthy | 14 | 10 | 24 | 58.3% | 69.0% | 0.352 |
| Infected | 8 | 26 | 34 | 76.5% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 63.6% | 72.2% | ||||||
| BPNN | Healthy | 13 | 11 | 24 | 54.2% | 65.5% | 0.280 | |
| Infected | 9 | 25 | 34 | 73.5% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 59.1% | 69.4% | ||||||
| KNN | Healthy | 12 | 12 | 24 | 50.0% | 62.1% | 0.208 | |
| Infected | 10 | 24 | 34 | 70.1% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 54.5% | 66.7% | ||||||
| April 02 | SVM | Healthy | 15 | 8 | 23 | 65.2% | 74.1% | 0.456 |
| Infected | 7 | 28 | 35 | 80.0% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 68.2% | 77.8% | ||||||
| BPNN | Healthy | 14 | 9 | 23 | 60.9% | 70.7% | 0.382 | |
| Infected | 8 | 27 | 35 | 77.1% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 63.6% | 75.0% | ||||||
| KNN | Healthy | 13 | 11 | 24 | 54.2% | 65.5% | 0.280 | |
| Infected | 9 | 25 | 34 | 73.5% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 59.1% | 69.4% | ||||||
| April 22 | SVM | Healthy | 17 | 7 | 24 | 70.8% | 79.3% | 0.568 |
| Infected | 5 | 29 | 34 | 85.3% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 77.3% | 80.6% | ||||||
| BPNN | Healthy | 16 | 8 | 24 | 66.7% | 75.9% | 0.496 | |
| Infected | 6 | 28 | 34 | 82.4% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 72.7% | 77.8% | ||||||
| KNN | Healthy | 15 | 10 | 25 | 60.0% | 70.7% | 0.394 | |
| Infected | 7 | 26 | 33 | 78.8% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 68.2% | 72.2% | ||||||
| April 27 | SVM | Healthy | 18 | 4 | 22 | 81.8% | 86.2% | 0.707 |
| Infected | 4 | 32 | 36 | 88.9% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 81.8% | 88.9% | ||||||
| BPNN | Healthy | 17 | 6 | 23 | 73.9% | 81.0% | 0.601 | |
| Infected | 5 | 30 | 35 | 85.7% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 77.3% | 83.3% | ||||||
| KNN | Healthy | 16 | 7 | 23 | 69.6% | 77.6% | 0.528 | |
| Infected | 6 | 29 | 35 | 82.9% | ||||
| Sum | 22 | 36 | 58 | |||||
| PA | 72.7% | 80.6% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruan, C.; Dong, Y.; Huang, W.; Huang, L.; Ye, H.; Ma, H.; Guo, A.; Ren, Y. Prediction of Wheat Stripe Rust Occurrence with Time Series Sentinel-2 Images. Agriculture 2021, 11, 1079. https://doi.org/10.3390/agriculture11111079
Ruan C, Dong Y, Huang W, Huang L, Ye H, Ma H, Guo A, Ren Y. Prediction of Wheat Stripe Rust Occurrence with Time Series Sentinel-2 Images. Agriculture. 2021; 11(11):1079. https://doi.org/10.3390/agriculture11111079
Chicago/Turabian StyleRuan, Chao, Yingying Dong, Wenjiang Huang, Linsheng Huang, Huichun Ye, Huiqin Ma, Anting Guo, and Yu Ren. 2021. "Prediction of Wheat Stripe Rust Occurrence with Time Series Sentinel-2 Images" Agriculture 11, no. 11: 1079. https://doi.org/10.3390/agriculture11111079
APA StyleRuan, C., Dong, Y., Huang, W., Huang, L., Ye, H., Ma, H., Guo, A., & Ren, Y. (2021). Prediction of Wheat Stripe Rust Occurrence with Time Series Sentinel-2 Images. Agriculture, 11(11), 1079. https://doi.org/10.3390/agriculture11111079








