A Pilot Study on MicroRNA Profile in Tear Fluid to Predict Response to Anti-VEGF Treatments for Diabetic Macular Edema
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Treatment of DME and Assessment of Treatment Response
2.3. Collection of Patient Specimens
2.4. RNA Extraction and Reverse Transcription
2.5. MicroRNA Expression Quantification Using qPCR Assay
2.6. Bioinformatics and Statistical Analysis of miRNA Expression
3. Results
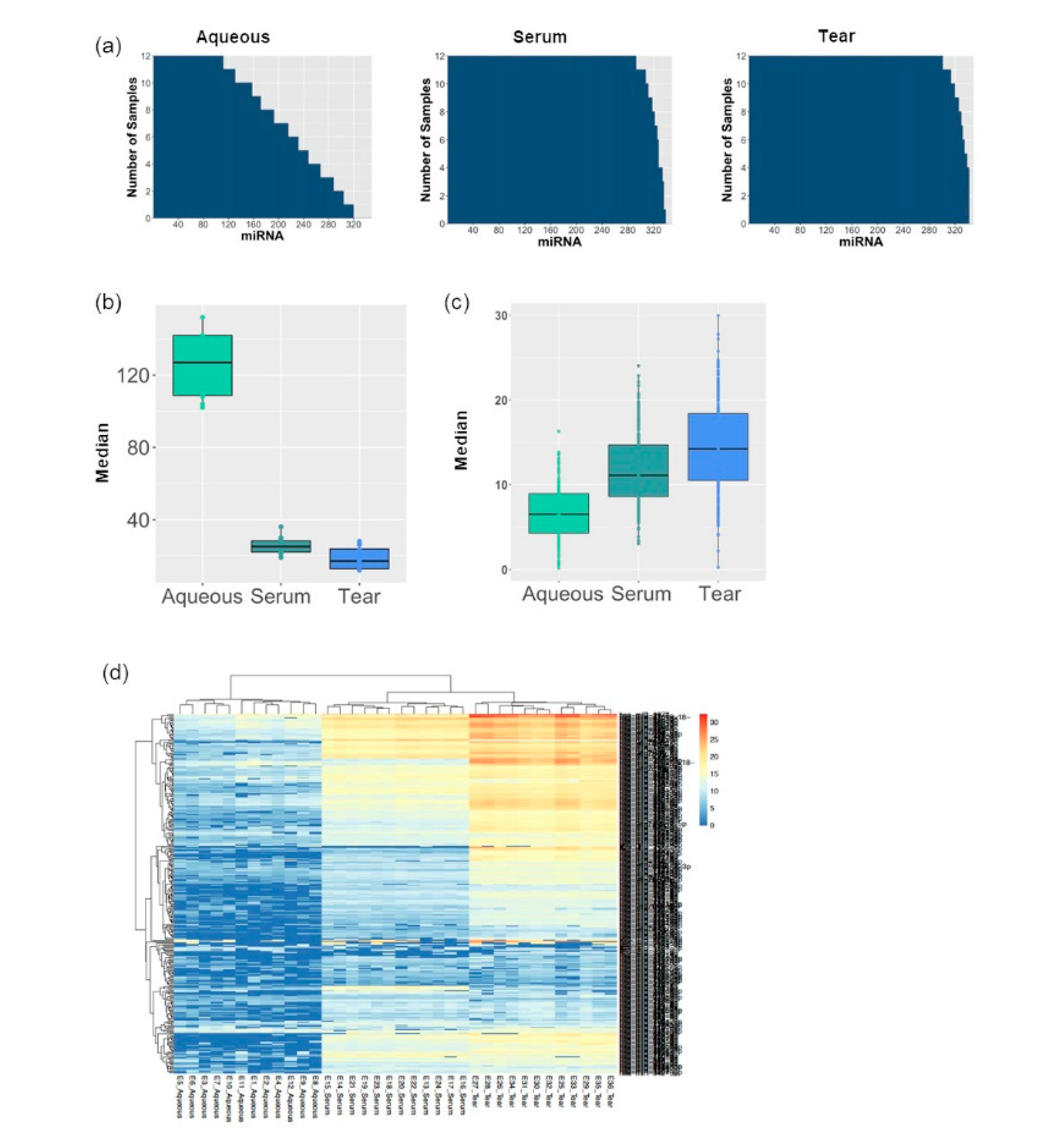
3.1. MiRNA Abundancy Was Significantly Higher in Tears Compared to Aqueous Humor
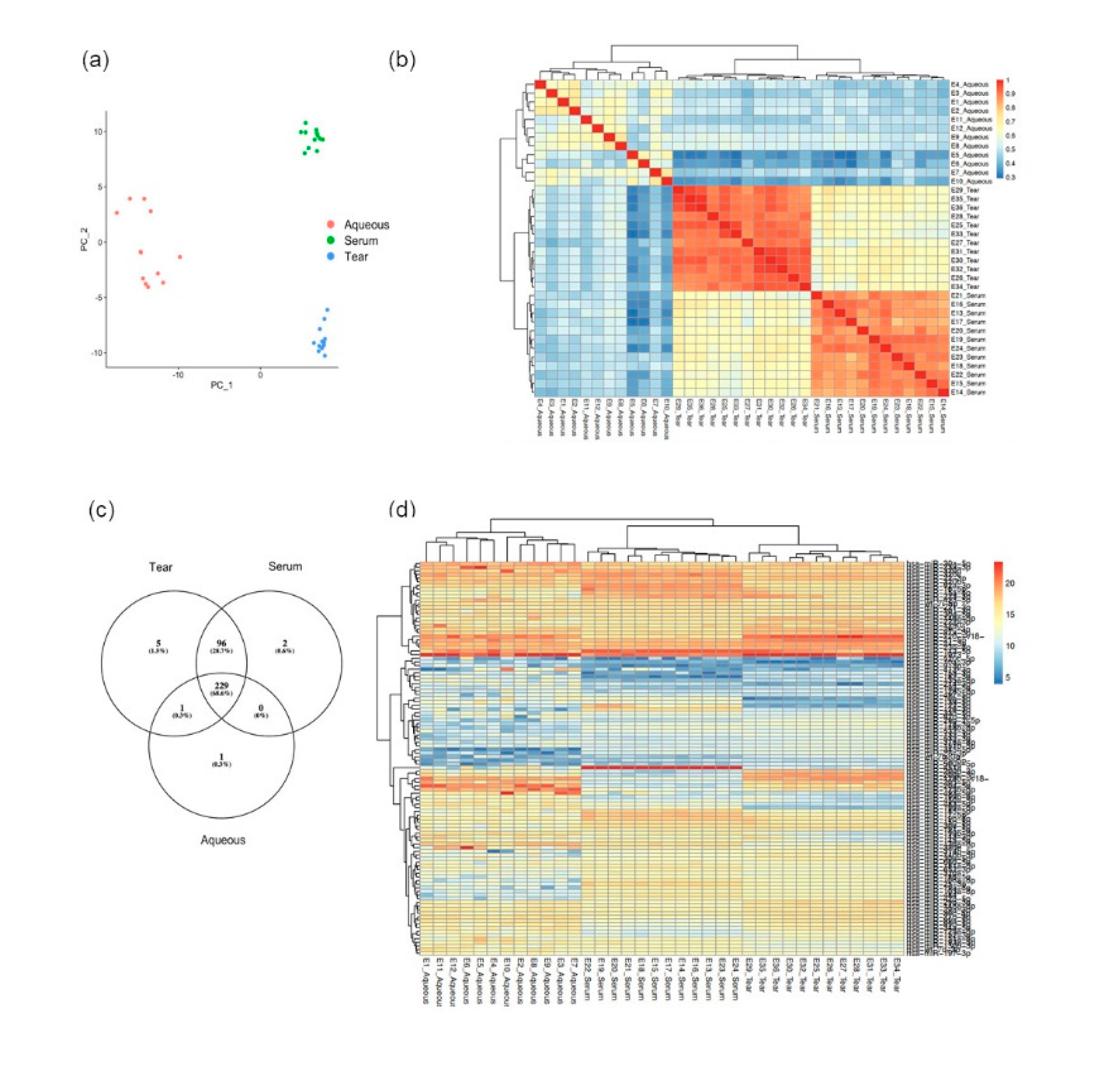
3.2. Tear Fluids miRNA Profiling Were More Consistent and Reproducible Compared to Aqueous Humor
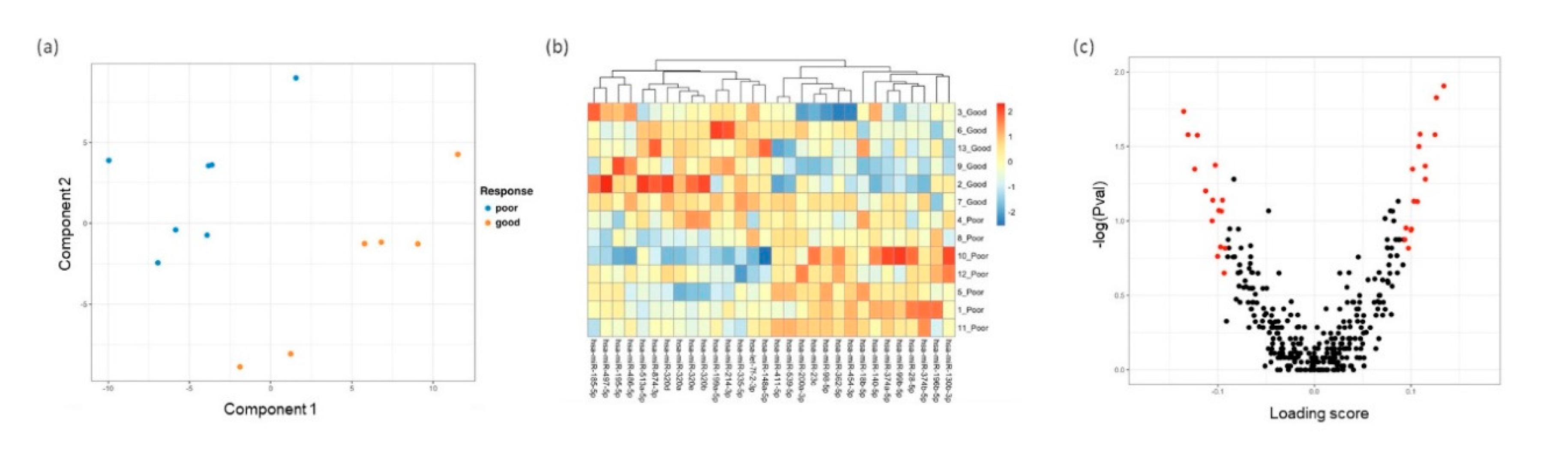
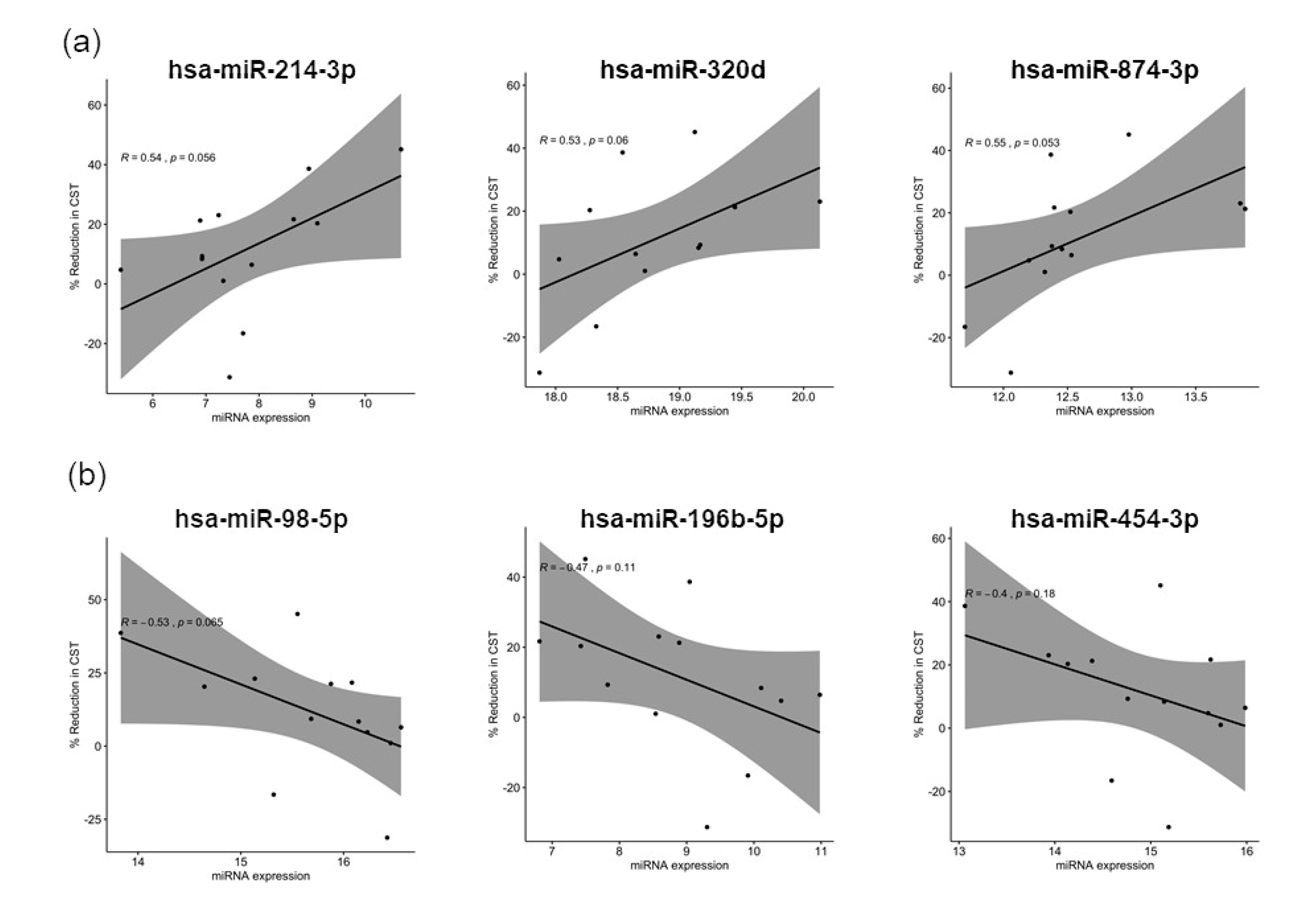
3.3. Distinct miRNA Expression Signatures in Tear Were Associated with Patient Response to Anti-VEGF Treatment
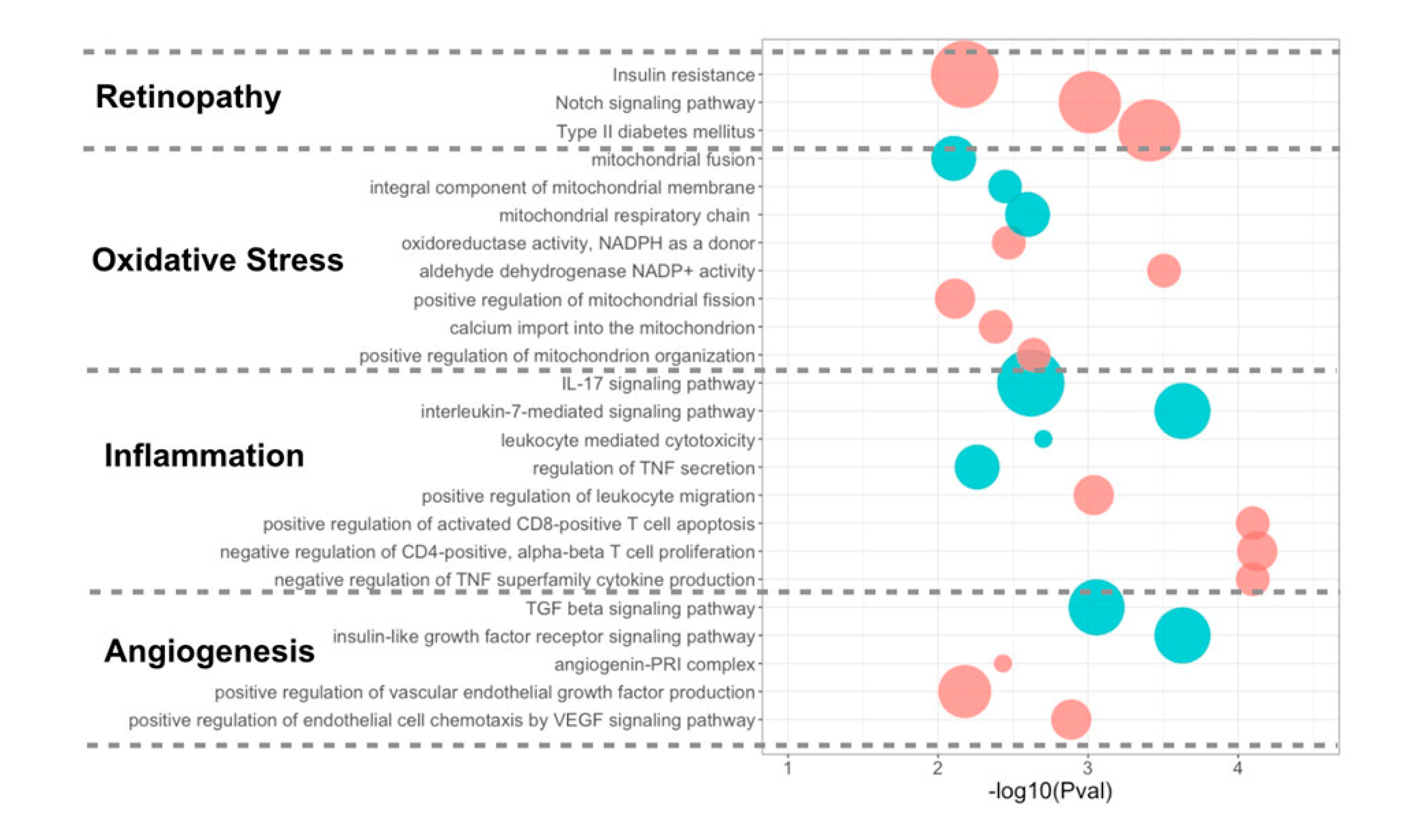
3.4. Differentially Expressed miRNA Targeting Genes and Functional Analysis
4. Discussion
4.1. Extracellular miRNAs as a Biomarker in DME
4.2. Comparison of the miRNA Expression Profile of Tear Fluid, Aqueous Humor, and Serum
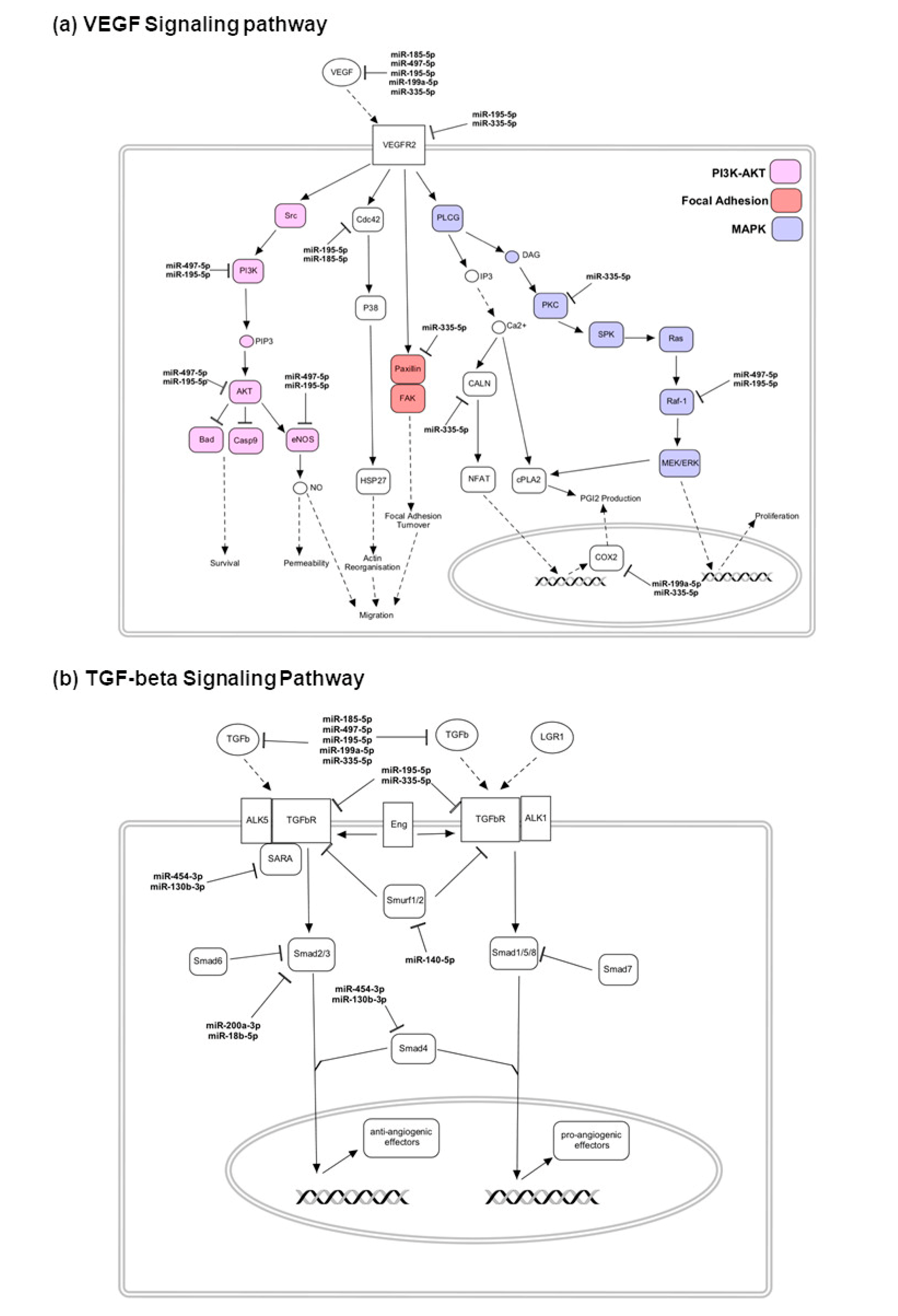
4.3. Upregulated miRNAs and the Targeting Pathways in Good Responders
4.4. Upregulated miRNAs and the Targeting Pathways in Poor Responders
4.5. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Varma, R.; Bressler, N.M.; Doan, Q.V.; Gleeson, M.; Danese, M.; Bower, J.K.; Selvin, E.; Dolan, C.; Fine, J.; Colman, S.; et al. Prevalence of and risk factors for diabetic macular edema in the United States. JAMA Ophthalmol. 2014, 132, 1334–1340. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.S.; Cheung, N.; Simo, R.; Cheung, G.C.; Wong, T.Y. Diabetic macular oedema. Lancet Diabetes Endocrinol. 2017, 5, 143–155. [Google Scholar] [CrossRef]
- Klein, R.; Klein, B.E.; Moss, S.E.; Cruickshanks, K.J. The Wisconsin Epidemiologic Study of Diabetic Retinopathy. XV. The long-term incidence of macular edema. Ophthalmology 1995, 102, 7–16. [Google Scholar] [CrossRef]
- Yau, J.W.; Rogers, S.L.; Kawasaki, R.; Lamoureux, E.L.; Kowalski, J.W.; Bek, T.; Chen, S.J.; Dekker, J.M.; Fletcher, A.; Grauslund, J.; et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care 2012, 35, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.D.; Brown, D.M.; Marcus, D.M.; Boyer, D.S.; Patel, S.; Feiner, L.; Gibson, A.; Sy, J.; Rundle, A.C.; Hopkins, J.J.; et al. Ranibizumab for diabetic macular edema: Results from 2 phase III randomized trials: RISE and RIDE. Ophthalmology 2012, 119, 789–801. [Google Scholar] [CrossRef]
- Michaelides, M.; Kaines, A.; Hamilton, R.D.; Fraser-Bell, S.; Rajendram, R.; Quhill, F.; Boos, C.J.; Xing, W.; Egan, C.; Peto, T.; et al. A prospective randomized trial of intravitreal bevacizumab or laser therapy in the management of diabetic macular edema (BOLT study) 12-month data: Report 2. Ophthalmology 2010, 117, 1078–1086.e1072. [Google Scholar] [CrossRef]
- Bressler, S.B.; Ayala, A.R.; Bressler, N.M.; Melia, M.; Qin, H.; Ferris, F.L., III; Flaxel, C.J.; Friedman, S.M.; Glassman, A.R.; Jampol, L.M.; et al. Persistent Macular Thickening After Ranibizumab Treatment for Diabetic Macular Edema With Vision Impairment. JAMA Ophthalmol. 2016, 134, 278–285. [Google Scholar] [CrossRef]
- Ashraf, M.; Souka, A.; Adelman, R. Predicting outcomes to anti-vascular endothelial growth factor (VEGF) therapy in diabetic macular oedema: A review of the literature. Br. J. Ophthalmol. 2016, 100, 1596–1604. [Google Scholar] [CrossRef]
- Ting, D.S.; Tan, K.A.; Phua, V.; Tan, G.S.; Wong, C.W.; Wong, T.Y. Biomarkers of Diabetic Retinopathy. Curr. Diab. Rep. 2016, 16, 125. [Google Scholar] [CrossRef]
- Busch, C.; Zur, D.; Fraser-Bell, S.; Lains, I.; Santos, A.R.; Lupidi, M.; Cagini, C.; Gabrielle, P.H.; Couturier, A.; Mane-Tauty, V.; et al. Shall we stay, or shall we switch? Continued anti-VEGF therapy versus early switch to dexamethasone implant in refractory diabetic macular edema. Acta Diabetol. 2018, 55, 789–796. [Google Scholar] [CrossRef]
- Sivaprasad, S.; Crosby-Nwaobi, R.; Heng, L.Z.; Peto, T.; Michaelides, M.; Hykin, P. Injection frequency and response to bevacizumab monotherapy for diabetic macular oedema (BOLT Report 5). Br. J. Ophthalmol. 2013, 97, 1177–1180. [Google Scholar] [CrossRef]
- Sassen, S.; Miska, E.A.; Caldas, C. MicroRNA: Implications for cancer. Virchows Arch. Int. J. Pathol. 2008, 452, 1–10. [Google Scholar] [CrossRef]
- Sempere, L.F. Celebrating 25 Years of MicroRNA Research: From Discovery to Clinical Application. Int. J. Mol. Sci. 2019, 20, 1987. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, K.; Cooper, N.G. MicroRNAs in the Neural Retina. Int. J. Genom. 2014, 2014, 165897. [Google Scholar] [CrossRef]
- Dunmire, J.J.; Lagouros, E.; Bouhenni, R.A.; Jones, M.; Edward, D.P. MicroRNA in aqueous humor from patients with cataract. Exp. Eye. Res. 2013, 108, 68–71. [Google Scholar] [CrossRef]
- Ragusa, M.; Caltabiano, R.; Russo, A.; Puzzo, L.; Avitabile, T.; Longo, A.; Toro, M.D.; Di Pietro, C.; Purrello, M.; Reibaldi, M. MicroRNAs in vitreus humor from patients with ocular diseases. Mol. Vis. 2013, 19, 430–440. [Google Scholar]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Bauer, A.; Elsharawy, A.; Haas, J.; Backes, C.; Wendschlag, A.; Giese, N.; Tjaden, C.; Ott, K.; et al. Toward the blood-borne miRNome of human diseases. Nat. Methods 2011, 8, 841–843. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.G.; Oliveira-Fernandes, M.; Lavker, R.M. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 2006, 12, 1175–1184. [Google Scholar] [PubMed]
- Xu, S.; Witmer, P.D.; Lumayag, S.; Kovacs, B.; Valle, D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007, 282, 25053–25066. [Google Scholar] [CrossRef] [PubMed]
- Busskamp, V.; Krol, J.; Nelidova, D.; Daum, J.; Szikra, T.; Tsuda, B.; Juttner, J.; Farrow, K.; Scherf, B.G.; Alvarez, C.P.; et al. miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron 2014, 83, 586–600. [Google Scholar] [CrossRef] [PubMed]
- Damiani, D.; Alexander, J.J.; O’Rourke, J.R.; McManus, M.; Jadhav, A.P.; Cepko, C.L.; Hauswirth, W.W.; Harfe, B.D.; Strettoi, E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J. Neurosci. 2008, 28, 4878–4887. [Google Scholar] [CrossRef]
- Xu, S. microRNA expression in the eyes and their significance in relation to functions. Prog. Retin. Eye Res. 2009, 28, 87–116. [Google Scholar] [CrossRef]
- McArthur, K.; Feng, B.; Wu, Y.; Chen, S.; Chakrabarti, S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes 2011, 60, 1314–1323. [Google Scholar] [CrossRef]
- Murray, A.R.; Chen, Q.; Takahashi, Y.; Zhou, K.K.; Park, K.; Ma, J.X. MicroRNA-200b downregulates oxidation resistance 1 (Oxr1) expression in the retina of type 1 diabetes model. Invest. Ophthalmol. Vis. Sci. 2013, 54, 1689–1697. [Google Scholar] [CrossRef]
- Feng, B.; Chen, S.; McArthur, K.; Wu, Y.; Sen, S.; Ding, Q.; Feldman, R.D.; Chakrabarti, S. miR-146a-Mediated extracellular matrix protein production in chronic diabetes complications. Diabetes 2011, 60, 2975–2984. [Google Scholar] [CrossRef]
- Bressler, N.M.; Beaulieu, W.T.; Maguire, M.G.; Glassman, A.R.; Blinder, K.J.; Bressler, S.B.; Gonzalez, V.H.; Jampol, L.M.; Melia, M.; Sun, J.K.; et al. Early Response to Anti-Vascular Endothelial Growth Factor and Two-Year Outcomes Among Eyes With Diabetic Macular Edema in Protocol T. Am. J. Ophthalmol. 2018, 195, 93–100. [Google Scholar] [CrossRef]
- KA, L.C.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; KA, L.C. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Khaleeq, Q.T.; Meese, E.; Keller, A. miEAA: microRNA enrichment analysis and annotation. Nucleic Acids Res. 2016, 44, W110–W116. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-C.-D.; Li, J.; Huang, K.-Y.; Shrestha, S.; Hong, H.-C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2019, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein. Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef]
- Kovacs, B.; Lumayag, S.; Cowan, C.; Xu, S. MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4402–4409. [Google Scholar] [CrossRef]
- Wu, J.H.; Gao, Y.; Ren, A.J.; Zhao, S.H.; Zhong, M.; Peng, Y.J.; Shen, W.; Jing, M.; Liu, L. Altered microRNA expression profiles in retinas with diabetic retinopathy. Ophthalmic. Res. 2012, 47, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Hindle, A.G.; Thoonen, R.; Jasien, J.V.; Grange, R.M.H.; Amin, K.; Wise, J.; Ozaki, M.; Ritch, R.; Malhotra, R.; Buys, E.S. Identification of Candidate miRNA Biomarkers for Glaucoma. Investig. Ophthalmol. Vis. Sci. 2019, 60, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Lobov, I.; Mikhailova, N. The Role of Dll4/Notch Signaling in Normal and Pathological Ocular Angiogenesis: Dll4 Controls Blood Vessel Sprouting and Vessel Remodeling in Normal and Pathological Conditions. J. Ophthalmol. 2018, 2018, 3565292. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.O. Vascular endothelial growth factors and vascular permeability. Cardiovasc. Res. 2010, 87, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J.; Ronnback, L.; Hansson, E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 2006, 7, 41–53. [Google Scholar] [CrossRef]
- Antonetti, D.A.; Barber, A.J.; Khin, S.; Lieth, E.; Tarbell, J.M.; Gardner, T.W. Vascular permeability in experimental diabetes is associated with reduced endothelial occludin content: Vascular endothelial growth factor decreases occludin in retinal endothelial cells. Penn State Retina Research Group. Diabetes 1998, 47, 1953–1959. [Google Scholar] [CrossRef]
- Harhaj, N.S.; Felinski, E.A.; Wolpert, E.B.; Sundstrom, J.M.; Gardner, T.W.; Antonetti, D.A. VEGF activation of protein kinase C stimulates occludin phosphorylation and contributes to endothelial permeability. Investig. Ophthalmol. Vis. Sci. 2006, 47, 5106–5115. [Google Scholar] [CrossRef]
- Miloudi, K.; Oubaha, M.; Ménard, C.; Dejda, A.; Guber, V.; Cagnone, G.; Wilson, A.M.; Tétreault, N.; Mawambo, G.; Binet, F.; et al. NOTCH1 signaling induces pathological vascular permeability in diabetic retinopathy. Proc. Nat. Acad. Sci. USA 2019, 116, 4538. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, W.; Lilly, B. Evaluation of Notch3 Deficiency in Diabetes-Induced Pericyte Loss in the Retina. J. Vasc. Res. 2018, 55, 308–318. [Google Scholar] [CrossRef]
- Arboleda-Velasquez, J.F.; Primo, V.; Graham, M.; James, A.; Manent, J.; D’Amore, P.A. Notch signaling functions in retinal pericyte survival. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5191–5199. [Google Scholar] [CrossRef]
- Shapiro, R.; Vallee, B.L. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc. Natl. Acad. Sci. USA 1987, 84, 2238–2241. [Google Scholar] [CrossRef] [PubMed]
- Neubauer-Geryk, J.; Kozera, G.M.; Wolnik, B.; Szczyrba, S.; Nyka, W.M.; Bieniaszewski, L. Angiogenin in middle-aged type 1 diabetes patients. Microvasc. Res. 2012, 84, 387–389. [Google Scholar] [CrossRef] [PubMed]
- Malamitsi-Puchner, A.; Sarandakou, A.; Dafogianni, C.; Tziotis, J.; Bartsocas, C.S. Serum angiogenin levels in children and adolescents with insulin-dependent diabetes mellitus. Pediatr. Res. 1998, 43, 798–800. [Google Scholar] [CrossRef] [PubMed]
- Siebert, J.; Reiwer-Gostomska, M.; Mysliwska, J.; Marek, N.; Raczynska, K.; Glasner, L. Glycemic control influences serum angiogenin concentrations in patients with type 2 diabetes. Diabetes Care 2010, 33, 1829–1830. [Google Scholar] [CrossRef]
- Marek, N.; Raczynska, K.; Siebert, J.; Mysliwiec, M.; Zorena, K.; Mysliwska, J.; Reiwer-Gostomska, M.; Trzonkowski, P. Decreased angiogenin concentration in vitreous and serum in proliferative diabetic retinopathy. Microvasc. Res. 2011, 82, 1–5. [Google Scholar] [CrossRef]
- Liang, Y.Z.; Li, J.J.; Xiao, H.B.; He, Y.; Zhang, L.; Yan, Y.X. Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: A systematic review and meta-analysis. J. Diabetes 2018. [Google Scholar] [CrossRef]
- Kim, M.; Zhang, X. The Profiling and Role of miRNAs in Diabetes Mellitus. J. Diabetes Clin. Res. 2019, 1, 5–23. [Google Scholar] [CrossRef]
- Shen, J.; Yang, X.; Xie, B.; Chen, Y.; Swaim, M.; Hackett, S.F.; Campochiaro, P.A. MicroRNAs regulate ocular neovascularization. Mol. Ther. J. Am. Soc. Gene Ther. 2008, 16, 1208–1216. [Google Scholar] [CrossRef]
- Yang, S.; Fei, X.; Lu, Y.; Xu, B.; Ma, Y.; Wan, H. miRNA-214 suppresses oxidative stress in diabetic nephropathy via the ROS/Akt/mTOR signaling pathway and uncoupling protein 2. Exp. Med. 2019, 17, 3530–3538. [Google Scholar] [CrossRef]
- Van Balkom, B.W.; de Jong, O.G.; Smits, M.; Brummelman, J.; den Ouden, K.; de Bree, P.M.; van Eijndhoven, M.A.; Pegtel, D.M.; Stoorvogel, W.; Würdinger, T.; et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood 2013, 121, 3997–4006. [Google Scholar] [CrossRef]
- Hirota, K.; Keino, H.; Inoue, M.; Ishida, H.; Hirakata, A. Comparisons of microRNA expression profiles in vitreous humor between eyes with macular hole and eyes with proliferative diabetic retinopathy. Graefes Arch. Clin. Exp. Ophthalmol. 2015, 253, 335–342. [Google Scholar] [CrossRef]
- Li, R.; Yuan, H.; Zhao, T.; Yan, Y.; Liu, Z.; Cai, J.; Qiu, C.; Li, C. Mir-874 Ameliorates Retinopathy in Diabetic Rats by Nf-Κb Signaling Pathway; Research Square: Durham, NC, USA, 2020. [Google Scholar] [CrossRef]
- Yao, T.; Zha, D.; Gao, P.; Shui, H.; Wu, X. MiR-874 alleviates renal injury and inflammatory response in diabetic nephropathy through targeting toll-like receptor-4. J. Cell. Physiol. 2018, 234, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Sun, X.; Wang, S.; Man, H. Upregulation of miR-874-3p decreases cerebral ischemia/reperfusion injury by directly targeting BMF and BCL2L13. Biomed. Pharm. 2019, 117, 108941. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, X.; Ma, S.; Guo, Y.; Li, Y. MicroRNA-98-5p ameliorates oxygen-glucose deprivation/reoxygenation (OGD/R)-induced neuronal injury by inhibiting Bach1 and promoting Nrf2/ARE signaling. Mol. Biochem. Biophys. Res. Commun. 2018, 507, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Yahya, S.M.M.; Yahya, S.M.M. The Effect of miR-98 and miR-214 on Apoptotic and Angiogenic Pathways in Hepatocellular Carcinoma HepG2 Cells. Indian J. Clin. Biochem. 2020, 35, 353–358. [Google Scholar] [CrossRef]
- Hu, D.; Yu, Y.; Wang, C.; Li, D.; Tai, Y.; Fang, L. microRNA-98 mediated microvascular hyperpermeability during burn shock phase via inhibiting FIH-1. Eur. J. Med. Res. 2015, 20, 51. [Google Scholar] [CrossRef][Green Version]
- Chen, F.Z.; Zhao, Y.; Chen, H.Z. MicroRNA-98 reduces amyloid β-protein production and improves oxidative stress and mitochondrial dysfunction through the Notch signaling pathway via HEY2 in Alzheimer’s disease mice. Int. J. Mol. Med. 2019, 43, 91–102. [Google Scholar] [CrossRef]
- Rebucci, M.; Sermeus, A.; Leonard, E.; Delaive, E.; Dieu, M.; Fransolet, M.; Arnould, T.; Michiels, C. miRNA-196b inhibits cell proliferation and induces apoptosis in HepG2 cells by targeting IGF2BP1. Mol. Cancer 2015, 14, 79. [Google Scholar] [CrossRef]
- Erener, S.; Marwaha, A.; Tan, R.; Panagiotopoulos, C.; Kieffer, T.J. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight 2017, 2, e89656. [Google Scholar] [CrossRef]
- Zampetaki, A.; Willeit, P.; Burr, S.; Yin, X.; Langley, S.R.; Kiechl, S.; Klein, R.; Rossing, P.; Chaturvedi, N.; Mayr, M. Angiogenic microRNAs Linked to Incidence and Progression of Diabetic Retinopathy in Type 1 Diabetes. Diabetes 2016, 65, 216–227. [Google Scholar] [CrossRef]
- Fan, Y.; Shi, C.; Li, T.; Kuang, T. microRNA-454 shows anti-angiogenic and anti-metastatic activity in pancreatic ductal adenocarcinoma by targeting LRP6. Am. J. Cancer Res. 2017, 7, 139–147. [Google Scholar]
- Gonsalves, C.S.; Li, C.; Malik, P.; Tahara, S.M.; Kalra, V.K. Peroxisome proliferator-activated receptor-α-mediated transcription of miR-301a and miR-454 and their host gene SKA2 regulates endothelin-1 and PAI-1 expression in sickle cell disease. Biosci. Rep. 2015, 35. [Google Scholar] [CrossRef] [PubMed]
- Jampol, L.M.; Glassman, A.R.; Bressler, N.M.; Wells, J.A.; Ayala, A.R.; Diabetic Retinopathy Clinical Research Network. Anti-Vascular Endothelial Growth Factor Comparative Effectiveness Trial for Diabetic Macular Edema: Additional Efficacy Post Hoc Analyses of a Randomized Clinical Trial. JAMA Ophthalmol. 2016, 134, 1429–1434. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Batch I | Batch II | |
|---|---|---|---|
| Treatment | Bevacizumab (n = 6) | Bevacizumab (n = 7) | Aflibercept (n = 11) |
| Age, years Mean ± SD | 59 ± 8.7 | 57.6 ± 7.7 | 59 ± 10.0 |
| Gender | |||
| Male (%) | 2 (33.3%) | 6 (85.7%) | 8 (72.7%) |
| Female (%) | 4 (66.7%) | 1 (14.3%) | 3 (27.3%) |
| HbA1c Mean ± SD | 8.0 ± 1.2 | 9.2 ± 2.0 | 7.5 ± 1.3 |
| Central subfield thickness | |||
| Baseline mean ± SD | 349.7 ± 41.3 | 404.4 ± 94.6 | 458.3 ± 87.4 |
| Post-treatment mean ± SD | 324.2 ± 65.0 | 376.3 ± 106.3 | 369.3 ± 56.3 |
| Response | |||
| Good (%) | 0 (0.0%) | 2 (28.6%) | 4 (36.4%) |
| Partial (%) | 3 (50.0%) | 2 (28.6%) | 3 (27.2%) |
| Poor (%) | 3 (50.0%) | 3 (42.9%) | 4 (36.4%) |
| miRNA | p-Value | R-Value | miRNA | p-Value | R-Value |
|---|---|---|---|---|---|
| hsa-miR-148A-5p | 0.48 | 0.2 | hsa-miR-130b-3p | 0.49 | −0.12 |
| hsa- let-7f-2-3p | 0.15 | 0.34 | hsa-miR-140-5p | 0.78 | 0.83 |
| hsa-miR-185-5p | 0.2 | 0.4 | hsa-mIR-18b-5p | 0.097 | −0.39 |
| hsa-miR-195-5p | 0.9 | 0.1 | hsa-miR-196b-5p | 0.12 | −0.47 |
| hsa-mIR-199a-5p | 0.29 | 0.34 | hsa-miR-200a-3p | 0.61 | −0.2 |
| ho-miR-214-3p | 0.13 | 0.54 | hsa-miR-23c | 0.25 | −0.33 |
| hsa-miR-320a | 0.11 | 0.42 | hsa-miR-28-5p | 0.23 | −0.35 |
| hsa-miR-320b | 0.06 | 0.51 | hsa-miR-362-5p | 0.19 | −0.34 |
| hsa-miR-320d | 0.038 | 0.53 | hsa-mIR-374a-5p | 0.46 | −0.25 |
| hsa-miR-320e | 0.055 | 0.52 | hsa-miR-374b-5p | 0.75 | −0.13 |
| hsa-mIR-335-5p | 0.19 | 0.38 | hsa-miR-411-5p | 0.18 | −0.38 |
| hsa-miR-486-5p | 0.15 | 0.48 | hsa-miR-454-3p | 0.15 | −0.4 |
| hsa-miR-497-5p | 0.23 | 0.35 | hsa-mIR-539-5p | 0.25 | −0.34 |
| hsa-miR-513a-5p | 0.11 | 0.46 | hsa-miR-98-5p | 0.049 | −0.53 |
| hsa-mIR-874-3p | 0.048 | 0.55 | hsa-miR-99b-5p | 0.27 | −0.34 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, H.W.; Yang, B.; Wong, W.; Blakeley, P.; Seah, I.; Tan, Q.S.W.; Wang, H.; Bhargava, M.; Lin, H.A.; Chai, C.H.; et al. A Pilot Study on MicroRNA Profile in Tear Fluid to Predict Response to Anti-VEGF Treatments for Diabetic Macular Edema. J. Clin. Med. 2020, 9, 2920. https://doi.org/10.3390/jcm9092920
Chan HW, Yang B, Wong W, Blakeley P, Seah I, Tan QSW, Wang H, Bhargava M, Lin HA, Chai CH, et al. A Pilot Study on MicroRNA Profile in Tear Fluid to Predict Response to Anti-VEGF Treatments for Diabetic Macular Edema. Journal of Clinical Medicine. 2020; 9(9):2920. https://doi.org/10.3390/jcm9092920
Chicago/Turabian StyleChan, Hwei Wuen, Binxia Yang, Wendy Wong, Paul Blakeley, Ivan Seah, Queenie Shu Woon Tan, Haofei Wang, Mayuri Bhargava, Hazel Anne Lin, Charmaine HC Chai, and et al. 2020. "A Pilot Study on MicroRNA Profile in Tear Fluid to Predict Response to Anti-VEGF Treatments for Diabetic Macular Edema" Journal of Clinical Medicine 9, no. 9: 2920. https://doi.org/10.3390/jcm9092920
APA StyleChan, H. W., Yang, B., Wong, W., Blakeley, P., Seah, I., Tan, Q. S. W., Wang, H., Bhargava, M., Lin, H. A., Chai, C. H., Mangunkusumo, E. A., Thet, N., Yuen, Y. S., Sethi, R., Wang, S., Hunziker, W., Lingam, G., & Su, X. (2020). A Pilot Study on MicroRNA Profile in Tear Fluid to Predict Response to Anti-VEGF Treatments for Diabetic Macular Edema. Journal of Clinical Medicine, 9(9), 2920. https://doi.org/10.3390/jcm9092920





