Where to Biopsy to Detect Helicobacter pylori and How Many Biopsies Are Needed to Detect Antibiotic Resistance in a Human Stomach
Abstract
:1. Introduction
2. Methods
2.1. Patients
2.2. Collection of Gastric Biopsies
2.3. Analysis of Gastric Biopsies
2.3.1. Culture and AST
2.3.2. DNA Extraction and Molecular Biology Analyses
2.4. Statistical Analysis
2.5. Ethics
3. Results
3.1. Characteristics of Included Patients
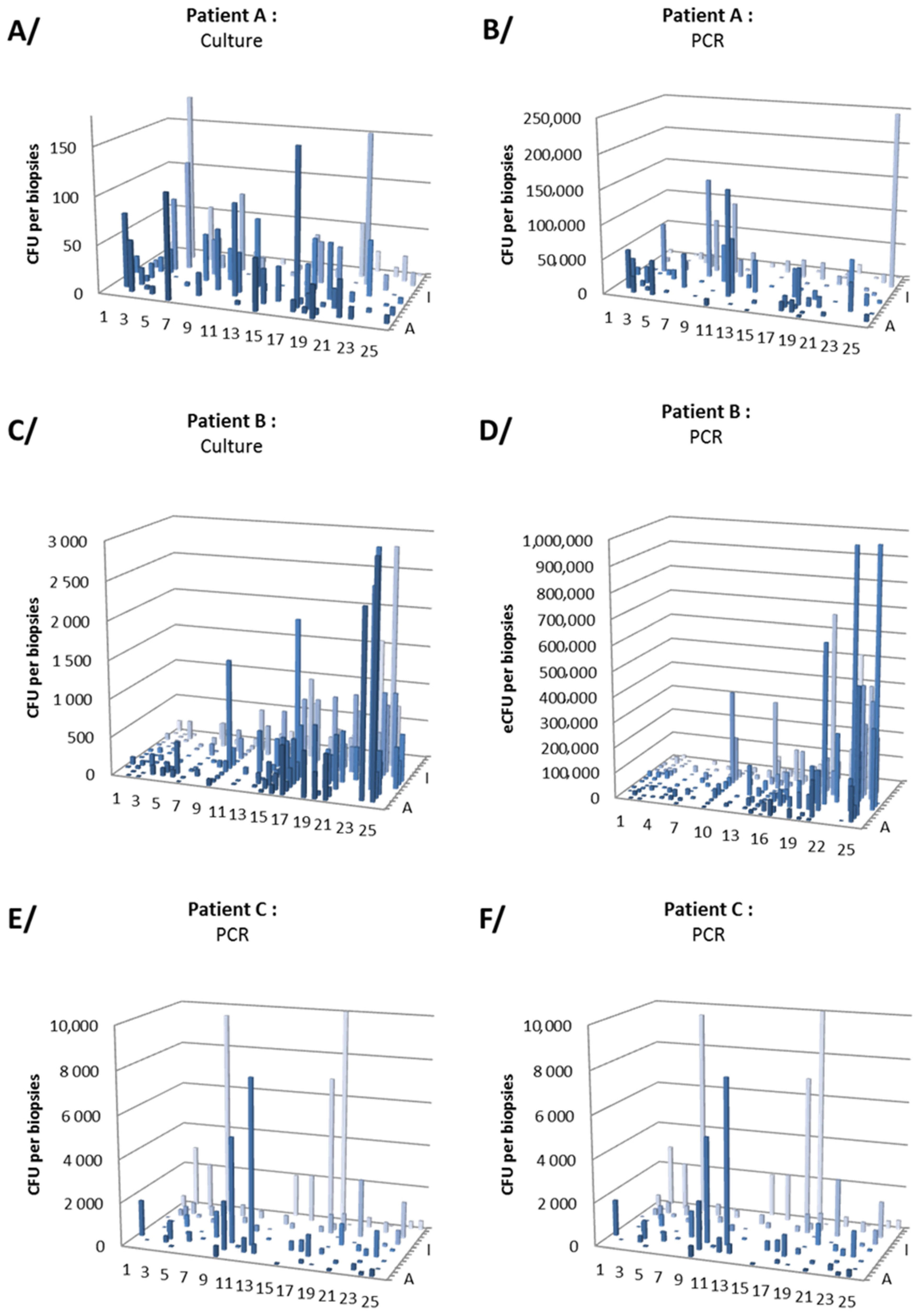
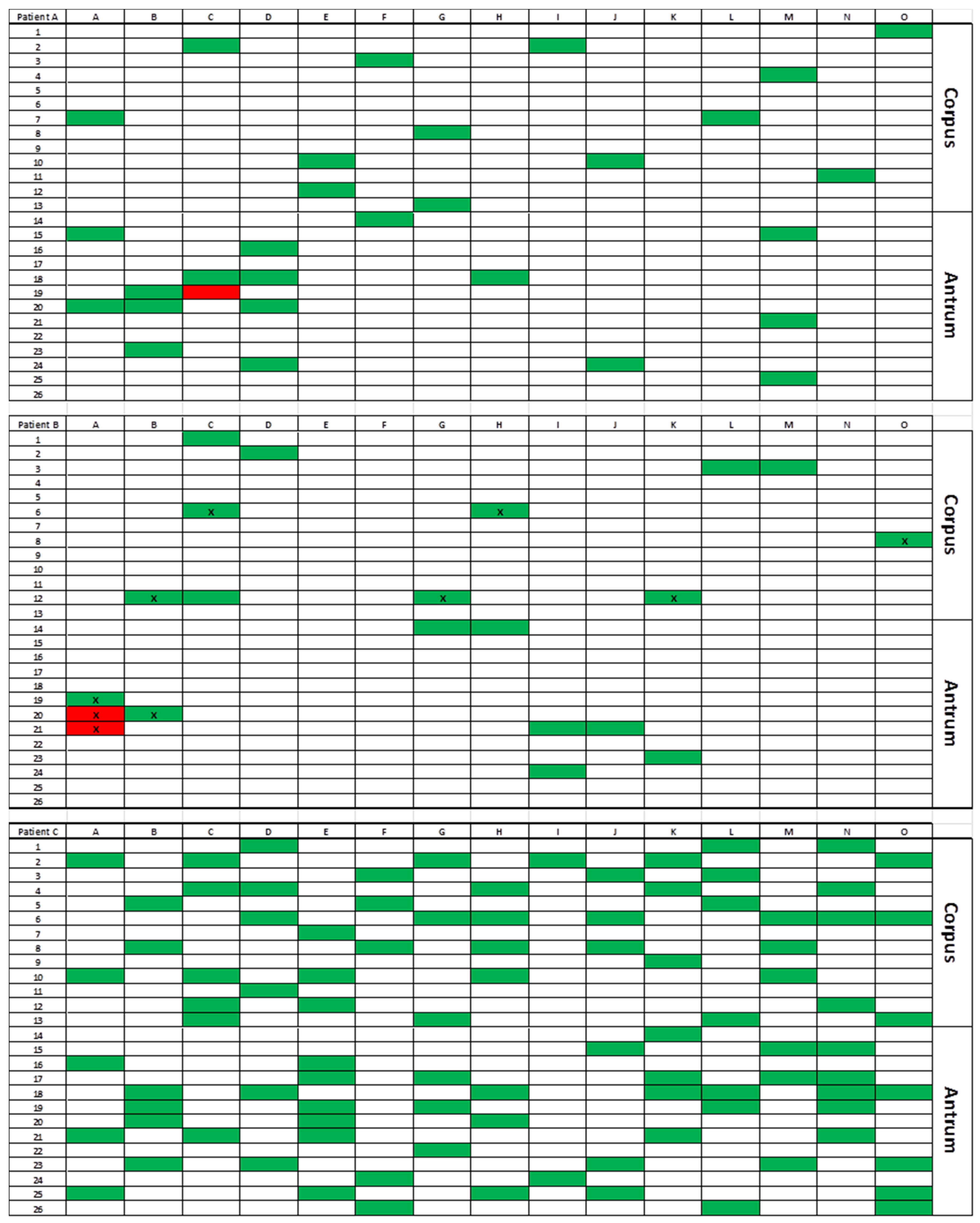
3.2. Gastric Distribution and Density of H. pylori
3.3. Antibiotic Resistance
3.4. RAPD Genotyping and CagA Detection
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Suerbaum, S.; Michetti, P. Helicobacter pylori infection. N. Engl. J. Med. 2002, 347, 1175–1186. [Google Scholar] [CrossRef] [Green Version]
- Atherton, J.C.; Blaser, M.J. Coadaptation of Helicobacter pylori and humans: Ancient history, modern implications. J. Clin. Investig. 2009, 119, 2475–2487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malaty, H.M.; El-Kasabany, A.; Graham, D.Y.; Miller, C.C.; Reddy, S.G.; Srinivasan, S.R.; Yamaoka, Y.; Berenson, G.S. Age at acquisition of Helicobacter pylori infection: A follow-up study from infancy to adulthood. Lancet 2002, 359, 931–935. [Google Scholar] [CrossRef]
- Fung, C.; Tan, S.; Nakajima, M.; Skoog, E.C.; Camarillo-Guerrero, L.F.; Klein, J.A.; Lawley, T.D.; Solnick, J.V.; Fukami, T.; Amieva, M.R. High-resolution mapping reveals that microniches in the gastric glands control Helicobacter pylori colonization of the stomach. PLoS Biol. 2019, 17, e3000231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichon, M.; Burucoa, C. Impact of the Gastro-Intestinal Bacterial Microbiome on Helicobacter-Associated Diseases. Healthcare 2019, 7, 34. [Google Scholar] [CrossRef] [Green Version]
- Tegtmeyer, N.; Wessler, S.; Backert, S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011, 278, 1190–1202. [Google Scholar] [CrossRef] [PubMed]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-Analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382. [Google Scholar] [CrossRef] [Green Version]
- Bouihat, N.; Burucoa, C.; Benkirane, A.; Seddik, H.; Sentissi, S.; Al Bouzidi, A.; Elouennas, M.; Benouda, A. Helicobacter pylori Primary Antibiotic Resistance in 2015 in Morocco: A Phenotypic and Genotypic Prospective and Multicenter Study. Microb. Drug Resist. 2017, 23, 727–732. [Google Scholar] [CrossRef]
- Djennane-Hadibi, F.; Bachtarzi, M.; Layaida, K.; Ali Arous, N.; Nakmouche, M.; Saadi, B.; Tazir, M.; Ramdani-Bouguessa, N.; Burucoa, C. High-Level Primary Clarithromycin Resistance of Helicobacter pylori in Algiers, Algeria: A Prospective Multicenter Molecular Study. Microb. Drug Resist. 2016, 22, 223–226. [Google Scholar] [CrossRef]
- Raymond, J.; Lamarque, D.; Kalach, N.; Chaussade, S.; Burucoa, C. High level of antimicrobial resistance in French Helicobacter pylori isolates. Helicobacter 2010, 15, 21–27. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management of Helicobacter pylori infection—The Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenzhen, Y.; Yumin, L.; Quanlin, G.; Kehu, Y.; Lei, J.; Donghai, W.; Lijuan, Y. Is antimicrobial susceptibility testing necessary before first-line treatment for Helicobacter pylori infection? Meta-analysis of randomized controlled trials. Intern. Med. 2010, 49, 1103–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elitsur, Y.; Lawrence, Z.; Triest, W.E. Distribution of Helicobacter pylori organisms in the stomachs of children with H. pylori infection. Hum. Pathol. 2002, 33, 1133–1135. [Google Scholar] [CrossRef] [PubMed]
- Genta, R.M.; Graham, D.Y. Comparison of biopsy sites for the histopathologic diagnosis of Helicobacter pylori: A topographic study of H. pylori density and distribution. Gastrointest. Endosc. 1994, 40, 342–345. [Google Scholar] [CrossRef]
- Goodwin, C.S.; Blincow, E.D.; Warren, J.R.; Waters, T.E.; Sanderson, C.R.; Easton, L. Evaluation of cultural techniques for isolating Campylobacter pyloridis from endoscopic biopsies of gastric mucosa. J. Clin. Pathol. 1985, 38, 1127–1131. [Google Scholar] [CrossRef] [Green Version]
- Khulusi, S.; Mendall, M.A.; Patel, P.; Levy, J.; Badve, S.; Northfield, T.C. Helicobacter pylori infection density and gastric inflammation in duodenal ulcer and non-ulcer subjects. Gut 1995, 37, 319–324. [Google Scholar] [CrossRef]
- Misra, V.; Misra, S.; Dwivedi, M.; Singh, U.P.; Bhargava, V.; Gupta, S.C. A topographic study of Helicobacter pylori density, distribution and associated gastritis. J. Gastroenterol. Hepatol. 2000, 15, 737–743. [Google Scholar] [CrossRef]
- Morris, A.; Ali, M.R.; Brown, P.; Lane, M.; Patton, K. Campylobacter pylori infection in biopsy specimens of gastric antrum: Laboratory diagnosis and estimation of sampling error. J. Clin. Pathol. 1989, 42, 727–732. [Google Scholar] [CrossRef]
- Ben Mansour, K.; Fendri, C.; Battikh, H.; Garnier, M.; Zribi, M.; Jlizi, A.; Burucoa, C. Multiple and mixed Helicobacter pylori infections: Comparison of two epidemiological situations in Tunisia and France. Infect. Genet. Evol. 2016, 37, 43–48. [Google Scholar] [CrossRef]
- Selgrad, M.; Tammer, I.; Langner, C.; Bornschein, J.; Meißle, J.; Kandulski, A.; Varbanova, M.; Wex, T.; Schlüter, D.; Malfertheiner, P. Different antibiotic susceptibility between antrum and corpus of the stomach, a possible reason for treatment failure of Helicobacter pylori infection. World J. Gastroenterol. 2014, 20, 16245–16251. [Google Scholar] [CrossRef]
- Seo, J.W.; Park, J.Y.; Shin, T.S.; Kim, J.G. The analysis of virulence factors and antibiotic resistance between Helicobacter pylori strains isolated from gastric antrum and body. BMC Gastroenterol. 2019, 19, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ailloud, F.; Didelot, X.; Woltemate, S.; Pfaffinger, G.; Overmann, J.; Bader, R.C.; Schulz, C.; Malfertheiner, P.; Suerbaum, S. Within-host evolution of Helicobacter pylori shaped by niche-specific adaptation, intragastric migrations and selective sweeps. Nat. Commun. 2019, 10, 2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gumbs, A.A.; Gagner, M.; Dakin, G.; Pomp, A. Sleeve gastrectomy for morbid obesity. Obes. Surg. 2007, 17, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Mustapha, P.; Paris, I.; Garcia, M.; Tran, C.T.; Cremniter, J.; Garnier, M.; Faure, J.-P.; Barthes, T.; Boneca, I.G.; Morel, F.; et al. Chemokines and antimicrobial peptides have a cag-dependent early response to Helicobacter pylori infection in primary human gastric epithelial cells. Infect. Immun. 2014, 82, 2881–2889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.; Chomel, J.C.; Mustapha, P.; Tran, C.T.; Garnier, M.; Paris, I.; Quellard, N.; Godet, J.; Cremniter, J.; Bennaceur-Griscelli, A.; et al. In vitro culture and phenotypic and molecular characterization of gastric stem cells from human stomach. Helicobacter 2017, 22, e12351. [Google Scholar] [CrossRef] [PubMed]
- Tran, C.T.; Garcia, M.; Garnier, M.; Burucoa, C.; Bodet, C. Inflammatory signaling pathways induced by Helicobacter pylori in primary human gastric epithelial cells. Innate Immun. 2017, 23, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Francaise de Microbiologie. CASFM/EUCAST: Société Française de Microbiologie, 2017th ed.; SFM: Paris, France, 2017. [Google Scholar]
- Cremniter, J.; Bodet, C.; Tougeron, D.; Dray, X.; Guilhot, J.; Jégou, J.F.; Morel, F.; Lecron, J.C.; Silvain, C.; Burucoa, C. Th-17 response and antimicrobial peptide expression are uniformly expressed in gastric mucosa of Helicobacter pylori-infected patients independently of their clinical outcomes. Helicobacter 2018, 23, e12479. [Google Scholar] [CrossRef]
- Burucoa, C.; Garnier, M.; Silvain, C.; Fauchère, J.L. Quadruplex real-time PCR assay using allele-specific scorpion primers for detection of mutations conferring clarithromycin resistance to Helicobacter pylori. J. Clin. Microbiol. 2008, 46, 2320–2326. [Google Scholar] [CrossRef] [Green Version]
- Burucoa, C.; Lhomme, V.; Fauchere, J.L. Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: Experimental results and meta-analysis. J. Clin. Microbiol. 1999, 37, 4071–4080. [Google Scholar] [CrossRef] [Green Version]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef] [Green Version]
- Helander, H.F.; Fändriks, L. Surface area of the digestive tract-revisited. Scand. J. Gastroenterol. 2014, 49, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Tummala, S.; Sheth, S.G.; Goldsmith, J.D.; Goldar-Najafi, A.; Murphy, C.K.; Osburne, M.S.; Mullin, S.; Buxton, D.; Wagner, D.A.; Kelly, C.P. Quantifying gastric Helicobacter pylori infection: A comparison of quantitative culture, urease breath testing, and histology. Dig. Dis. Sci. 2007, 52, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Bayerdörffer, E.; Oertel, H.; Lehn, N.; Kasper, G.; Mannes, G.A.; Sauerbruch, T.; Stolte, M. Topographic association between active gastritis and Campylobacter pylori colonisation. J. Clin. Pathol. 1989, 42, 834–839. [Google Scholar] [CrossRef] [Green Version]
- Atherton, J.C.; Tham, K.T.; Peek, R.M.; Cover, T.L.; Blaser, M.J. Density of Helicobacter pylori infection in vivo as assessed by quantitative culture and histology. J. Infect. Dis. 1996, 174, 552–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engstrand, L.; Rosberg, K.; Hübinette, R.; Berglindh, T.; Rolfsen, W.; Gustavsson, S. Topographic mapping of Helicobacter pylori colonization in long-term-infected pigs. Infect. Immun. 1992, 60, 653–656. [Google Scholar] [CrossRef] [Green Version]
- Grasso, G.M.; Sammarco, M.L.; Ripabelli, G.; Ruberto, A.; Iannitto, G. In situ mapping of urease-positive areas in porcine gastric mucosa. Microbios 1995, 82, 245–249. [Google Scholar]
- Yousfi, M.M.; Reddy, R.; Osato, M.S.; Graham, D.Y. Is antrum or corpus the best site for culture of Helicobacter pylori? Helicobacter 1996, 1, 88–91. [Google Scholar] [CrossRef]
- Nedenskov-Sørensen, P.; Aase, S.; Bjørneklett, A.; Fausa, O.; Bukholm, G. Sampling efficiency in the diagnosis of Helicobacter pylori infection and chronic active gastritis. J. Clin. Microbiol. 1991, 29, 672–675. [Google Scholar] [CrossRef] [Green Version]
- Lascols, C.; Lamarque, D.; Costa, J.M.; Copie-Bergman, C.; Le Glaunec, J.M.; Deforges, L.; Soussy, C.J.; Petit, J.C.; Delchier, J.C.; Tankovic, J. Fast and accurate quantitative detection of Helicobacter pylori and identification of clarithromycin resistance mutations in H. pylori isolates from gastric biopsy specimens by real-time PCR. J. Clin. Microbiol. 2003, 41, 4573–4577. [Google Scholar] [CrossRef] [Green Version]
- Pichon, M.; Pichard, B.; Barrioz, T.; Plouzeau, C.; Croquet, V.; Fotsing, G.; Chéron, A.; Vuillemin, É.; Wangermez, M.; Haineaux, P.A.; et al. Diagnostic Accuracy of a Noninvasive Test for Detection of Helicobacter pylori and Resistance to Clarithromycin in Stool by the Amplidiag, H. pylori+ClariR Real-Time PCR Assay. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Furuta, T.; Kaneko, E.; Suzuki, M.; Arai, H.; Futami, H. Quantitative study of Helicobacter pylori in gastric mucus by competitive PCR using synthetic DNA fragments. J. Clin. Microbiol. 1996, 34, 2421–2425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, L.P.; Rasmussen, L. Helicobacter pylori-coccoid forms and biofilm formation. FEMS Immunol. Med. Microbiol. 2009, 56, 112–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dzierzanowska-Fangrat, K.; Crabtree, J.E.; Rozynek, E.; Dura, W.; Celiñska-Cedro, D.; Wojda, U.; Dzierzanowska, D. Helicobacter pylori cagA genotype and density of colonization in relation to gastric inflammation in children. Eur. J. Gastroenterol. Hepatol. 2002, 14, 1303–1307. [Google Scholar] [CrossRef]
- Belda, S.; Saez, J.; Santibáñez, M.; Rodríguez, J.C.; Galiana, A.; Sola-Vera, J.; Ruiz-García, M.; Brotons, A.; López-Girona, E.; Royo, G. Quantification of Helicobacter pylori in gastric mucosa by real-time polymerase chain reaction: Comparison with traditional diagnostic methods. Diagn. Microbiol. Infect. Dis. 2012, 74, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Bastos, J.; Peleteiro, B.; Barros, R.; Alves, L.; Severo, M.; de Fátima Pina, M.; Pinto, H.; Carvalho, S.; Marinho, A.; Guimarães, J.T.; et al. Sociodemographic determinants of prevalence and incidence of Helicobacter pylori infection in Portuguese adults. Helicobacter 2013, 18, 413–422. [Google Scholar] [CrossRef] [Green Version]
- Satoh, K.; Kimura, K.; Taniguchi, Y.; Kihira, K.; Takimoto, T.; Saifuku, K.; Kawata, H.; Tokumaru, K.; Kojima, T.; Seki, M.; et al. Biopsy sites suitable for the diagnosis of Helicobacter pylori infection and the assessment of the extent of atrophic gastritis. Am. J. Gastroenterol. 1998, 93, 569–573. [Google Scholar] [CrossRef]
- Masuda, H.; Hiyama, T.; Yoshihara, M.; Tanaka, S.; Shimamoto, F.; Haruma, K.; Chayama, K. Necessity of multiple gastric biopsies from different sites for detection of clarithromycin-resistant Helicobacter pylori strains. Scand. J. Gastroenterol. 2003, 38, 942–946. [Google Scholar]

| Patient | Age (years) | Sex | BMI (kg/m2) | Place of Birth | Digestive Symptom | Results on Biopsies Collected Two Months before Sleeve Gastrectomy | |
|---|---|---|---|---|---|---|---|
| Histology | AST | ||||||
| A | 29 | F | 43 | France | No | Chronic active gastritis | S |
| B | 42 | F | 49 | Portugal | No | Chronic active gastritis | S |
| C | 29 | F | 48 | France | No | Chronic active gastritis | Clari R-Mtz R |
| Patient A | Patient B | Patient C | Total | |
|---|---|---|---|---|
| Culture (culture-positive biopsies/analyzed biopsies (mean CFU/biopsy)) | ||||
| Antrum | 38/42 (36) | 90/90 (553) | 52/52 (349) | 180/184 (377) |
| Corpus | 42/46 (25) | 85/86 (102) | 47/49 (399) | 174/181 (171) |
| Total | 80/88 (30) | 175/176 (333) | 99/101 (373) | 354/365 (271) |
| qPCR (PCR-positive biopsies/analyzed biopsies (log mean eCFU/biopsy)) | ||||
| Antrum | 43/44 (4.41) | 87/90 (5.18) | 47/53 (3.00) | 177/187 (4.89) |
| Corpus | 46/46 (4.23) | 86/87 (4.28) | 42/50 (3.08) | 174/183 (4.11) |
| Total | 89/90 (4.32) | 173/177 (4.94) | 89/103 (3.04) | 351/370 (4.67) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pichon, M.; Tran, C.T.; Motillon, G.; Debiais, C.; Gautier, S.; Aballea, M.; Cremniter, J.; Vasseur, P.; Tougeron, D.; Garcia, M.; et al. Where to Biopsy to Detect Helicobacter pylori and How Many Biopsies Are Needed to Detect Antibiotic Resistance in a Human Stomach. J. Clin. Med. 2020, 9, 2812. https://doi.org/10.3390/jcm9092812
Pichon M, Tran CT, Motillon G, Debiais C, Gautier S, Aballea M, Cremniter J, Vasseur P, Tougeron D, Garcia M, et al. Where to Biopsy to Detect Helicobacter pylori and How Many Biopsies Are Needed to Detect Antibiotic Resistance in a Human Stomach. Journal of Clinical Medicine. 2020; 9(9):2812. https://doi.org/10.3390/jcm9092812
Chicago/Turabian StylePichon, Maxime, Cong Tri Tran, Gaëtan Motillon, Charlotte Debiais, Sylvain Gautier, Marie Aballea, Julie Cremniter, Philippe Vasseur, David Tougeron, Magali Garcia, and et al. 2020. "Where to Biopsy to Detect Helicobacter pylori and How Many Biopsies Are Needed to Detect Antibiotic Resistance in a Human Stomach" Journal of Clinical Medicine 9, no. 9: 2812. https://doi.org/10.3390/jcm9092812






