The Role of the Liquid Biopsy in Decision-Making for Patients with Non-Small Cell Lung Cancer
Abstract
:1. Introduction: State of the Art and Uses of Liquid Biopsy in Lung Cancer
2. Available Assays and Target Genomic Alterations in Lung Cancer
2.1. Liquid Biopsy Assays
2.1.1. Real-Time PCR
2.1.2. Digital Droplet PCR
2.1.3. Next-Generation Sequencing
2.2. Target Genomic Alterations
2.2.1. EGFR Del19 and p. L858R Mutations
2.2.2. EGFR p. T790M Mutation
2.2.3. EGFR p.C797S Mutation
2.2.4. ALK
2.2.5. MET
2.2.6. BRAF
2.2.7. ROS1
2.2.8. RET
2.2.9. KRAS
3. Case Series: Challenges of the Use of Liquid Biopsy in Daily Management of Lung Cancer Patients
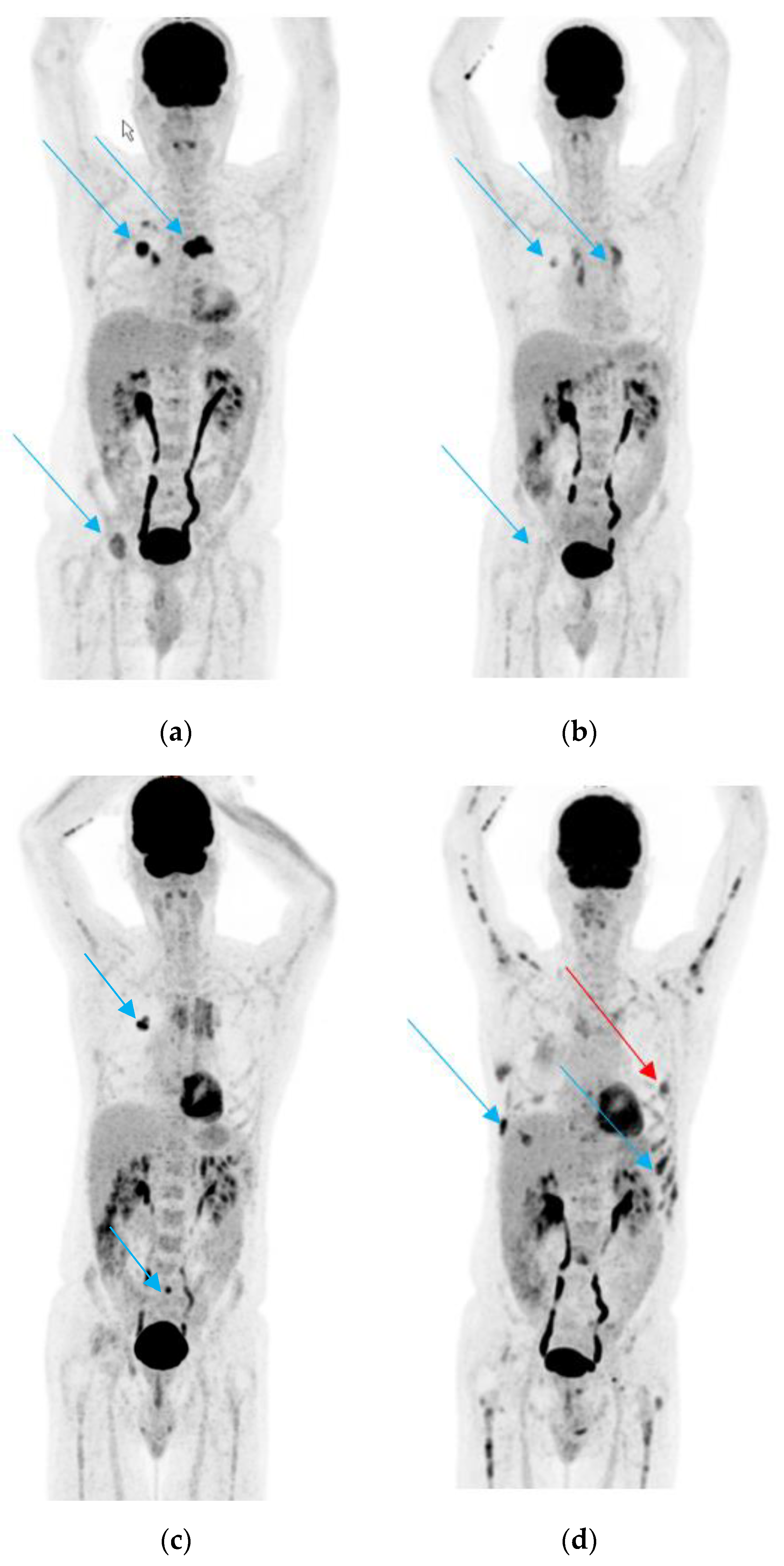
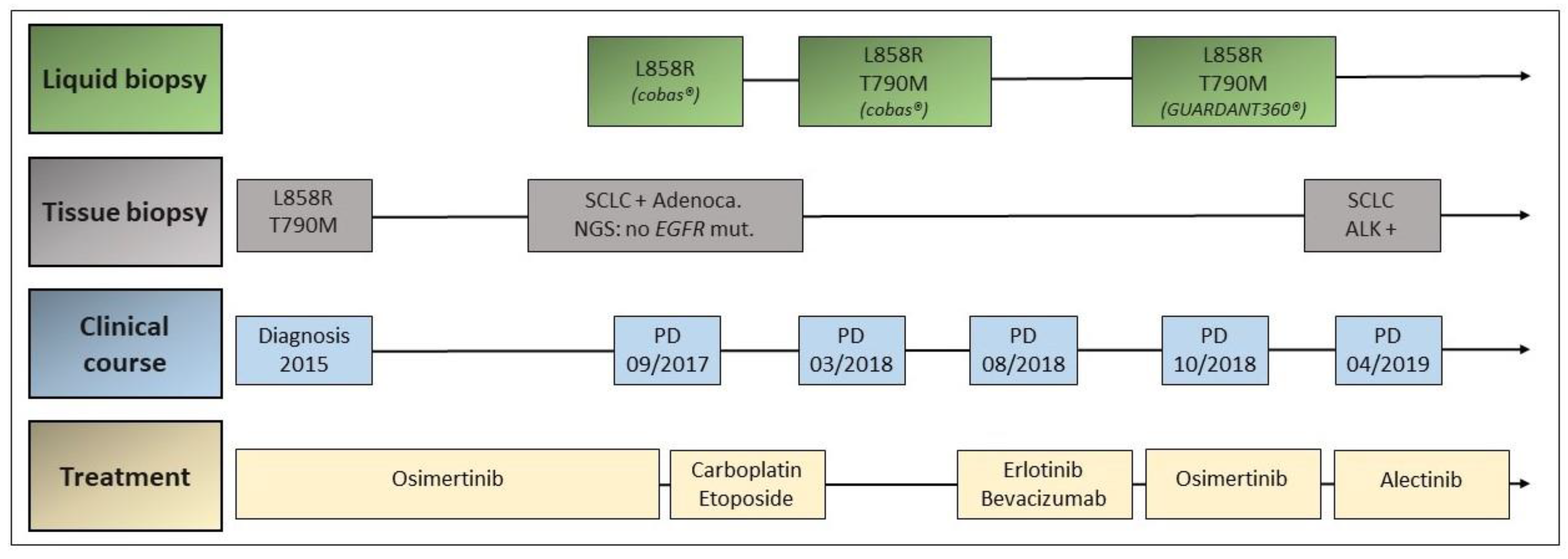
3.1. Case 1
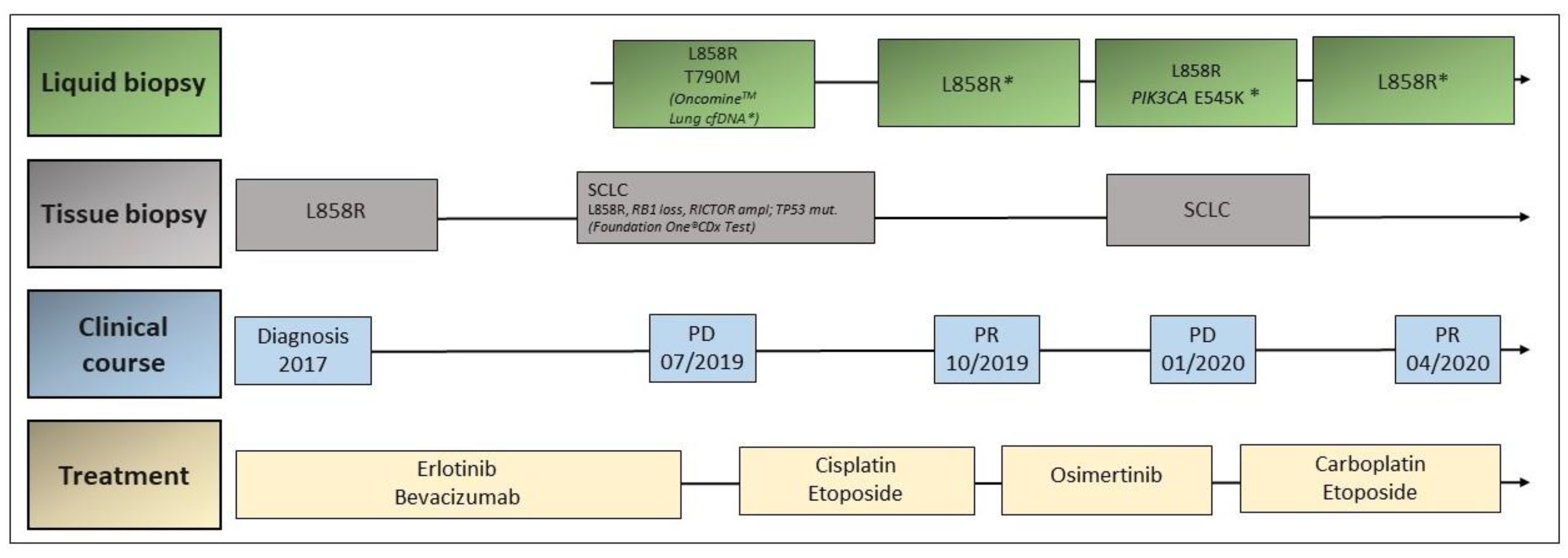
3.2. Case 2
3.3. Case 3
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, Y.-C.; Zhou, Q.; Wu, Y.-L. The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. J. Hematol. Oncol. 2017, 10, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revelo, A.E.; Martin, A.; Velasquez, R.; Kulandaisamy, P.C.; Bustamante, J.; Keshishyan, S.; Otterson, G. Liquid biopsy for lung cancers: An update on recent developments. Ann. Transl. Med. 2019, 7, 349. [Google Scholar] [CrossRef] [PubMed]
- Santarpia, M.; Liguori, A.; D’Aveni, A.; Karachaliou, N.; Gonzalez-Cao, M.; Daffinà, M.G.; Lazzari, C.; Altavilla, G.; Rosell, R. Liquid biopsy for lung cancer early detection. J. Thorac. Dis. 2018, 10, S882–S897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofman, P. Liquid biopsy for early detection of lung cancer. Curr. Opin. Oncol. 2017, 29, 73–78. [Google Scholar] [CrossRef]
- Postel, M.; Roosen, A.; Laurent-Puig, P.; Taly, V.; Wang-Renault, S.-F. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: A cancer diagnostic perspective. Expert Rev. Mol. Diagn. 2018, 18, 7–17. [Google Scholar] [CrossRef]
- Buttitta, F.; Felicioni, L.; Di Lorito, A.; Cortellini, A.; Irtelli, L.; Brocco, D.; Di Marino, P.; Traisci, D.; D’Ostilio, N.; Di Paolo, A.; et al. Early prediction of resistance to tyrosine kinase inhibitors by plasma monitoring of EGFR mutations in NSCLC: A new algorithm for patient selection and personalized treatment. Oncotarget 2020, 11, 982–991. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Cole, R.; McWalter, G.; Walker, J.; Dearden, S.; Webster, A.; Milenkova, T.; et al. Gefitinib Treatment in EGFR Mutated Caucasian NSCLC: Circulating-Free Tumor DNA as a Surrogate for Determination of EGFR Status. J. Thorac. Oncol. 2014, 9, 1345–1353. [Google Scholar] [CrossRef] [Green Version]
- Veldore, V.H.; Choughule, A.; Routhu, T.; Mandloi, N.; Noronha, V.; Joshi, A.; Dutt, A.; Gupta, R.; Ramprasad, V.L.; Prabhash, K. Validation of liquid biopsy: Plasma cell-free DNA testing in clinical management of advanced non-small cell lung cancer. Lung Cancer Targets Ther. 2018, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Shin, S.; Lee, K.-A. A Comparative Study for Detection of EGFR Mutations in Plasma Cell-Free DNA in Korean Clinical Diagnostic Laboratories. BioMed Res. Int. 2018, 2018, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Xu, T.; Wang, S.; Chang, H.; Yu, T.; Zhu, Y.; Chen, J. Liquid Biopsy Applications in the Clinic. Mol. Diagn. Ther. 2020, 24, 125–132. [Google Scholar] [CrossRef]
- Schwarz, G.; Bäumler, S.; Block, A.; Felsenstein, F.G.; Wenzel, G. Determination of detection and quantification limits for SNP allele frequency estimation in DNA pools using real time PCR. Nucleic Acids Res. 2004, 32, e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Zhou, C.; Liam, C.-K.; Wu, G.; Liu, X.; Zhong, Z.; Lu, S.; Cheng, Y.; Han, B.; Chen, L.; et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: Analyses from the phase III, randomized, open-label, ENSURE study. Ann. Oncol. 2015, 26, 1883–1889. [Google Scholar] [CrossRef] [PubMed]
- Goss, G.; Tsai, C.-M.; Shepherd, F.A.; Bazhenova, L.; Lee, J.S.; Chang, G.-C.; Crino, L.; Satouchi, M.; Chu, Q.; Hida, T.; et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): A multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 2016, 17, 1643–1652. [Google Scholar] [CrossRef]
- Jenkins, S.; Yang, J.C.-H.; Ramalingam, S.S.; Yu, K.; Patel, S.; Weston, S.; Hodge, R.; Cantarini, M.; Jänne, P.A.; Mitsudomi, T.; et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1061–1070. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-L.; Sequist, L.V.; Hu, C.-P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Schuler, M.; Mok, T.; et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: Analysis of LUX-Lung 3 and 6. Br. J. Cancer 2017, 116, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Cui, X.; Hu, J.; Li, Z.; Choi, J.R.; Yang, Q.; Lin, M.; Hui, L.Y.; Xu, F. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens. Bioelectron. 2017, 90, 459–474. [Google Scholar] [CrossRef]
- Hussung, S.; Follo, M.; Klar, R.F.; Michalczyk, S.; Fritsch, K.; Nollmann, F.; Hipp, J.; Duyster, J.; Scherer, F.; Von Bubnoff, N.; et al. Development and Clinical Validation of Discriminatory Multitarget Digital Droplet PCR Assays for the Detection of Hot Spot KRAS and NRAS Mutations in Cell-Free DNA. J. Mol. Diagn. 2020, 22, 943–956. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Paweletz, C.P.; Kuang, Y.; Mach, S.L.; O’Connell, A.; Messineo, M.M.; Luke, J.J.; Butaney, M.; Kirschmeier, P.; Jackman, D.M.; et al. Noninvasive Detection of Response and Resistance in EGFR-Mutant Lung Cancer Using Quantitative Next-Generation Genotyping of Cell-Free Plasma DNA. Clin. Cancer Res. 2014, 20, 1698–1705. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material for KRAS mutation detection. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Supplee, J.G.; Milan, M.S.; Lim, L.P.; Potts, K.T.; Sholl, L.M.; Oxnard, G.R.; Paweletz, C.P. Sensitivity of next-generation sequencing assays detecting oncogenic fusions in plasma cell-free DNA. Lung Cancer 2019, 134, 96–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vollbrecht, C.; Lehmann, A.; Lenze, D.; Hummel, M. Validation and comparison of two NGS assays for the detection of EGFR T790M resistance mutation in liquid biopsies of NSCLC patients. Oncotarget 2018, 9, 18529–18539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, Y.K.; Davis, A.A.; Jain, S.; Santa-Maria, C.; Flaum, L.; Beaubier, N.; Platanias, L.C.; Gradishar, W.; Giles, F.J.; Cristofanilli, M. Concordance of Genomic Alterations by Next-Generation Sequencing in Tumor Tissue versus Circulating Tumor DNA in Breast Cancer. Mol. Cancer Ther. 2017, 16, 1412–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, J.E.; Okamoto, I.; Sriuranpong, V.; Vansteenkiste, J.; Imamura, F.; Lee, J.S.; Pang, Y.-K.; Cobo, M.; Kasahara, K.; Cheng, Y.; et al. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 6644–6652. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Wei, S.; Song, Y. T790M and acquired resistance of EGFR TKI: A literature review of clinical reports. J. Thorac. Dis. 2011, 3, 10–18. [Google Scholar]
- Wu, S.-G.; Liu, Y.-N.; Tsai, M.-F.; Chang, Y.-L.; Yu, C.-J.; Yang, P.-C.; Yang, J.C.-H.; Wen, Y.-F.; Shih, J.-Y. The mechanism of acquired resistance to irreversible EGFR tyrosine kinase inhibitor-afatinib in lung adenocarcinoma patients. Oncotarget 2016, 7, 12404–12413. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-L.; Jenkins, S.; Ramalingam, S.; Han, J.-Y.; Delmonte, A.; Hsia, T.-C.; Laskin, J.; Kim, S.-W.; He, Y.; Patel, S.; et al. MA08.03 Osimertinib vs. Platinum-Pemetrexed for T790M-Mutation Positive Advanced NSCLC (AURA3): Plasma ctDNA Analysis. J. Thorac. Oncol. 2017, 12, S386. [Google Scholar] [CrossRef] [Green Version]
- Thress, K.S.; Brant, R.; Carr, T.H.; Dearden, S.; Jenkins, S.; Brown, H.; Hammett, T.; Cantarini, M.; Barrett, J.C. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015, 90, 509–515. [Google Scholar] [CrossRef] [Green Version]
- Helman, E.; Nguyen, M.; Karlovich, C.A.; DesPain, D.; Choquette, A.K.; Spira, A.I.; Yu, H.A.; Camidge, D.R.; Harding, T.C.; Lanman, R.B.; et al. Cell-Free DNA Next-Generation Sequencing Prediction of Response and Resistance to Third-Generation EGFR Inhibitor. Clin. Lung Cancer 2018, 19, 518–530.e7. [Google Scholar] [CrossRef] [Green Version]
- Passiglia, F.; Rizzo, S.; Di Maio, M.; Galvano, A.; Badalamenti, G.; Listì, A.; Gulotta, L.; Castiglia, M.; Fulfaro, F.; Bazan, V.; et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: A systematic review and meta-analysis. Sci. Rep. 2018, 8, 13379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madic, J.; Jovelet, C.; Lopez, J.; André, B.; Fatien, J.; Miran, I.; Honoré, A.; Mezquita, L.; Besse, B.; Lacroix, L.; et al. EGFR C797S, EGFR T790M and EGFR sensitizing mutations in non-small cell lung cancer revealed by six-color crystal digital PCR. Oncotarget 2018, 9, 37393–37406. [Google Scholar] [CrossRef] [PubMed]
- Thress, K.S.; Paweletz, C.P.; Felip, E.; Cho, B.C.; Stetson, D.; Dougherty, B.; Lai, Z.; Markovets, A.; Vivancos, A.P.; Kuang, Y.; et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat. Med. 2015, 21, 560–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mileham, K.F.; Zhang, Q.; Farhangfar, C.J.; Haggstrom, D.E.; Fairclough, S.; Zill, O.A.; Carrizosa, D.R.; Lanman, R.B.; Kim, E.S. Abstract C80: Development of EGFR C797S mutation in serial liquid biopsy assessments in the clinical practice setting. Drug Resist. Modif. 2015, 14. [Google Scholar] [CrossRef]
- Chabon, J.J.; Simmons, A.D.; Lovejoy, A.F.; Esfahani, M.S.; Newman, A.M.; Haringsma, H.J.; Kurtz, D.M.; Stehr, H.; Scherer, F.; Karlovich, C.A.; et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat. Commun. 2016, 7, 11815. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aggarwal, C.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 1.2020. J. Natl. Compr. Cancer Netw. 2019, 17, 1464–1472. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.J.A.; Karachaliou, N.; Berenguer, J.; Gimenez-Capitan, A.; Schellen, P.; Teixido, C.; Tannous, J.; Kuiper, J.L.; Drees, E.; Grabowska, M.; et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016, 7, 1066–1075. [Google Scholar] [CrossRef] [Green Version]
- Park, C.-K.; Kim, J.-E.; Kim, M.-S.; Kho, B.-G.; Park, H.-Y.; Kim, T.-O.; Shin, H.-J.; Cho, H.-J.; Choi, Y.-D.; Oh, I.-J.; et al. Feasibility of liquid biopsy using plasma and platelets for detection of anaplastic lymphoma kinase rearrangements in non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 2071–2082. [Google Scholar] [CrossRef] [Green Version]
- McCoach, C.E.; Blakely, C.M.; Banks, K.C.; Levy, B.M.; Chue, B.M.; Raymond, V.M.; Le, A.T.; Lee, C.E.; Diaz, J.; Waqar, S.N.; et al. Clinical Utility of Cell-Free DNA for the Detection ofALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 2758–2770. [Google Scholar] [CrossRef] [Green Version]
- Gadgeel, S.M.; Mok, T.S.K.; Peters, S.; Alexander, J.A.A.; Leighl, N.B.; Sriuranpong, V.; Perol, M.; De Castro, G., Jr.; Nadal, E.; De Marinis, F.; et al. LBA81_PR-Phase II/III blood first assay screening trial (BFAST) in patients (pts) with treatment-naïve NSCLC: Initial results from the ALK+ cohort. Ann. Oncol. 2019, 30, v918. [Google Scholar] [CrossRef]
- Caparica, R.; Yen, C.T.; Coudry, R.; Ou, S.-H.I.; Varella-Garcia, M.; Camidge, D.R.; De Castro, G. Responses to Crizotinib Can Occur in High-Level MET -Amplified Non-Small Cell Lung Cancer Independent of MET Exon 14 Alterations. J. Thorac. Oncol. 2017, 12, 141–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drilon, A.; Clark, J.W.; Weiss, J.; Ou, S.-H.I.; Camidge, D.R.; Solomon, B.J.; Otterson, G.A.; Villaruz, L.C.; Riely, G.J.; Heist, R.S.; et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat. Med. 2020, 26, 47–51. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves First Targeted Therapy to Treat Aggressive Form of Lung Cancer. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-treat-aggressive-form-lung-cancer (accessed on 12 November 2020).
- Wolf, J.; Seto, T.; Han, J.-Y.; Reguart, N.; Garon, E.; Groen, H.J.; Tan, D.; Hida, T.; De Jonge, M.; Orlov, S.; et al. Results of the GEOMETRY mono-1 phase II study for evaluation of the MET inhibitor capmatinib (INC280) in patients (pts) with METΔex14 mutated advanced non-small cell lung cancer (NSCLC). Ann. Oncol. 2018, 29, viii741–viii742. [Google Scholar] [CrossRef]
- FDA Grants Accelerated Approval to Capmatinib for Metastatic Non-Small Cell Lung Cancer. Available online: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-grants-accelerated-approval-capmatinib-metastatic-non-small-cell-lung-cancer (accessed on 12 November 2020).
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Reungwetwattana, T.; Liang, Y.; Zhu, V.; Ou, S.-H.I. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: The Why, the How, the Who, the Unknown, and the Inevitable. Lung Cancer 2017, 103, 27–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Mondelo-Macía, P.; Rodríguez, C.; Valiña, L.; Aguín, S.; León-Mateos, L.; García-González, J.; Abalo, A.; Rapado-González, Ó.; Suárez-Cunqueiro, M.M.; Díaz-Lagares, A.; et al. Detection of MET Alterations Using Cell Free DNA and Circulating Tumor Cells from Cancer Patients. Cells 2020, 9, 522. [Google Scholar] [CrossRef] [Green Version]
- Schrock, A.B.; Welsh, A.; Chung, J.H.; Pavlick, D.; Bernicker, E.; Creelan, B.C.; Forcier, B.; Ross, J.S.; Stephens, P.J.; Ali, S.M.; et al. Hybrid Capture-Based Genomic Profiling of Circulating Tumor DNA from Patients with Advanced Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2019, 14, 255–264. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, S.; Schwaederle, M.; Mohindra, M.; Jardim, D.L.F.; Kurzrock, R. MET alterations detected in blood-derived circulating tumor DNA correlate with bone metastases and poor prognosis. J. Hematol. Oncol. 2018, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Planchard, D.; Kim, T.M.; Mazieres, J.; Quoix, E.; Riely, G.; Barlesi, F.; Souquet, P.-J.; Smit, E.F.; Groen, H.J.M.; Kelly, R.J.; et al. Dabrafenib in patients with BRAFV600E-positive advanced non-small-cell lung cancer: A single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 642–650. [Google Scholar] [CrossRef] [Green Version]
- Herbreteau, G.; Vallée, A.; Charpentier, S.; Normanno, N.; Hofman, P.; Denis, M.G. Circulating free tumor DNA in non-small cell lung cancer (NSCLC): Clinical application and future perspectives. J. Thorac. Dis. 2019, 11, S113–S126. [Google Scholar] [CrossRef] [PubMed]
- Guibert, N.; Pradines, A.; Casanova, A.; Farella, M.; Keller, L.; Soria, J.-C.; Favre, G.; Mazieres, J. Detection and Monitoring of the BRAF Mutation in Circulating Tumor Cells and Circulating Tumor DNA in BRAF-Mutated Lung Adenocarcinoma. J. Thorac. Oncol. 2016, 11, e109–e112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Shen, X.; Li, R.; Shen, J.; Zhang, H.; Yu, L.; Liu, B.; Wang, L. The detection and significance of EGFR and BRAF in cell-free DNA of peripheral blood in NSCLC. Oncotarget 2017, 8, 49773–49782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solassol, J.; Vendrell, J.; Senal, R.; Audran, P.; Leenhardt, F.; Quantin, X. Challenging BRAF/EGFR co-inhibition in NSCLC using sequential liquid biopsies. Lung Cancer 2019, 133, 45–47. [Google Scholar] [CrossRef] [PubMed]
- Gainor, J.F.; Shaw, A.T. Novel Targets in Non-Small Cell Lung Cancer:ROS1andRETFusions. Oncology 2013, 18, 865–875. [Google Scholar] [CrossRef] [Green Version]
- Shaw, A.T.; Ou, S.-H.I.; Bang, Y.-J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.B.; et al. Crizotinib in ROS1-Rearranged Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheffler, M.; Schultheis, A.; Teixido, C.; Michels, S.; Morales-Espinosa, D.; Viteri, S.; Hartmann, W.; Merkelbach-Bruse, S.; Fischer, R.; Schildhaus, H.-U.; et al. ROS1 rearrangements in lung adenocarcinoma: Prognostic impact, therapeutic options and genetic variability. Oncotarget 2015, 6, 10577–10585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, H.Y.; Li, Q.; Engstrom, L.D.; West, M.; Appleman, V.; Wong, K.A.; McTigue, M.; Deng, Y.-L.; Liu, W.; Brooun, A.; et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc. Natl. Acad. Sci. USA 2015, 112, 3493–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drilon, A.; Siena, S.; Ou, S.I.; Patel, M.; Ahn, M.J.; Lee, J.; Bauer, T.M.; Farago, A.F.; Wheler, J.J.; Liu, S.V.; et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 2017, 7, 400–409. [Google Scholar] [CrossRef] [Green Version]
- Morris, T.A.; Khoo, C.; Solomon, B. Targeting ROS1 Rearrangements in Non-Small Cell Lung Cancer: Crizotinib and Newer Generation Tyrosine Kinase Inhibitors. Drugs 2019, 79, 1277–1286. [Google Scholar] [CrossRef]
- Mezquita, L.; Swalduz, A.; Jovelet, C.; Ortiz-Cuaran, S.; Howarth, K.; Planchard, D.; Avrillon, V.; Recondo, G.; Marteau, S.; Benitez, J.C.; et al. Clinical Relevance of an Amplicon-Based Liquid Biopsy for Detecting ALK and ROS1 Fusion and Resistance Mutations in Patients with Non-Small-Cell Lung Cancer. JCO Precis. Oncol. 2020, 2020, 272–282. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nat. Cell Biol. 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Drilon, A.; Oxnard, G.R.; Tan, D.S.; Loong, H.H.; Johnson, M.; Gainor, J.; McCoach, C.E.; Gautschi, O.; Besse, B.; Cho, B.C.; et al. Efficacy of Selpercatinib in RET Fusion-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Subbiah, V.; Gainor, J.; Taylor, M.; Zhu, V.; Doebele, R.; Lopes, G.; Baik, C.; Garralda, E.; Gadgeel, S.; et al. Treatment with pralsetinib (formerly BLU-667), a potent and selective RET inhibitor, provides rapid clearance of ctDNA in patients with RET-altered non-small cell lung cancer (NSCLC) and medullary thyroid cancer (MTC). Ann. Oncol. 2019, 30, ix122. [Google Scholar] [CrossRef]
- Rich, T.A.; Reckamp, K.L.; Chae, Y.K.; Doebele, R.C.; Iams, W.T.; Oh, M.; Raymond, V.M.; Lanman, R.B.; Riess, J.W.; Stinchcombe, T.E.; et al. Analysis of Cell-Free DNA from 32,989 Advanced Cancers Reveals Novel Co-occurring Activating RET Alterations and Oncogenic Signaling Pathway Aberrations. Clin. Cancer Res. 2019, 25, 5832–5842. [Google Scholar] [CrossRef] [Green Version]
- Kempf, E.; Rousseau, B.; Besse, B.; Paz-Ares, L. KRAS oncogene in lung cancer: Focus on molecularly driven clinical trials. Eur. Respir. Rev. 2016, 25, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Nacchio, M.; Sgariglia, R.; Gristina, V.; Pisapia, P.; Pepe, F.; De Luca, C.; Migliatico, I.; Clery, E.; Greco, L.; Vigliar, E.; et al. KRAS mutations testing in non-small cell lung cancer: The role of Liquid biopsy in the basal setting. J. Thorac. Dis. 2020, 12, 3836–3843. [Google Scholar] [CrossRef]
- AMG 510 First to Inhibit “Undruggable” KRAS. Cancer Discov. 2019, 9, 988–989. [CrossRef]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Hlth, D.; Sacher, A.; et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- Li, S.; Li, L.; Zhu, Y.; Huang, C.; Qin, Y.; Liu, H.; Renheidenreich, L.; Shi, B.; Ren, H.; Chu, X.; et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: A comprehensive mutation profiling from 5125 Chinese cohorts. Br. J. Cancer 2014, 110, 2812–2820. [Google Scholar] [CrossRef]
- Benesova, L.; Minarik, M.; Jancarikova, D.; Belsanova, B.; Pešek, M. Multiplicity of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC) patients treated with tyrosine kinase inhibitors. Anticancer. Res. 2010, 30, 1667–1671. [Google Scholar] [PubMed]
- Rachiglio, A.M.; Fenizia, F.; Piccirillo, M.C.; Galetta, D.; Crino, L.; Vincenzi, B.; Barletta, E.; Pinto, C.; Ferraù, F.; Lambiase, M.; et al. The Presence of Concomitant Mutations Affects the Activity of EGFR Tyrosine Kinase Inhibitors in EGFR-Mutant Non-Small Cell Lung Cancer (NSCLC) Patients. Cancers 2019, 11, 341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guibert, N.; Pradines, A.; Farella, M.; Casanova, A.; Gouin, S.; Keller, L.; Favre, G.; Mazieres, J. Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS -mutated lung adenocarcinoma. Lung Cancer 2016, 100, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Karo, A.; Buckingham, L.; Basu, S.; Borgia, J.A.; Bonomi, P.; Batus, M.; Fidler, M.J. Association of KRAS mutations detected via liquid biopsy in metastatic non-small cell lung cancer patients with high levels of FDG-PET SUV. J. Clin. Oncol. 2017, 35, e20594. [Google Scholar] [CrossRef]
- Rosell, R.; Dafni, U.; Felip, E.; Curioni-Fontecedro, A.; Gautschi, O.; Budczies, J.; Massutí, B.; Palmero, R.; Aix, S.P.; Carcereny, E.; et al. Erlotinib and bevacizumab in patients with advanced non-small-cell lung cancer and activating EGFR mutations (BELIEF): An international, multicentre, single-arm, phase 2 trial. Lancet Respir. Med. 2017, 5, 435–444. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, Z.; Shi, Q. Liquid Biopsy Based Single-Cell Transcriptome Profiling Characterizes Heterogeneity of Disseminated Tumor Cells from Lung Adenocarcinoma. Proteomics 2020, 20, e1900224. [Google Scholar] [CrossRef]
- Iacono, D.; Osman, G.A.; Migliorino, M.R.; Grillo, L.; Remotti, D.; Nunnari, J.; Ricciardi, S.; Rossi, A.; Mancuso, A.; Graziano, P.; et al. Intrapatient Molecular and Histologic Heterogeneity After First-generation or Second-generation TKI Therapy of NSCLC Patients. Am. J. Clin. Oncol. 2019, 42, 845–850. [Google Scholar] [CrossRef]
- Vojnic, M.; Kubota, D.; Kurzatkowski, C.; Offin, M.; Suzawa, K.; Benayed, R.; Schoenfeld, A.J.; Plodkowski, A.J.; Poirier, J.T.; Rudin, C.M.; et al. Acquired BRAF Rearrangements Induce Secondary Resistance to EGFR therapy in EGFR-Mutated Lung Cancers. J. Thorac. Oncol. 2019, 14, 802–815. [Google Scholar] [CrossRef]
- Ho, C.-C.; Liao, W.-Y.; Lin, C.-A.; Shih, J.-Y.; Yu, C.-J.; Yang, J.C. Acquired BRAF V600E Mutation as Resistant Mechanism after Treatment with Osimertinib. J. Thorac. Oncol. 2017, 12, 567–572. [Google Scholar] [CrossRef] [Green Version]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef] [Green Version]
- Soria-Comes, T.; Palomar-Abril, V.; Ureste, M.M.; Guerola, M.T.; Maiques, I.C.M. Real-World Data of the Correlation between EGFR Determination by Liquid Biopsy in Non-squamous Non-small Cell Lung Cancer (NSCLC) and the EGFR Profile in Tumor Biopsy. Pathol. Oncol. Res. 2019, 26, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.; Wu, Y.-L.; Lee, J.S.; Yu, C.-J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, B.; Meldgaard, P.; Hager, H.; Wu, L.; Wei, W.; Tsai, J.; Khalil, A.A.; Nexo, E.; Sorensen, B.S. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014, 14, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paweletz, C.P.; Sacher, A.G.; Raymond, C.K.; Alden, R.S.; O’Connell, A.; Mach, S.L.; Kuang, Y.; Gandhi, L.; Kirschmeier, P.; English, J.M.; et al. Bias-Corrected Targeted Next-Generati on Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin. Cancer Res. 2015, 22, 915–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.; Blau, S.; Collisson, E.A.; Divers, S.G.; Hoon, D.S.B.; Kopetz, E.S.; et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar] [CrossRef]
- Tan, C.-S.; Kumarakulasinghe, N.B.; Huang, Y.-Q.; Ang, Y.L.E.; Choo, J.R.-E.; Goh, B.-C.; Soo, R.A. Third generation EGFR TKIs: Current data and future directions. Mol. Cancer 2018, 17, 1–14. [Google Scholar] [CrossRef]
- Gu, Y.; Zhu, X.; Cao, B.; Wu, X.; Tong, X.; Shao, Y.W.; Liang, L. Transformation to small cell lung cancer and activation of KRAS during long-term erlotinib maintenance in a patient with non-small cell lung cancer: A case report. Oncol. Lett. 2019, 17, 5219–5223. [Google Scholar] [CrossRef]
- Chang, S.; Hur, J.Y.; Choi, Y.-L.; Lee, C.H.; Kim, W.S. Current status and future perspectives of liquid biopsy in non-small cell lung cancer. J. Pathol. Transl. Med. 2020, 54, 204–212. [Google Scholar] [CrossRef]
- Minari, R.; Bordi, P.; Del Re, M.; Facchinetti, F.; Mazzoni, F.; Barbieri, F.; Camerini, A.; Comin, C.E.; Gnetti, L.; Azzoni, C.; et al. Primary resistance to osimertinib due to SCLC transformation: Issue of T790M determination on liquid re-biopsy. Lung Cancer 2018, 115, 21–27. [Google Scholar] [CrossRef]

| Assay Type | Advantages | Main Limitations |
|---|---|---|
| Real-time PCR | -Rapid results -Cost effectiveness | -Limited number of DNA sequences -Allele frequency of at least 1% for some assays (Cobas® EGFR Mutation Test v2, Therascreen) |
| Digital droplet PCR | -High sensitivity (detection of low allele frequency, 0.005–0.1%) | -Limited number of DNA sequences |
| Next-generation sequencing | -Larger number of DNA sequences -Detection of coexisting mutations | -Higher allele frequency required compared to ddPCR (0.1%) -Higher costs -More complex data analysis and interpretation |
| Target Genomic Alteration | Most Frequent Liquid Biopsy Assays | Relevant Features | ||
|---|---|---|---|---|
| EGFR | Del19 and p. L858R | RT-PCR | -Cobas® EGFR Mutation Test v2 -IdyllaTM ctEGFR Mutation Test -EntroGen ctEGFR Mutation Detection Kit | -Cobas®: 79% concordance with positive tissue test (FLAURA trial) |
| p. T790M | RT-PCR | -Cobas® EGFR Mutation Test v2 -IdyllaTM ctEGFR Mutation Test -EntroGen ctEGFR Mutation Detection Kit | -Cobas®: 51% concordance with positive tissue test (AURA3 trial) -Relevance of tissue rebiopsies in case of negative plasma test | |
| Droplet Digital™ PCR | -71% vs. 41% sensitivity, 83% vs. 100% specificity as compared to Cobas®, and 74% concordance with tissue test (AURA trial, 38 samples) | |||
| BEAMing digital PCR | -81% vs. 73% sensitivity, 58% vs. 67% specificity as compared to Cobas® (AURA trial, 72 samples) | |||
| NGS * | -For GUARDANT360® 70-gene NGS: 85% concordance with positive tissue test (Therascreen EGFR RGQ PCR Kit, Qiagen) | |||
| p. C797S | RT-PCR (EntroGen ctEGFR Mutation Detection Kit) | |||
| ddPCR or NGS* | ||||
| ALK | Fusions and mutations | RT-PCR | -From plasma or platelets (65–79% sensitivity, higher for platelets; 89–100% specificity) | |
| NGS * | -Allows identification of distinct fusion partners | |||
| MET | Exon 14 skipping and amplifications | ddPCR or NGS * | -ddPCR for MET amplification: 86% sensitivity and 100% specificity as compared with positive tissue FISH | |
| BRAF | Mutations | RT-PCR | -Competitive Allele-Specific TaqMan® PCR | -29% sensitivity, 93% specificity, and 89% concordance as compared with positive tissue test |
| -IdyllaTM ctBRAF Mutation Test | ||||
| ddPCR or NGS * | ||||
| ROS1 | Fusions and mutations | NGS * | -InVisionFirst®-Lung: 67% sensitivity for ROS1/ALK fusions at diagnosis | |
| RET | Fusions and mutations | NGS * | -Allows identification of distinct fusion partners | |
| KRAS | Mutations | RT-PCR, ddPCR, or NGS * | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhoundova, D.; Mosquera Martinez, J.; Musmann, L.E.; Britschgi, C.; Rütsche, C.; Rechsteiner, M.; Nadal, E.; Garcia Campelo, M.R.; Curioni-Fontecedro, A. The Role of the Liquid Biopsy in Decision-Making for Patients with Non-Small Cell Lung Cancer. J. Clin. Med. 2020, 9, 3674. https://doi.org/10.3390/jcm9113674
Akhoundova D, Mosquera Martinez J, Musmann LE, Britschgi C, Rütsche C, Rechsteiner M, Nadal E, Garcia Campelo MR, Curioni-Fontecedro A. The Role of the Liquid Biopsy in Decision-Making for Patients with Non-Small Cell Lung Cancer. Journal of Clinical Medicine. 2020; 9(11):3674. https://doi.org/10.3390/jcm9113674
Chicago/Turabian StyleAkhoundova, D., J. Mosquera Martinez, L. E. Musmann, C. Britschgi, C. Rütsche, M. Rechsteiner, E. Nadal, M. R. Garcia Campelo, and A. Curioni-Fontecedro. 2020. "The Role of the Liquid Biopsy in Decision-Making for Patients with Non-Small Cell Lung Cancer" Journal of Clinical Medicine 9, no. 11: 3674. https://doi.org/10.3390/jcm9113674
APA StyleAkhoundova, D., Mosquera Martinez, J., Musmann, L. E., Britschgi, C., Rütsche, C., Rechsteiner, M., Nadal, E., Garcia Campelo, M. R., & Curioni-Fontecedro, A. (2020). The Role of the Liquid Biopsy in Decision-Making for Patients with Non-Small Cell Lung Cancer. Journal of Clinical Medicine, 9(11), 3674. https://doi.org/10.3390/jcm9113674





