TMPRSS4: A Novel Tumor Prognostic Indicator for the Stratification of Stage IA Tumors and a Liquid Biopsy Biomarker for NSCLC Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Cohort of Patients for Immunohistochemical Analysis of TMPRSS4
2.3. Cohort of Patients for Methylation Analysis of TMPRSS4
2.4. Evaluation of TMPRSS4 Expression by Immunohistochemistry
2.5. DNA Isolation and Bisulfite Conversion
2.6. Digital Droplet PCR to Detect the Methylation Status of TMPRSS4 and SHOX2
2.7. Statistical Analyses
3. Results
3.1. Prognostic Value of TMPRSS4 Protein Expression in NSCLC
3.2. Development of TMPRSS4 and SHOX2 Methylation Assays by ddPCR
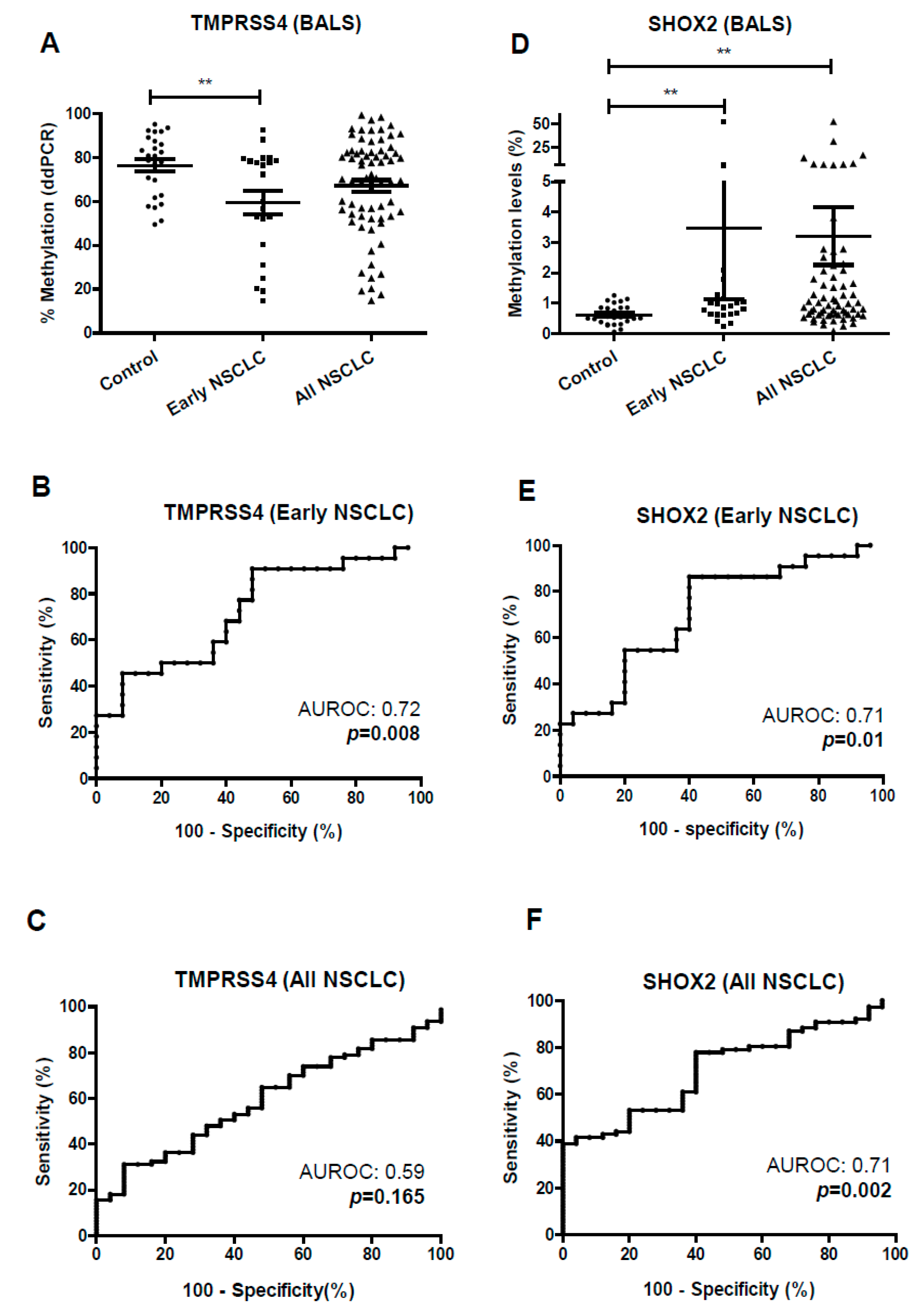
3.3. Diagnostic Potential of TMPRSS4 and SHOX2 Methylation in Liquid Biopsy Evaluated by ddPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Uramoto, H.; Tanaka, F. Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res. 2014, 3, 242–249. [Google Scholar] [PubMed]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Burotto, M.; Thomas, A.; Subramaniam, D.; Giaccone, G.; Rajan, A. Biomarkers in Early-Stage Non–Small-Cell Lung Cancer: Current Concepts and Future Directions. J. Thorac. Oncol. 2014, 9, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Exposito, F.; Villalba, M.; Redrado, M.; de Aberasturi, A.L.; Cirauqui, C.; Redin, E.; Guruceaga, E.; de Andrea, C.; Vicent, S.; Ajona, D.; et al. Targeting of TMPRSS4 sensitizes lung cancer cells to chemotherapy by impairing the proliferation machinery. Cancer Lett. 2019, 453, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-H.; Guo, Z.-Y.; Chen, Z.-T.; Zhi, X.-T.; Li, D.-K.; Dong, Z.-R.; Chen, Z.Q.; Hu, S.Y.; Li, T. TMPRSS4 facilitates epithelial-mesenchymal transition of hepatocellular carcinoma and is a predictive marker for poor prognosis of patients after curative resection. Sci. Rep. 2015, 5, 12366. [Google Scholar] [CrossRef]
- Villalba, M.; Diaz-Lagares, A.; Redrado, M.; de Aberasturi, A.L.; Segura, V.; Bodegas, M.E.; Pajares, M.J.; Pio, R.; Freire, J.; Gomez-Roman, J.; et al. Epigenetic alterations leading to TMPRSS4 promoter hypomethylation and protein overexpression predict poor prognosis in squamous lung cancer patients. Oncotarget 2016, 7, 22752–22769. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, K.; Geng, L.; Sun, J.; Xu, W.; Liu, D.; Gong, S.; Zhu, Y. Identification of candidate diagnostic and prognostic biomarkers for pancreatic carcinoma. EBioMedicine 2019, 40, 382–393. [Google Scholar] [CrossRef]
- Liang, B.; Wu, M.; Bu, Y.; Zhao, A.; Xie, F. Prognostic value of TMPRSS4 expression in patients with breast cancer. Med. Oncol. 2013, 30, 497. [Google Scholar] [CrossRef]
- Cheng, D.; Kong, H.; Li, Y. TMPRSS4 as a Poor Prognostic Factor for Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2013, 14, 14659–14668. [Google Scholar] [CrossRef]
- Liu, G.T.; Shen, C.; Ren, X.H.; Yang, L.; Yu, Y.M.; Xiu, Y.X.; Li, R.H.; Jiang, L.; Zhang, C.L.; Li, Y.W. Relationship between transmembrane serine protease expression and prognosis of esophageal squamous cell carcinoma. J. Biol. Regul. Homeost. Agents 2017, 31, 1067–1072. [Google Scholar] [PubMed]
- Kim, S.; Ko, D.; Lee, Y.; Jang, S.; Lee, Y.; Lee, I.Y.; Kim, S. Anti-cancer activity of the novel 2-hydroxydiarylamide derivatives IMD-0354 and KRT1853 through suppression of cancer cell invasion, proliferation, and survival mediated by TMPRSS4. Sci. Rep. 2019, 9, 10003. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Malapelle, U.; Troncone, G. Liquid Biopsy and Lung Cancer. Acta Cytol. 2018, 9, 1–8. [Google Scholar]
- Alegre, E.; Fusco, J.P.; Restituto, P.; Salas-Benito, D.; Rodríguez-Ruiz, M.E.; Andueza, M.P.; Pajares, M.J.; Patiño-García, A.; Pio, R.; Lozano, M.D.; et al. Total and mutated EGFR quantification in cell-free DNA from non-small cell lung cancer patients detects tumor heterogeneity and presents prognostic value. Tumor Biol. 2016, 37, 13687–13694. [Google Scholar] [CrossRef] [PubMed]
- Kneip, C.; Schmidt, B.; Seegebarth, A.; Weickmann, S.; Fleischhacker, M.; Liebenberg, V.; Field, J.K.; Dietrich, D. SHOX2 DNA Methylation Is a Biomarker for the Diagnosis of Lung Cancer in Plasma. J. Thorac. Oncol. 2011, 6, 1632–1638. [Google Scholar] [CrossRef]
- Konecny, M.; Markus, J.; Waczulikova, I.; Dolesova, L.; Kozlova, R.; Repiska, V.; Novosadova, H.; Majer, I. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma 2016, 63, 246–253. [Google Scholar] [CrossRef]
- Schmidt, B.; Beyer, J.; Dietrich, D.; Bork, I.; Liebenberg, V.; Fleischhacker, M. Quantification of cell-free mSHOX2 Plasma DNA for therapy monitoring in advanced stage non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients. PLoS ONE Public Libr. Sci. 2015, 10, e0118195. [Google Scholar] [CrossRef]
- Schmidt, B.; Liebenberg, V.; Dietrich, D.; Schlegel, T.; Kneip, C.; Seegebarth, A.; Field, J.K.; Dietrich, D. SHOX2 DNA Methylation is a Biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer 2010, 10, 600. [Google Scholar] [CrossRef]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and Elaboration. PLoS Med. 2012, 9, e1001216. [Google Scholar] [CrossRef]
- Villalba, M.; Lopez, L.; Redrado, M.; Ruiz, T.; de Aberasturi, A.L.; de la Roja, N.; Garcia, D.; Exposito, F.; de Andrea, C.; Alvarez-Fernandez, E.; et al. Development of biological tools to assess the role of TMPRSS4 and identification of novel tumor types with high expression of this prometastatic protein. Histol. Histopathol. 2017, 32, 929–940. [Google Scholar]
- Martínez-Terroba, E.; Behrens, C.; de Miguel, F.J.; Agorreta, J.; Monsó, E.; Millares, L.; Sainz, C.; Mesa-Guzman, M.; Pérez-Gracia, J.L.; Lozano, M.D.; et al. A novel protein-based prognostic signature improves risk stratification to guide clinical management in early-stage lung adenocarcinoma patients. J. Pathol. 2018, 245, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Roselli, M.; Mariotti, S.; Ferroni, P.; Laudisi, A.; Mineo, D.; Pompeo, E.; Mineo, T.C. Postsurgical chemotherapy in stage IB nonsmall cell lung cancer: Long-term survival in a randomized study. Int. J. Cancer 2006, 119, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, C.; Lee, H.; Baek, H.; Zo, J.; Shim, Y. Postoperative adjuvant chemotherapy for stage I non-small cell lung cancer. Eur. J. Cardio Thorac. Surg. 2005, 27, 1086–1091. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Park, H.S.; Cha, Y.J.; Lee, S.; Jeung, H.-C.; Cho, J.Y.; Kim, H.J.; Byun, M.K. Efficacy of adjuvant chemotherapy for completely resected stage IB non-small cell lung cancer: A retrospective study. J. Thorac. Dis. 2018, 10, 2279–2287. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, N.; Lv, C.; Yan, S.; Yang, Y. Should patients with stage IB non-small cell lung cancer receive adjuvant chemotherapy? A comparison of survival between the 8th and 7th editions of the AJCC TNM staging system for stage IB patients. J. Cancer Res. Clin. Oncol. 2019, 145, 463–469. [Google Scholar] [CrossRef]
- Seijo, L.M.; Peled, N.; Ajona, D.; Boeri, M.; Field, J.K.; Sozzi, G.; Pio, R.; Zulueta, J.J.; Spira, A.; Massion, P.P.; et al. Biomarkers in Lung Cancer Screening: Achievements, Promises, and Challenges. J. Thorac. Oncol. 2019, 14, 343–357. [Google Scholar] [CrossRef]
- De Aberasturi, A.L.; Redrado, M.; Villalba, M.; Larzabal, L.; Pajares, M.J.; Garcia, J.; Evans, S.R.; Garcia-Ros, D.; Bodegas, M.E.; Lopez, L.; et al. TMPRSS4 induces cancer stem cell-like properties in lung cancer cells and correlates with ALDH expression in NSCLC patients. Cancer Lett. 2016, 370, 165–176. [Google Scholar] [CrossRef]
- Mari-Alexandre, J.; Diaz-Lagares, A.; Villalba, M.; Juan, O.; Crujeiras, A.B.; Calvo, A.; Sandoval, J. Translating cancer epigenomics into the clinic: Focus on lung cancer. Transl. Res. 2017, 189, 76–92. [Google Scholar] [CrossRef]
- Martinez, R.; Esteller, M. The DNA methylome of glioblastoma multiforme. Neurobiol. Dis. 2010, 39, 40–46. [Google Scholar] [CrossRef]
- Weiss, G.; Schlegel, A.; Kottwitz, D.; König, T.; Tetzner, R. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J. Thorac. Oncol. 2017, 12, 77–84. [Google Scholar] [CrossRef]
- Su, Y.; Fang, H.; Bin Jiang, F. An epigenetic classifier for early stage lung cancer. Clin. Epigenet. BioMed Cent. 2018, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Lagares, A.; Mendez-Gonzalez, J.; Hervas, D.; Saigi, M.; Pajares, M.J.; Garcia, D.; Crujerias, A.B.; Pio, R.; Montuenga, L.M.; Zulueta, J.; et al. A Novel Epigenetic Signature for Early Diagnosis in Lung Cancer. Clin. Cancer Res. 2016, 22, 3361–3371. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; McCrory, D.C. Performance characteristics of different modalities for diagnosis of suspected lung cancer: Summary of published evidence. Chest 2003, 123, 115S–128S. [Google Scholar] [CrossRef] [PubMed]
- Ilse, P.; Biesterfeld, S.; Pomjanski, N.; Wrobel, C.; Schramm, M. Analysis of SHOX2 methylation as an aid to cytology in lung cancer diagnosis. Cancer Genom. Proteom. 2014, 11, 251–258. [Google Scholar]





| MDA Cohort (n = 489) | CIMA-CUN Cohort (n = 95) | ||
|---|---|---|---|
| n (%) | n (%) | ||
| Age | <65 | 180 (36.8) | 47 (49.5) |
| ≥65 | 309 (63.2) | 48 (50.5) | |
| Gender | Female | 260 (53.2) | 13 (13.7) |
| Male | 229 (46.8) | 82 (86.3) | |
| Smoking habits | Current | 167 (34.2) | 21 (22.1) |
| Former | 255 (52.1) | 66 (69.5) | |
| Never | 67 (13.7) | 8 (8.4) | |
| Histology | ADC | 299 (61.1) | 45 (47.4) |
| SCC | 171 (35.0) | 46 (48.4) | |
| Other | 19 (3.9) | 4 (4.2) | |
| Stage (8th edition) | I | 282 (57.7) | 44 (46.3) |
| II | 106 (21.7) | 31 (32.6) | |
| III | 92 (18.8) | 18 (18.9) | |
| IV | 9 (1.8) | 2 (2.1) | |
| Adjuvant therapy | No | 317 (64.8) | 54 (56.8) |
| Yes | 119 (24.3) | 41 (43.2) | |
| NA * | 53 (10.8) |
| MDA Cohort (n = 489) | |||||||
|---|---|---|---|---|---|---|---|
| RFS | OS | ||||||
| HR | 95% CI | p | HR | 95% CI | p | ||
| Age | <65 | 1 | |||||
| ≥65 | 1.83 | (1.35–2.47) | <0.001 | ||||
| Gender | Female | 1 | |||||
| Male | 0.83 | (0.63–1.10) | 0.211 | ||||
| Smoking habits | Never | 1 | 1 | ||||
| Former | 1.66 | (0.88–3.15) | 0.115 | 1.22 | (0.74–2.02) | 0.416 | |
| Current | 1.99 | (1.02–3.87) | 0.041 | 1.62 | (0.96–2.73) | 0.066 | |
| Histology | ADC | 1 | |||||
| SCC | 1.4 | (1.05–1.08) | 0.02 | ||||
| Other | 1.35 | (0.66–2.78) | 0.406 | ||||
| Stage (8th edition) | I | 1 | 1 | ||||
| II | 1.99 | (1.29–3.08) | 0.002 | 2.15 | (1.55–2.97) | <0.001 | |
| III–IV | 2.73 | (1.70–4.37) | <0.001 | 2.68 | (1.92–3.74) | <0.001 | |
| Adjuvant therapy | No | 1 | |||||
| Yes | 0.72 | (0.47–1.09) | 0.122 | ||||
| TMPRSS4 | Low | 1 | 1 | ||||
| High | 1.82 | (1.28–2.60) | 0.001 | 1.44 | (1.07–1.94) | 0.014 | |
| MDA Cohort (n = 283) | |||||||
|---|---|---|---|---|---|---|---|
| RFS | OS | ||||||
| HR | 95% CI | p | HR | 95% CI | p | ||
| Age | <65 | 1 | 0.292 | ||||
| ≥65 | 1.28 | (0.80–2.04) | |||||
| Gender | Female | 1 | 0.076 | ||||
| Male | 0.67 | (0.43–1.04) | |||||
| Smoking habits | Current | 1 | 1 | ||||
| Former | 1.73 | (0.76–3.93) | 0.183 | 1.37 | (0.69–2.73) | 0.352 | |
| Never | 1.93 | (0.82–4.50) | 0.127 | 1.24 | (0.59–2.61) | 0.56 | |
| Histology | ADC | 1 | |||||
| SCC | 1.53 | (0.97–2.39) | 0.062 | ||||
| Other | 2.73 | (1.16–6.43) | 0.021 | ||||
| Stage (8th edition) | IA | 1 | |||||
| IB | 1.27 | (0.79–2.02) | 0.31 | ||||
| TMPRSS4 | Low | 1 | 1 | ||||
| High | 2.42 | (1.47–3.99) | <0.001 | 1.99 | (1.25-3.16) | 0.004 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalba, M.; Exposito, F.; Pajares, M.J.; Sainz, C.; Redrado, M.; Remirez, A.; Wistuba, I.; Behrens, C.; Jantus-Lewintre, E.; Camps, C.; et al. TMPRSS4: A Novel Tumor Prognostic Indicator for the Stratification of Stage IA Tumors and a Liquid Biopsy Biomarker for NSCLC Patients. J. Clin. Med. 2019, 8, 2134. https://doi.org/10.3390/jcm8122134
Villalba M, Exposito F, Pajares MJ, Sainz C, Redrado M, Remirez A, Wistuba I, Behrens C, Jantus-Lewintre E, Camps C, et al. TMPRSS4: A Novel Tumor Prognostic Indicator for the Stratification of Stage IA Tumors and a Liquid Biopsy Biomarker for NSCLC Patients. Journal of Clinical Medicine. 2019; 8(12):2134. https://doi.org/10.3390/jcm8122134
Chicago/Turabian StyleVillalba, Maria, Francisco Exposito, Maria Jose Pajares, Cristina Sainz, Miriam Redrado, Ana Remirez, Ignacio Wistuba, Carmen Behrens, Eloisa Jantus-Lewintre, Carlos Camps, and et al. 2019. "TMPRSS4: A Novel Tumor Prognostic Indicator for the Stratification of Stage IA Tumors and a Liquid Biopsy Biomarker for NSCLC Patients" Journal of Clinical Medicine 8, no. 12: 2134. https://doi.org/10.3390/jcm8122134
APA StyleVillalba, M., Exposito, F., Pajares, M. J., Sainz, C., Redrado, M., Remirez, A., Wistuba, I., Behrens, C., Jantus-Lewintre, E., Camps, C., Montuenga, L. M., Pio, R., Lozano, M. D., de Andrea, C., & Calvo, A. (2019). TMPRSS4: A Novel Tumor Prognostic Indicator for the Stratification of Stage IA Tumors and a Liquid Biopsy Biomarker for NSCLC Patients. Journal of Clinical Medicine, 8(12), 2134. https://doi.org/10.3390/jcm8122134








