Cellular Processes in Human Ovarian Follicles Are Regulated by Expression Profile of New Gene Markers—Clinical Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients Selection
2.2. Patients CC and GC Acquisition
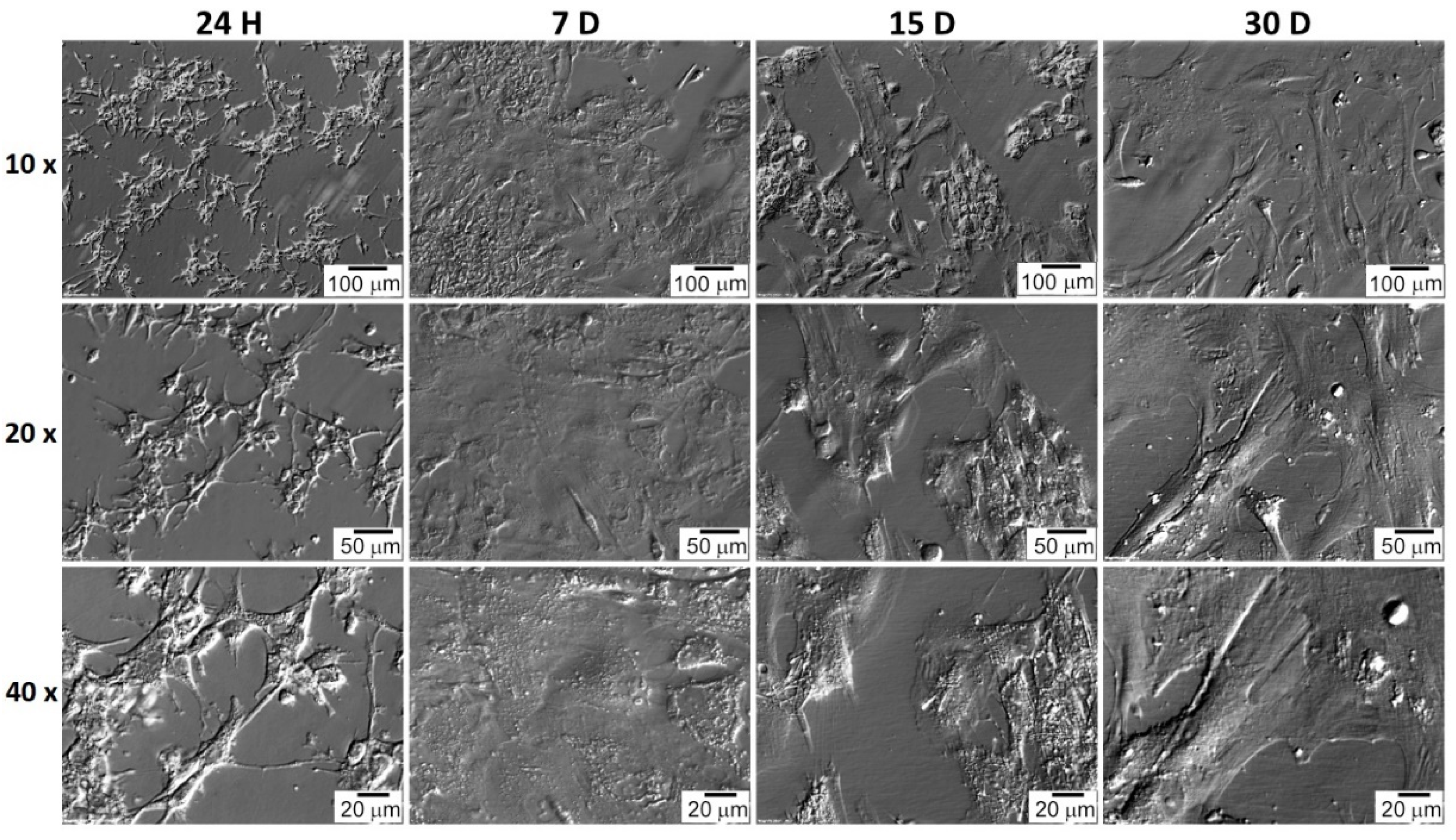
2.3. Cell In Vitro Culture
2.4. RNA Isolation
2.5. Microarray Expression Analysis and Statistics
2.6. RT-qPCR Analysis (Reverse Transcription Quantitative PCR)
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hummitzsch, K.; Anderson, R.A.; Wilhelm, D.; Wu, J.; Telfer, E.E.; Russell, D.L.; Robertson, S.A.; Rodgers, R.J. Stem cells, progenitor cells, and lineage decisions in the ovary. Endocr. Rev. 2015, 36, 65–91. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, X.; Shi, Y.; Wang, Z. The Signaling Pathways Involved in Ovarian Follicle Development. Front. Physiol. 2021, 12, 1610. [Google Scholar] [CrossRef]
- Turathum, B.; Gao, E.-M.; Chian, R.-C. The Function of Cumulus Cells in Oocyte Growth and Maturation and in Subsequent Ovulation and Fertilization. Cells 2021, 10, 2292. [Google Scholar] [CrossRef]
- Russell, D.L.; Gilchrist, R.; Brown, H.M.; Thompson, J.G. Bidirectional communication between cumulus cells and the oocyte: Old hands and new players? Theriogenology 2016, 86, 62–68. [Google Scholar] [CrossRef]
- Dumesic, D.A.; Meldrum, D.R.; Katz-Jaffe, M.G.; Krisher, R.L.; Schoolcraft, W.B. Oocyte environment: Follicular fluid and cumulus cells are critical for oocyte health. Fertil. Steril. 2015, 103, 303–316. [Google Scholar] [CrossRef]
- Diaz, F.; O’Brien, M.; Wigglesworth, K.; Eppig, J. The preantral granulosa cell to cumulus cell transition in the mouse ovary: Development of competence to undergo expansion. Dev. Biol. 2006, 299, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Vanderhyden, B.C. Loss of ovarian function and the risk of ovarian cancer. Cell Tissue Res. 2005, 322, 117–124. [Google Scholar] [CrossRef]
- Rimon-Dahari, N.; Yerushalmi-Heinemann, L.; Alyagor, L.; Dekel, N. Ovarian Folliculogenesis. Results Probl. Cell Differ. 2016, 58, 167–190. [Google Scholar] [CrossRef]
- Gondos, B. Surface epithelium of the developing ovary. Possible correlation with ovarian neoplasia. Am. J. Pathol. 1975, 81, 303–321. [Google Scholar]
- Sawyer, H.R.; Smith, P.; Heath, D.A.; Juengel, J.L.; Wakefield, S.J.; McNatty, K.P. Formation of ovarian follicles during fetal development in sheep. Biol. Reprod. 2002, 66, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zheng, T.; Hong, W.; Ye, H.; Hu, C.; Zheng, Y. Mechanism for the Decision of Ovarian Surface Epithelial Stem Cells to Undergo Neo-Oogenesis or Ovarian Tumorigenesis. Cell. Physiol. Biochem. 2018, 50, 214–232. [Google Scholar] [CrossRef]
- Hirshfield, A.N. Development of Follicles in the Mammalian Ovary. Int. Rev. Cytol. 1991, 124, 43–101. [Google Scholar] [CrossRef]
- Blaustein, A.; Lee, H. Surface cells of the ovary and pelvic peritoneum: A histochemical and ultrastructure comparison. Gynecol. Oncol. 1979, 8, 34–43. [Google Scholar] [CrossRef]
- Byskov, A.G. The Anatomy and Ultrastructure of the Rete System in the Fetal Mouse Ovary. Biol. Reprod. 1978, 19, 720–735. [Google Scholar] [CrossRef]
- Peters, H.; Pedersen, T. Origin of Follicle Cells in the Infant Mouse Ovary. Fertil. Steril. 1967, 18, 309–313. [Google Scholar] [CrossRef]
- Chermuła, B.; Kranc, W.; Jopek, K.; Budna-Tukan, J.; Hutchings, G.; Dompe, C.; Moncrieff, L.; Janowicz, K.; Józkowiak, M.; Jeseta, M.; et al. Human Cumulus Cells in Long-Term In Vitro Culture Reflect Differential Expression Profile of Genes Responsible for Planned Cell Death and Aging—A Study of New Molecular Markers. Cells 2020, 9, 1265. [Google Scholar] [CrossRef]
- Schmid, N.; Dietrich, K.-G.; Forne, I.; Burges, A.; Szymanska, M.; Meidan, R.; Mayr, D.; Mayerhofer, A. Sirtuin 1 and Sirtuin 3 in Granulosa Cell Tumors. Int. J. Mol. Sci. 2021, 22, 2047. [Google Scholar] [CrossRef] [PubMed]
- Dubeau, L. The Cell of Origin of Ovarian Epithelial Tumors and the Ovarian Surface Epithelium Dogma: Does the Emperor Have No Clothes? Gynecol. Oncol. 1999, 72, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.W.; Pejovic, T.; Lawson, M.; Jurevic, L.; Hobbs, T.; Stouffer, R.L. Ovulation in the Absence of the Ovarian Surface Epithelium in the Primate1. Biol. Reprod. 2010, 82, 599–605. [Google Scholar] [CrossRef][Green Version]
- Wright, J.W.; Pejovic, T.; Jurevic, L.; Bishop, C.; Hobbs, T.; Stouffer, R.L. Ovarian surface epitheliectomy in the non-human primate: Continued cyclic ovarian function and limited epithelial replacement. Hum. Reprod. 2011, 26, 1422–1430. [Google Scholar] [CrossRef] [PubMed]
- Chronowska, E. High-Throughput Analysis of Ovarian Granulosa Cell Transcriptome. BioMed Res. Int. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Dias, F.; Khan, M.I.R.; Sirard, M.A.; Adams, G.P.; Singh, J. Transcriptome analysis of granulosa cells after conventional vs long FSH-induced superstimulation in cattle. BMC Genom. 2018, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Stefańska, K.; Chamier-Gliszczyńska, A.; Jankowski, M.; Celichowski, P.; Kulus, M.; Rojewska, M.; Antosik, P.; Bukowska, D.; Bruska, M.; Nowicki, M.; et al. Epithelium morphogenesis and oviduct development are regulated by significant increase of expression of genes after long-term in vitro primary culture—A microarray assays. Med. J. Cell Biol. 2018, 6, 195–204. [Google Scholar] [CrossRef]
- Ożegowska, K.; Dyszkiewicz-Konwińska, M.; Celichowski, P.; Nawrocki, M.; Bryja, A.; Jankowski, M.; Kranc, W.; Brązert, M.; Knap, S.; Jeseta, M.; et al. Expression pattern of new genes regulating female sex differentiation and in vitro maturational status of oocytes in pigs. Theriogenology 2018, 121, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Kranc, W.; Brązert, M.; Celichowski, P.; Bryja, A.; Nawrocki, M.J.; Ożegowska, K.; Jankowski, M.; Jeseta, M.; Pawelczyk, L.; Bręborowicz, A.; et al. ‘Heart development and morphogenesis’ is a novel pathway for human ovarian granulosa cell differentiation during long-term in vitro cultivation-a microarray approach. Mol. Med. Rep. 2019, 19, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Kossowska-Tomaszczuk, K.; de Geyter, C.; de Geyter, M.; Martin, I.; Holzgreve, W.; Scherberich, A.; Zhang, H. The Multipotency of Luteinizing Granulosa Cells Collected from Mature Ovarian Follicles. Stem Cells 2009, 27, 210–219. [Google Scholar] [CrossRef]
- Galas, J.F. Primary Culture of Ovarian Cells for Research on Cell Interactions in the Hormonal Control of Steroidogenesis. Methods Mol. Biol. 2011, 806, 227–249. [Google Scholar] [CrossRef]
- Kranc, W.; Budna, J.; Kahan, R.; Chachuła, A.; Bryja, A.; Ciesiółka, S.; Borys, S.; Antosik, M.P.; Bukowska, D.; Brussow, K.P. Molecular basis of growth, proliferation, and differentiation of mammalian follicular granulosa cells. J. Biol. Regul. Homeost. Agents 2017, 31, 1–8. [Google Scholar]
- Chermuła, B.; Brązert, M.; Iżycki, D.; Ciesiółka, S.; Kranc, W.; Celichowski, P.; Ożegowska, K.; Nawrocki, M.J.; Jankowski, M.; Jeseta, M.; et al. New Gene Markers of Angiogenesis and Blood Vessels Development in Porcine Ovarian Granulosa Cells during Short-Term Primary Culture In Vitro. BioMed Res. Int. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Zhang, S.; Dolgalev, I.; Zhang, T.; Ran, H.; Levine, D.A.; Neel, B.G. Both fallopian tube and ovarian surface epithelium are cells-of-origin for high-grade serous ovarian carcinoma. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Ferraretti, A.P.; La Marca, A.; Fauser, B.C.J.M.; Tarlatzis, B.; Nargund, G.; Gianaroli, L.; on behalf of the ESHRE working group on Poor Ovarian Response Definition. ESHRE consensus on the definition of ‘poor response’ to ovarian stimulation for in vitro fertilization: The Bologna criteria. Hum. Reprod. 2011, 26, 1616–1624. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Ghosh, U.; Bose, S.; Ghosh, T.; Bandyopadhyay, T.K.; Basu, U. A monophasic solution for isolation of RNA devoid of polymerase chain reaction-detectable genomic DNA contamination. Anal. Biochem. 2015, 477, 50–52. [Google Scholar] [CrossRef] [PubMed]
- Szyszka, M.; Paschke, L.; Tyczewska, M.; Jopek, K.; Celichowski, P.; Milecka, P.; Sultanova, G.; Stelcer, E.; Malinska, A.; Malendowicz, L.K.; et al. Analysis of Transcriptome, Selected Intracellular Signaling Pathways, Proliferation and Apoptosis of LNCaP Cells Exposed to High Leptin Concentrations. Int. J. Mol. Sci. 2019, 20, 5412. [Google Scholar] [CrossRef] [PubMed]
- Jopek, K.; Celichowski, P.; Szyszka, M.; Tyczewska, M.; Milecka, P.; Malendowicz, L.K.; Rucinski, M. Transcriptome Profile of Rat Adrenal Evoked by Gonadectomy and Testosterone or Estradiol Replacement. Front. Endocrinol. 2017, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis: Figure 1. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Džafić, E.; Stimpfel, M.; Virant-Klun, I. Plasticity of granulosa cells: On the crossroad of stemness and transdifferentiation potential. J. Assist. Reprod. Genet. 2013, 30, 1255–1261. [Google Scholar] [CrossRef] [PubMed]
- Hoang, S.N.; Ho, Q.C.; Nguyen, T.T.P.; Doan, C.C.; Tran, D.H.; Le, L.T. Evaluation of stemness marker expression in bovine ovarian granulosa cells. Anim. Reprod. 2019, 16, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Combelles, C.M.; Fissore, R.A.; Albertini, D.F.; Racowsky, C. In vitro maturation of human oocytes and cumulus cells using a co-culture three-dimensional collagen gel system. Hum. Reprod. 2005, 20, 1349–1358. [Google Scholar] [CrossRef]
- Gilchrist, R.; Ritter, L.; Armstrong, D. Oocyte–somatic cell interactions during follicle development in mammals. Anim. Reprod. Sci. 2004, 82–83, 431–446. [Google Scholar] [CrossRef]
- Shomali, N.; Hemmatzadeh, M.; Yousefzadeh, Y.; Soltani-Zangbar, M.S.; Hamdi, K.; Mehdizadeh, A.; Yousefi, M. Exosomes: Emerging biomarkers and targets in folliculogenesis and endometriosis. J. Reprod. Immunol. 2020, 142, 103181. [Google Scholar] [CrossRef]
- McCoard, S.A.; Wise, T.H.; Ford, J.J. Germ cell development in Meishan and White Composite gilts. Anim. Reprod. Sci. 2003, 77, 85–105. [Google Scholar] [CrossRef][Green Version]
- Leung, C.C.; Wong, C.K. Effects of STC1 overexpression on tumorigenicity and metabolism of hepatocellular carcinoma. Oncotarget 2017, 9, 6852–6861. [Google Scholar] [CrossRef]
- Ma, X.; Gu, L.; Li, H.; Gao, Y.; Li, X.; Shen, D.; Gong, H.; Li, S.; Niu, S.; Zhang, Y.; et al. Hypoxia-induced overexpression of stanniocalcin-1 is associated with the metastasis of early stage clear cell renal cell carcinoma. J. Transl. Med. 2015, 13, 56. [Google Scholar] [CrossRef]
- Liu, G.; Yang, G.; Chang, B.; Mercado-Uribe, I.; Huang, M.; Zheng, J.; Bast, R.C.; Lin, S.-H.; Liu, J. Stanniocalcin 1 and Ovarian Tumorigenesis. J. Natl. Cancer Inst. 2010, 102, 812–827. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.C.-M.; Doherty, J.; Huschtscha, L.I.; Redvers, R.; Restall, C.; Reddel, R.R.; Anderson, R.L. STC1 expression is associated with tumor growth and metastasis in breast cancer. Clin. Exp. Metastasis 2014, 32, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, M.R.; Kløverpris, S.; Bøtkjær, J.A.; Wissing, M.L.; Andersen, C.Y.; Oxvig, C. The proteolytic activity of pregnancy-associated plasma protein-A is potentially regulated by stanniocalcin-1 and -2 during human ovarian follicle development. Hum. Reprod. 2016, 31, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Baioni, L.; Basini, G.; Bussolati, S.; Grasselli, F. Stanniocalcin 1 affects redox status of swine granulosa cells. Regul. Pept. 2011, 168, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Mamo, S.; Carter, F.; Lonergan, P.; Leal, C.L.; Al Naib, A.; McGettigan, P.; Mehta, J.P.; Evans, A.C.; Fair, T. Sequential analysis of global gene expression profiles in immature and in vitro matured bovine oocytes: Potential molecular markers of oocyte maturation. BMC Genom. 2011, 12, 151. [Google Scholar] [CrossRef]
- Xiang, Y.; Song, Y.; Li, Y.; Zhao, D.; Ma, L.; Tan, L. miR-483 is Down-Regulated in Polycystic Ovarian Syndrome and Inhibits KGN Cell Proliferation via Targeting Insulin-Like Growth Factor 1 (IGF1). Med. Sci. Monit. 2016, 22, 3383–3393. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Qin, N.; Xu, X.; Sun, X.; Chen, X.; Zhao, J.; Xu, R.; Mishra, B. Synergistic inhibition of csal1 and csal3 in granulosa cell proliferation and steroidogenesis of hen ovarian prehierarchical development. Biol. Reprod. 2019, 101, 986–1000. [Google Scholar] [CrossRef]
- Shimizu, T.; Hirai, Y.; Miyamoto, A. Expression of Cyclins and Cyclin-Dependent Kinase Inhibitors in Granulosa Cells from Bovine Ovary. Reprod. Domest. Anim. 2013, 48, e65–e69. [Google Scholar] [CrossRef]
- Andre, P.; Song, H.; Kim, W.; Kispert, A.; Yang, Y. Wnt5a and Wnt11 regulate mammalian anterior-posterior axis elongation. Development 2015, 142, 1516–1527. [Google Scholar] [CrossRef] [PubMed]
- Manuylov, N.L.; Smagulova, F.; Leach, L.; Tevosian, S.G. Ovarian development in mice requires the GATA4-FOG2 transcription complex. Development 2008, 135, 3731–3743. [Google Scholar] [CrossRef]
- Park, J.E.; Lee, D.H.; Lee, J.A.; Park, S.G.; Kim, N.-S.; Park, B.C.; Cho, S. Annexin A3 is a potential angiogenic mediator. Biochem. Biophys. Res. Commun. 2005, 337, 1283–1287. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Liu, B.; Zhou, L.; Wang, D.; He, X.; Xu, X.; Zhang, L.; Niu, C.; Liu, S. Downregulation of annexin A3 inhibits tumor metastasis and decreases drug resistance in breast cancer. Cell Death Dis. 2018, 9, 126. [Google Scholar] [CrossRef]
- Rougemont, A.-L.; Berczy, M.; Marq, N.L.; McKee, T.A.; Christinat, Y. Targeted RNA-sequencing identifies FBXW4 instead of MGEA5 as fusion partner of TGFBR3 in pleomorphic hyalinizing angiectatic tumor. Virchows Arch. 2019, 475, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.C.; Bernard, D.J.; Jelen, J.; Woodruff, T.K. Properties of inhibin binding to betaglycan, InhBP/p120 and the activin type II receptors. Mol. Cell. Endocrinol. 2002, 196, 79–93. [Google Scholar] [CrossRef]
- Matiller, V.; Hein, G.J.; Stassi, A.F.; Angeli, E.; Belotti, E.M.; Ortega, H.H.; Rey, F.; Salvetti, N.R. Expression of TGFBR1, TGFBR2, TGFBR3, ACVR1B and ACVR2B is altered in ovaries of cows with cystic ovarian disease. Reprod. Domest. Anim. 2018, 54, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-Q.; Wang, Y.-Q.; Xu, S.-M.; Liu, J.-F.; Bi, X.-Y.; Zhang, J.-P. The WNT/β-catenin signaling pathway may be involved in granulosa cell apoptosis from patients with PCOS in North China. J. Gynecol. Obstet. Hum. Reprod. 2017, 46, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Lee-Thacker, S.; Choi, Y.; Taniuchi, I.; Takarada, T.; Yoneda, Y.; Ko, C.; Jo, M. Core Binding Factor β Expression in Ovarian Granulosa Cells Is Essential for Female Fertility. Endocrinology 2018, 159, 2094–2109. [Google Scholar] [CrossRef]
- Hsieh, M.; Mulders, S.M.; Friis, R.R.; Dharmarajan, A.; Richards, J.S. Expression and Localization of Secreted Frizzled-Related Protein-4 in the Rodent Ovary: Evidence for Selective Up-Regulation in Luteinized Granulosa Cells. Endocrinology 2003, 144, 4597–4606. [Google Scholar] [CrossRef]
- Ożegowska, K.; Brązert, M.; Ciesiółka, S.; Nawrocki, M.; Kranc, W.; Celichowski, P.; Jankowski, M.; Bryja, A.; Jeseta, M.; Antosik, P.; et al. Genes Involved in the Processes of Cell Proliferation, Migration, Adhesion, and Tissue Development as New Potential Markers of Porcine Granulosa Cellular Processes In Vitro: A Microarray Approach. DNA Cell Biol. 2019, 38, 549–560. [Google Scholar] [CrossRef]
- Huang, K.; Dang, Y.; Zhang, P.; Shen, C.; Sui, X.; Xia, G.; Qin, Y.; Jiao, X.; Wang, C.; Huo, R.; et al. CAV1 regulates primordial follicle formation via the Notch2 signalling pathway and is associated with premature ovarian insufficiency in humans. Hum. Reprod. 2018, 33, 2087–2095. [Google Scholar] [CrossRef]
- Magnusson, K.; Gremel, G.; Rydén, L.; Pontén, V.; Uhlén, M.; Dimberg, A.; Jirström, K.; Pontén, F. ANLN is a prognostic biomarker independent of Ki-67 and essential for cell cycle progression in primary breast cancer. BMC Cancer 2016, 16, 1–13. [Google Scholar] [CrossRef]
- Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Arai, T.; Okato, A.; Kurozumi, A.; Kato, M.; Katada, K.; Okamoto, Y.; Seki, N. Regulation of ITGA 3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017, 108, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Shepel, E.; Grushka, N.; Makogon, N.; Sribna, V.; Pavlovych, S.; Yanchii, R. Changes in DNA integrity and gene expression in ovarian follicular cells of lipopolysaccharide-treated female mice. Pharmacol. Rep. 2018, 70, 1146–1149. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, L.J.; Pangas, S.A.; Carson, S.A.; Kovanci, E.; Cisneros, P.; Buster, J.E.; Amato, P.; Matzuk, M.M. Human cumulus granulosa cell gene expression: A predictor of fertilization and embryo selection in women undergoing IVF. Hum. Reprod. 2004, 19, 2869–2874. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.E.; Cook, D.; Collins, O.; Gamwell, L.F.; Dempster, H.A.; Wong, H.W.; McCloskey, C.W.; Garson, K.; Vuong, N.H.; Vanderhyden, B.C. COX2 is induced in the ovarian epithelium during ovulatory wound repair and promotes cell survival. Biol. Reprod. 2019, 101, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-J.; Park, J.-I.; Jeong, S.-E.; Seo, Y.-M.; Dam, P.T.M.; Seo, Y.-W.; Choi, B.-C.; Song, S.-J.; Chun, S.-Y.; Cho, M.-K. Regulation of interleukin-11 expression in ovulatory follicles of the rat ovary. Reprod. Fertil. Dev. 2017, 29, 2437–2445. [Google Scholar] [CrossRef]
- Xu, S.; Grande, F.; Garofalo, A.; Neamati, N.; Gu, D.; Liu, H.; Su, G.H.; Zhang, X.; Chin-Sinex, H.; Hanenberg, H.; et al. Discovery of a Novel Orally Active Small-Molecule gp130 Inhibitor for the Treatment of Ovarian Cancer. Mol. Cancer Ther. 2013, 12, 937–949. [Google Scholar] [CrossRef]
- Auersperg, N. Ovarian Surface Epithelium: Biology, Endocrinology, and Pathology. Endocr. Rev. 2001, 22, 255–288. [Google Scholar] [CrossRef]
- Mullany, L.K.; Fan, H.-Y.; Liu, Z.; White, L.D.; Marshall, A.; Gunaratne, P.; Anderson, M.L.; Creighton, C.J.; Xin, L.; Deavers, M.; et al. Molecular and functional characteristics of ovarian surface epithelial cells transformed by KrasG12D and loss of Pten in a mouse model in vivo. Oncogene 2011, 30, 3522–3536. [Google Scholar] [CrossRef]
- Fuller, P.J.; Chu, S.; Fikret, S.; Burger, H.G. Molecular pathogenesis of granulosa cell tumours. Mol. Cell. Endocrinol. 2002, 191, 89–96. [Google Scholar] [CrossRef]
- Eppig, J.J. Oocyte control of ovarian follicular development and function in mammals. Reproduction 2001, 122, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Pitman, J.L.; McNeilly, A.S.; McNeilly, J.R.; Hays, L.E.; Bagby, G.C., Jr.; Sawyer, H.R.; McNatty, K.P. The fate of granulosa cells following premature oocyte loss and the development of ovarian cancers. Int. J. Dev. Biol. 2012, 56, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Perales-Puchalt, A.; Svoronos, N.; Rutkowski, M.R.; Allegrezza, M.J.; Tesone, A.J.; Payne, K.; Wickramasinghe, J.; Nguyen, J.M.; O’Brien, S.W.; Gumireddy, K.; et al. Follicle-Stimulating Hormone Receptor Is Expressed by Most Ovarian Cancer Subtypes and Is a Safe and Effective Immunotherapeutic Target. Clin. Cancer Res. 2016, 23, 441–453. [Google Scholar] [CrossRef] [PubMed]
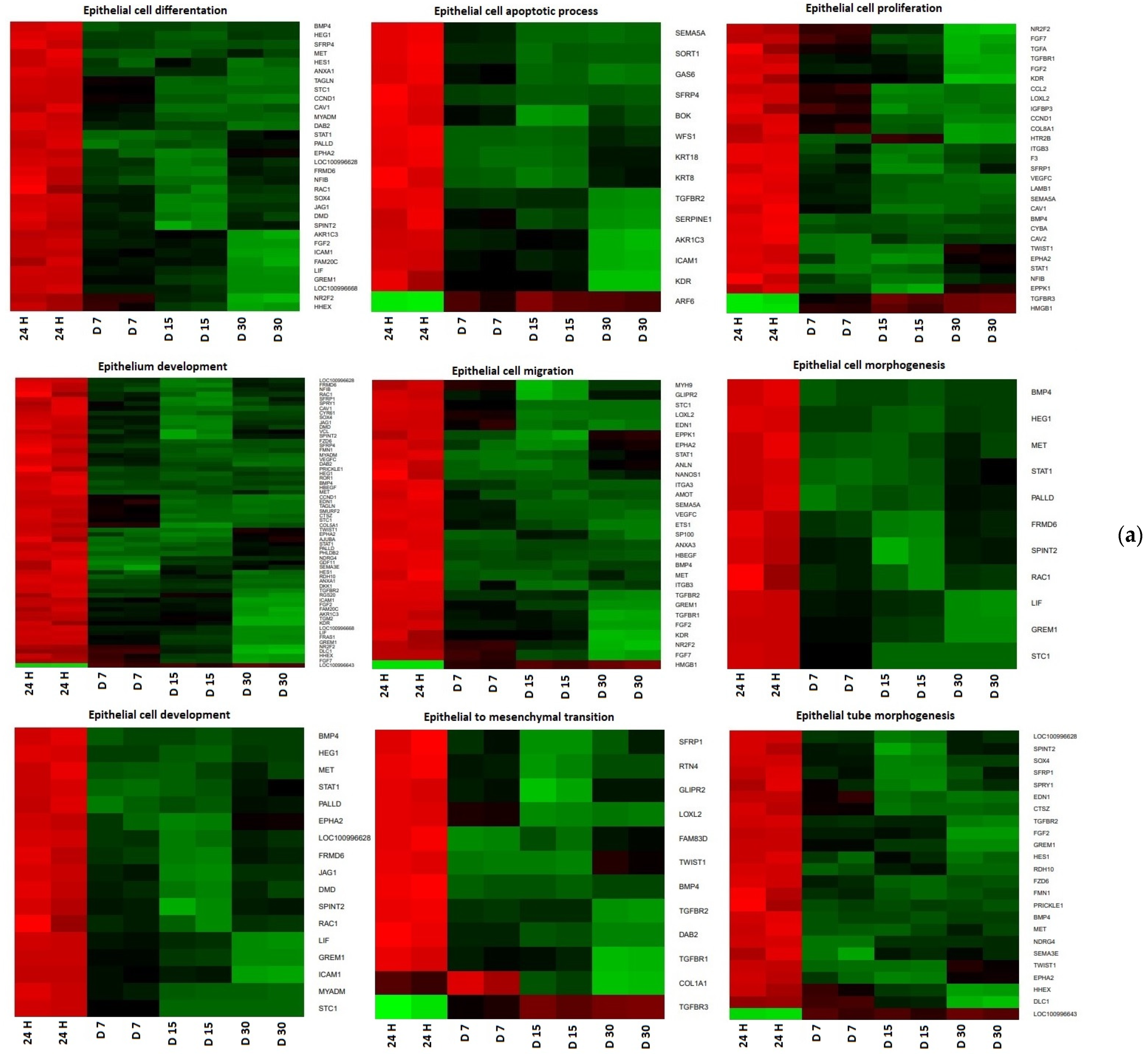
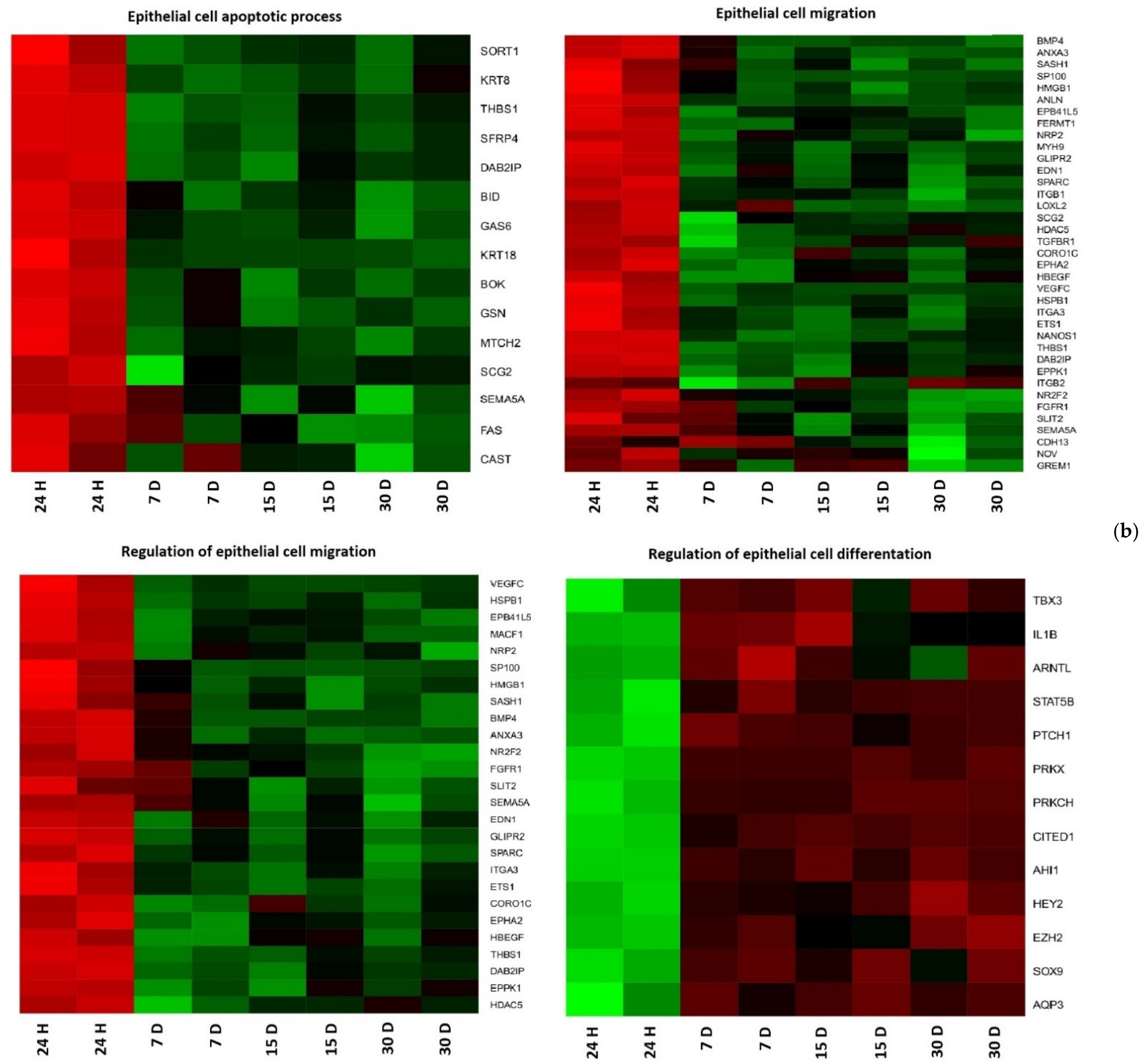
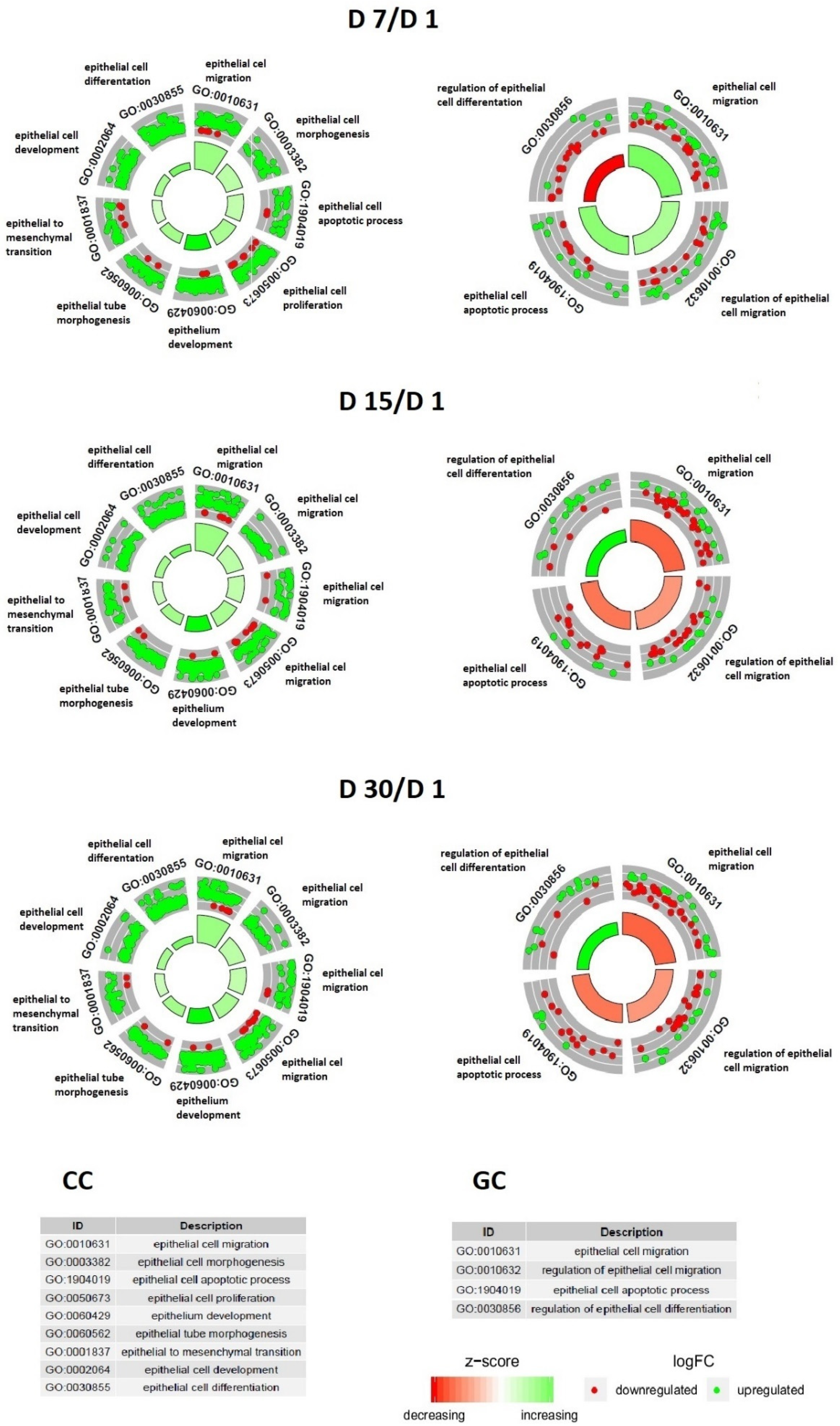
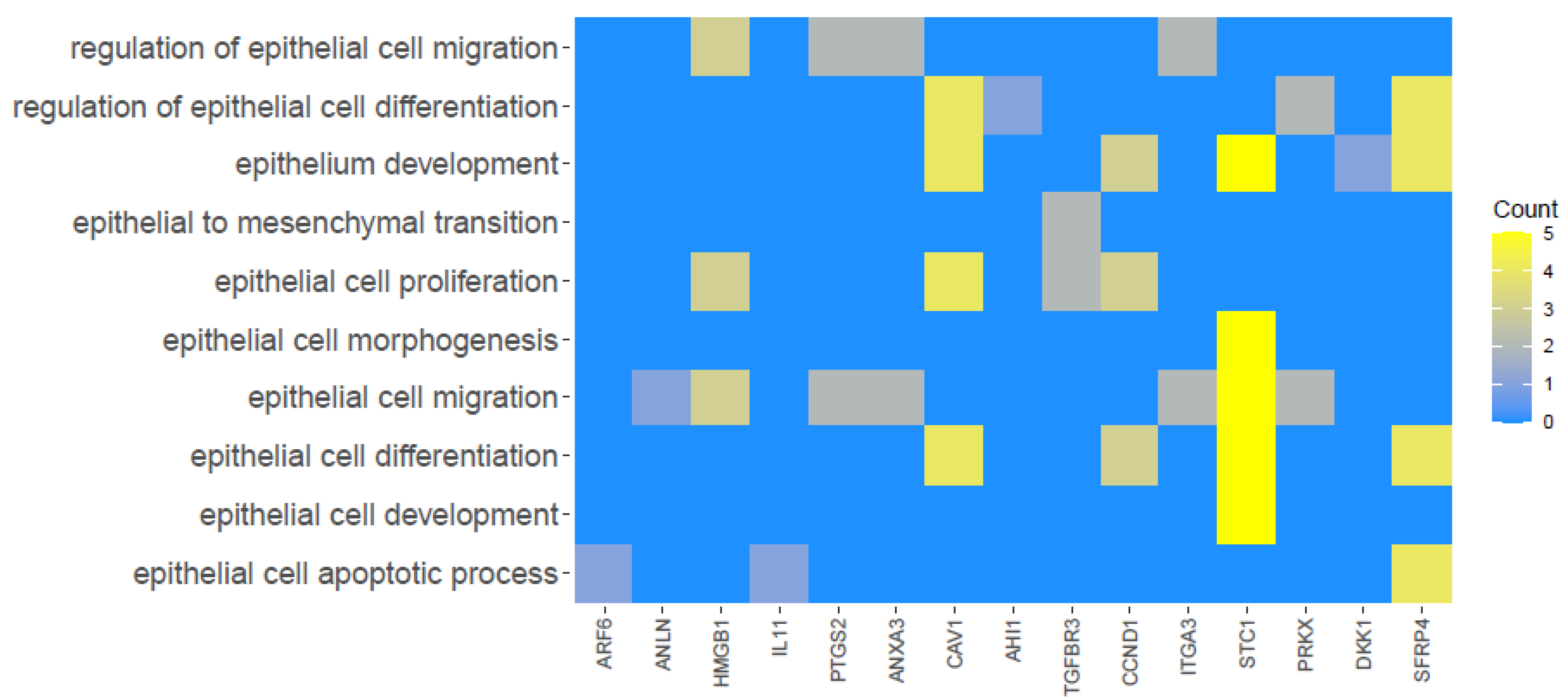
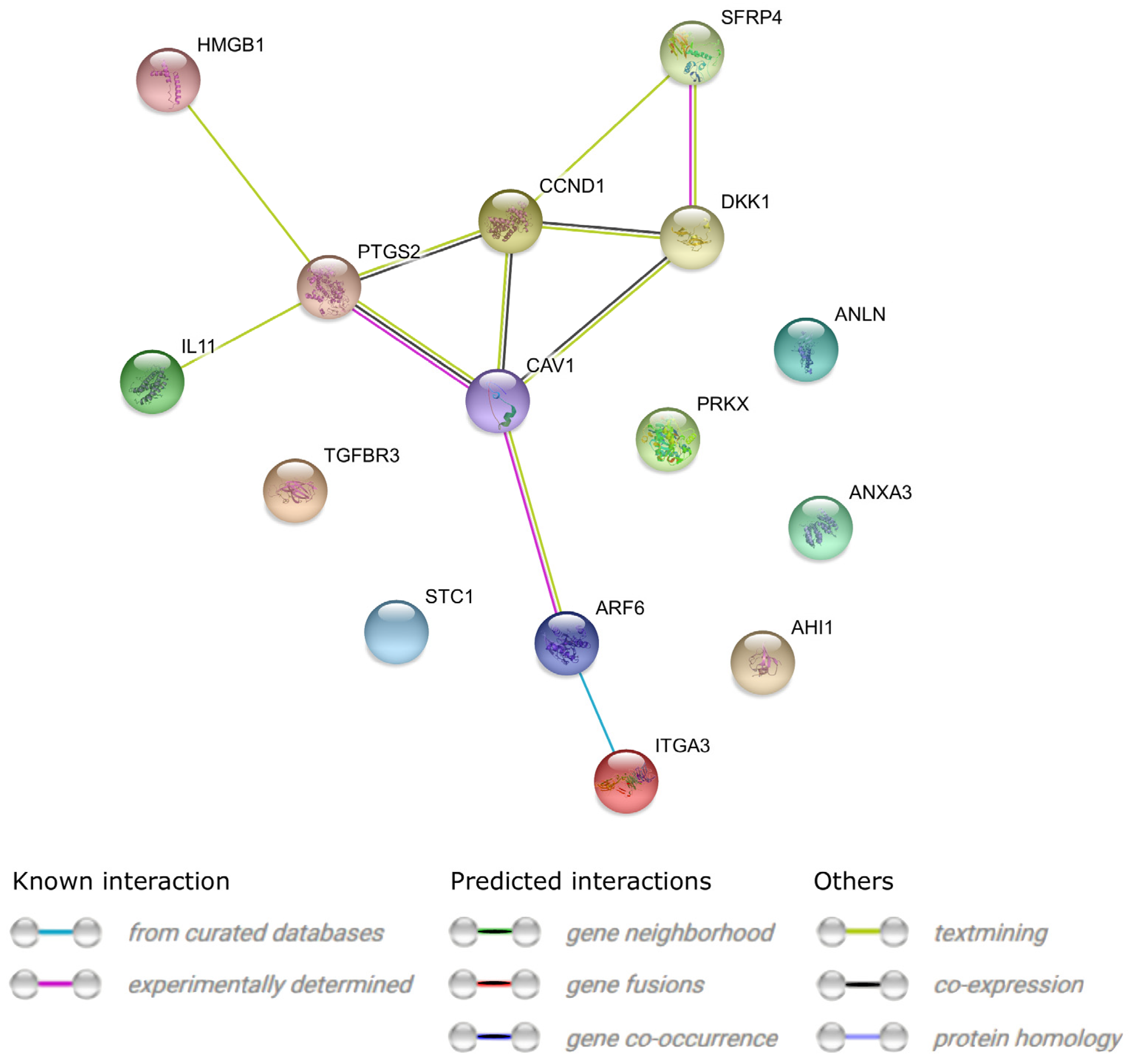
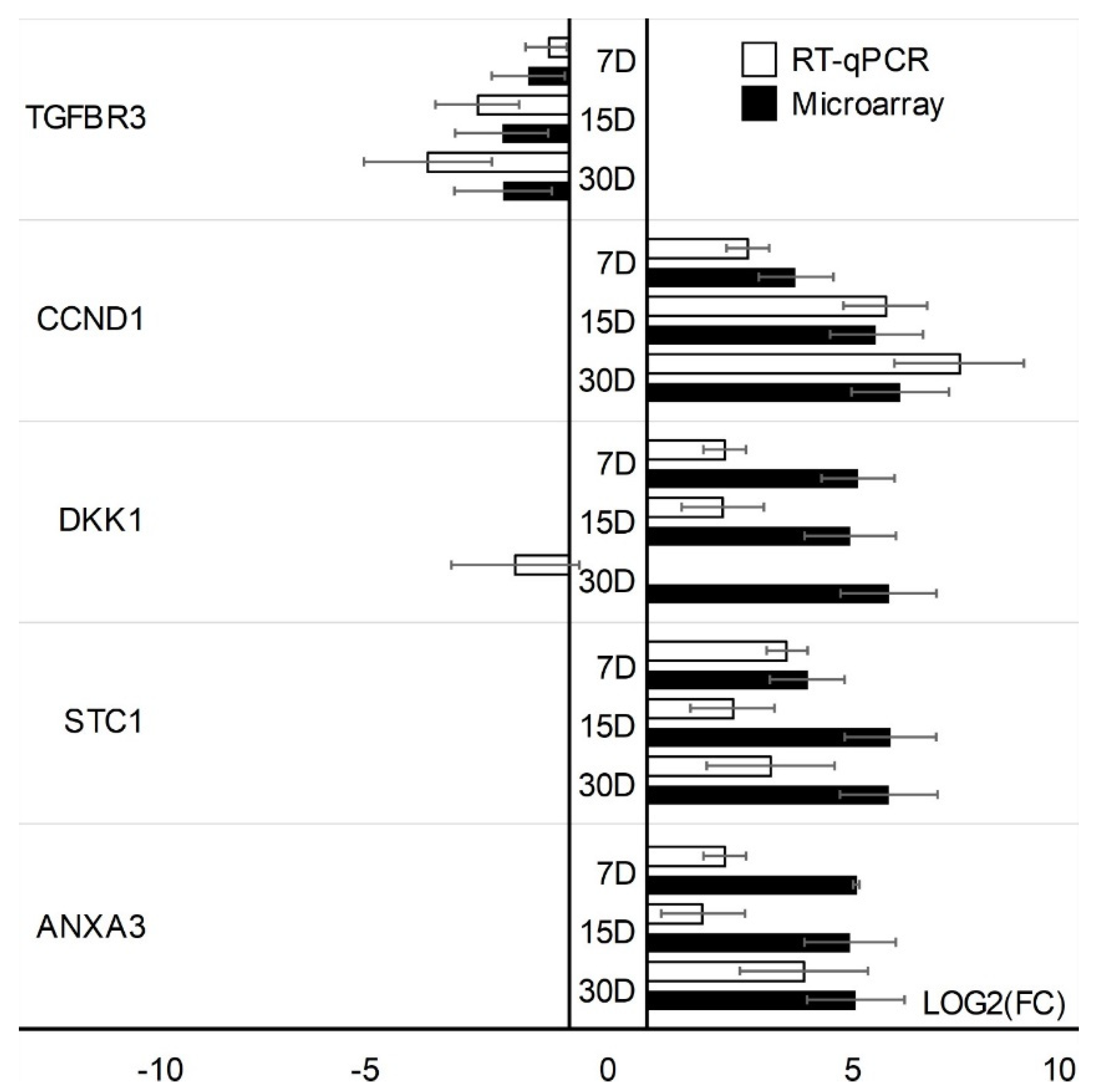
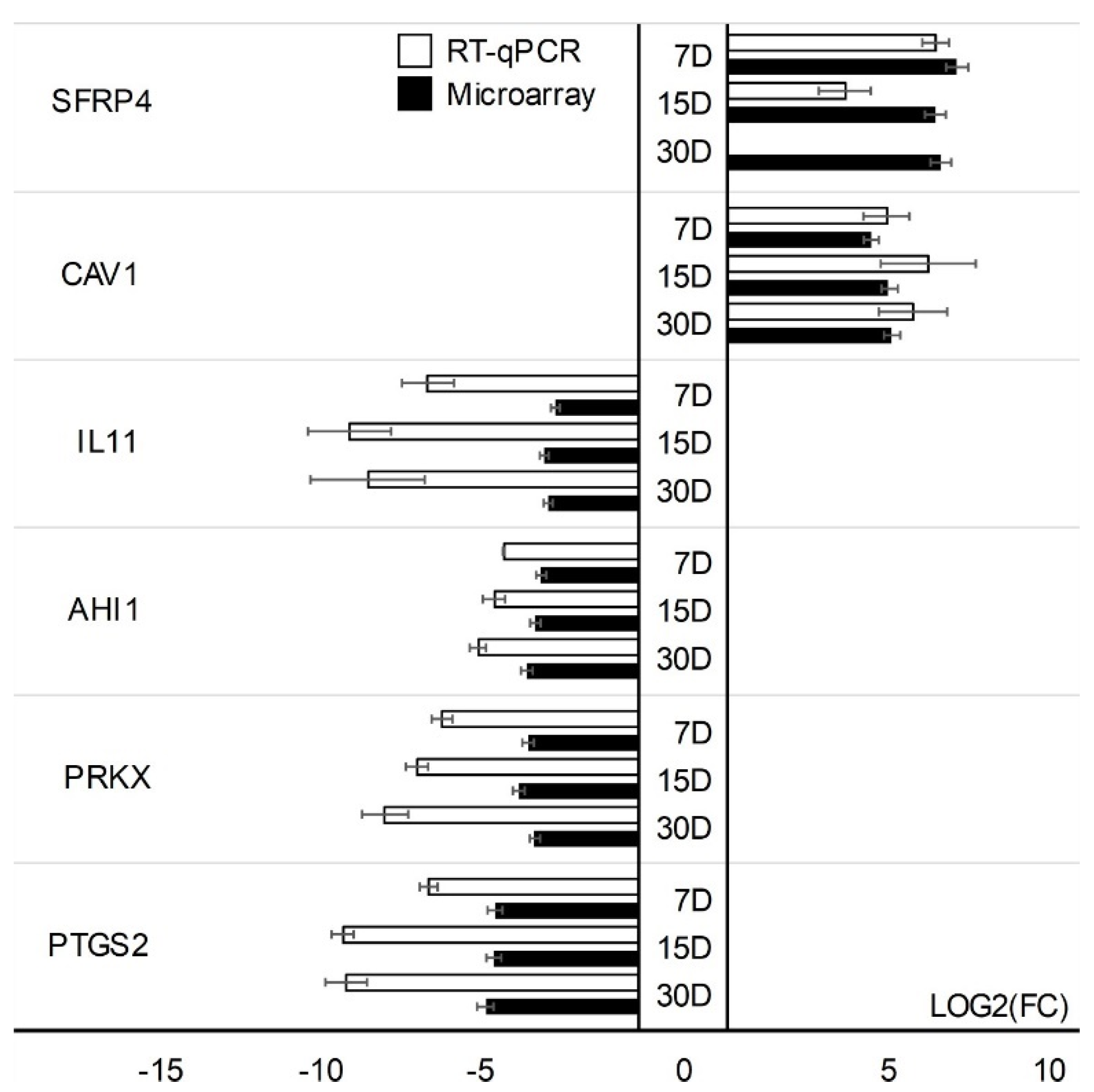
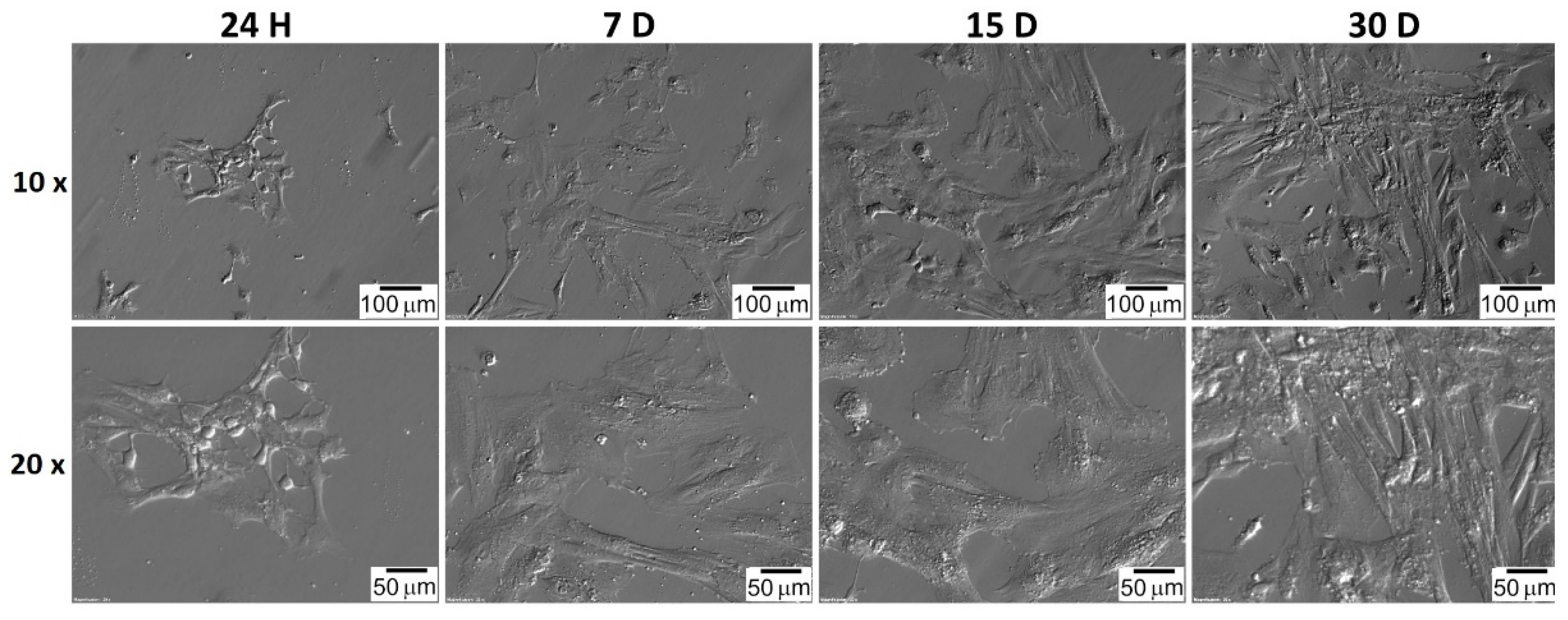
| Gene | Type of Cells | Primer Sequence (5′-3′) | Product Size (bp) | Entrez Gene ID (on Accession) |
|---|---|---|---|---|
| TGFBR3 | CCs | CCAAGATGAATGGCACACAC CCATCTGGCCAACCACTACT | 151 | 7049 |
| ANXA3 | CCs | GTTGGACACCGAGGAACAGT CACTAGGGCCACCATGAGAT | 249 | 306 |
| DKK1 | CCs | TCCGAGGAGAAATTGAGGAA CCTGAGGCACAGTCTGATGA | 157 | 22,943 |
| CCND1 | CCs | GAGGAAGAGGAGGAGGAGGA GAGATGGAAGGGGGAAAGAG | 236 | 595 |
| STC1 | CCs | TGATCAGTGCTTCTGCAACC GACGAATGCTTTTCCCTGAG | 242 | 6781 |
| PTGS2 | GCs | TGAGCATCTACGGTTTGCTG TGCTTGTCTGGAACAACTGC | 158 | 5743 |
| PRKX | GCs | CACGGGGCTCTTCTACTCTG CTACCAGCTTCTTGGCGAAC | 155 | 5613 |
| AHI1 | GCs | TTGGAACCCAGAAACAGGAG GGGCATCTTGACTTTGGTGT | 239 | 54,806 |
| IL11 | GCs | GCTGCACCTGACACTTGACT CACCCCTGCTCCTGAAATAA | 249 | 3859 |
| CAV1 | GCs | TCTCTACACCGTTCCCATCC CAATCTTGACCACGTCATCG | 164 | 857 |
| SFRP4 | GCs | GCCTGGGACAGCCTATGTAA TCTGTACCAAAGGGCAAACC | 160 | 6424 |
| GAPDH | CCs, GCs | TCAGCCGCATCTTCTTTTGC ACGACCAAATCCGTTGACTC | 90 | 2597 |
| ACTB | CCs, GCS | AAAGACCTGTACGCCAACAC CTCAGGAGGAGCAATGATCTTG | 132 | 60 |
| HPRT | CCs, GCs | TGGCGTCGTGATTAGTGATG ACATCTCGAGCAAGACGTTC | 141 | 3251 |
| Gene Symbol | Entrez Gene ID | Ratio 7 d/24 h | Ratio 15 d/24 h | Ratio 30 d/24 h | Adj. p-val. 7 d/24 h | Adj. p-val. 15 d/24 h | Adj. p-val. 30 d/24 h | Mean Ratio |
|---|---|---|---|---|---|---|---|---|
| CCs | ||||||||
| TGFBR3 | 7049 | −1.998 | −3.083 | −3.056 | 4.62 × 10−5 | 1.29 × 10−6 | 1.04 × 10−6 | −2.7125 |
| HMGB1 | 3146 | −2.054 | −2.261 | −2.526 | 6.96 × 10−6 | 2.04 × 10−6 | 6.84 × 10−7 | −2.2806 |
| ARF6 | 382 | −2.044 | −2.389 | −2.163 | 1.93 × 10−5 | 3.58 × 10−6 | 6.57 × 10−6 | −2.1993 |
| LOC100 996643 | 25902 | −2.042 | −2.005 | −2.167 | 1.04 × 10−5 | 8.36 × 10−6 | 3.38 × 10−6 | −2.0717 |
| CAV1 | 857 | 3.402 | 5.865 | 5.26 | 2.02 × 10−6 | 1.30 × 10−7 | 1.39 × 10−7 | 4.8427 |
| ANLN | 54443 | 5.724 | 5.910 | 3.1 | 4.27 × 10−7 | 2.57 × 10−7 | 4.09 × 10−6 | 4.912 |
| SFRP4 | 6424 | 22.184 | 28.775 | 27.230 | 5.34 × 10−8 | 1.93 × 10−8 | 1.44 × 10−8 | 26.0633 |
| ANXA3 | 306 | 34.272 | 30.545 | 33.593 | 5.51 × 10−9 | 3.16 × 10−9 | 1.57 × 10−9 | 32.8035 |
| DKK1 | 22943 | 34.811 | 30.492 | 58.943 | 5.58 × 10−9 | 3.43 × 10−9 | 9.21 × 10−10 | 41.4158 |
| CCND1 | 595 | 12.080 | 46.968 | 70.869 | 5.54 × 10−9 | 6.96 × 10−10 | 4.92 × 10−10 | 43.3059 |
| STC1 | 6781 | 14.962 | 60.004 | 58.346 | 5.58 × 10−9 | 6.96 × 10−10 | 4.92 × 10−10 | 44.4377 |
| GCs | ||||||||
| PTGS2 | 5743 | −22.653 | −23.248 | −27.601 | 9.56 × 10−4 | 8.93 × 10−4 | 6.17 × 10−4 | −24.5012 |
| PRKX | 5613 | −11.071 | −13.497 | −9.694 | 2.46 × 10−3 | 1.83 × 10−3 | 2.20 × 10−3 | −11.4212 |
| AHI1 | 54806 | −8.453 | −9.569 | −11.378 | 2.36 × 10−3 | 1.83 × 10−3 | 1.19 × 10−3 | −9.80055 |
| IL11 | 3589 | −6.124 | −7.771 | −7.132 | 2.11 × 10−3 | 1.35 × 10−3 | 1.19 × 10−3 | −7.00951 |
| ARF6 | 382 | −2.201 | −2.595 | −1.954 | 2.70 × 10−2 | 1.34 × 10−2 | 3.60 × 10−2 | −2.25027 |
| HMGB1 | 3146 | 1.889 | 2.106 | 1.967 | 4.38 × 10−2 | 2.45 × 10−2 | 2.96 × 10−2 | 1.98783 |
| ANXA3 | 306 | 13.404 | 19.427 | 21.684 | 2.57 × 10−2 | 1.52 × 10−2 | 1.21 × 10−2 | 18.1721 |
| ITGA3 | 3675 | 16.927 | 18.842 | 22.47 | 2.77 × 10−2 | 2.23 × 10−2 | 1.65 × 10−2 | 19.4133 |
| CAV1 | 857 | 22.409 | 32.071 | 34.618 | 1.95 × 10−3 | 1.33 × 10−3 | 9.44 × 10−4 | 29.6998 |
| ANLN | 54443 | 59.051 | 63.936 | 59.669 | 1.57 × 10−3 | 1.34 × 10−3 | 1.09 × 10−3 | 60.8857 |
| SFRP4 | 6424 | 141.911 | 89.682 | 101.05 | 5.14 × 10−3 | 6.31 × 10−3 | 5.01 × 10−3 | 110.8814 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chermuła, B.; Kranc, W.; Celichowski, P.; Stelmach, B.; Piotrowska-Kempisty, H.; Mozdziak, P.; Pawelczyk, L.; Spaczyński, R.Z.; Kempisty, B. Cellular Processes in Human Ovarian Follicles Are Regulated by Expression Profile of New Gene Markers—Clinical Approach. J. Clin. Med. 2022, 11, 73. https://doi.org/10.3390/jcm11010073
Chermuła B, Kranc W, Celichowski P, Stelmach B, Piotrowska-Kempisty H, Mozdziak P, Pawelczyk L, Spaczyński RZ, Kempisty B. Cellular Processes in Human Ovarian Follicles Are Regulated by Expression Profile of New Gene Markers—Clinical Approach. Journal of Clinical Medicine. 2022; 11(1):73. https://doi.org/10.3390/jcm11010073
Chicago/Turabian StyleChermuła, Błażej, Wiesława Kranc, Piotr Celichowski, Bogusława Stelmach, Hanna Piotrowska-Kempisty, Paul Mozdziak, Leszek Pawelczyk, Robert Zygmunt Spaczyński, and Bartosz Kempisty. 2022. "Cellular Processes in Human Ovarian Follicles Are Regulated by Expression Profile of New Gene Markers—Clinical Approach" Journal of Clinical Medicine 11, no. 1: 73. https://doi.org/10.3390/jcm11010073
APA StyleChermuła, B., Kranc, W., Celichowski, P., Stelmach, B., Piotrowska-Kempisty, H., Mozdziak, P., Pawelczyk, L., Spaczyński, R. Z., & Kempisty, B. (2022). Cellular Processes in Human Ovarian Follicles Are Regulated by Expression Profile of New Gene Markers—Clinical Approach. Journal of Clinical Medicine, 11(1), 73. https://doi.org/10.3390/jcm11010073








