Quality Assessment of Untargeted Analytical Data in a Large-Scale Metabolomic Study
Abstract
1. Introduction
2. Materials and Methods
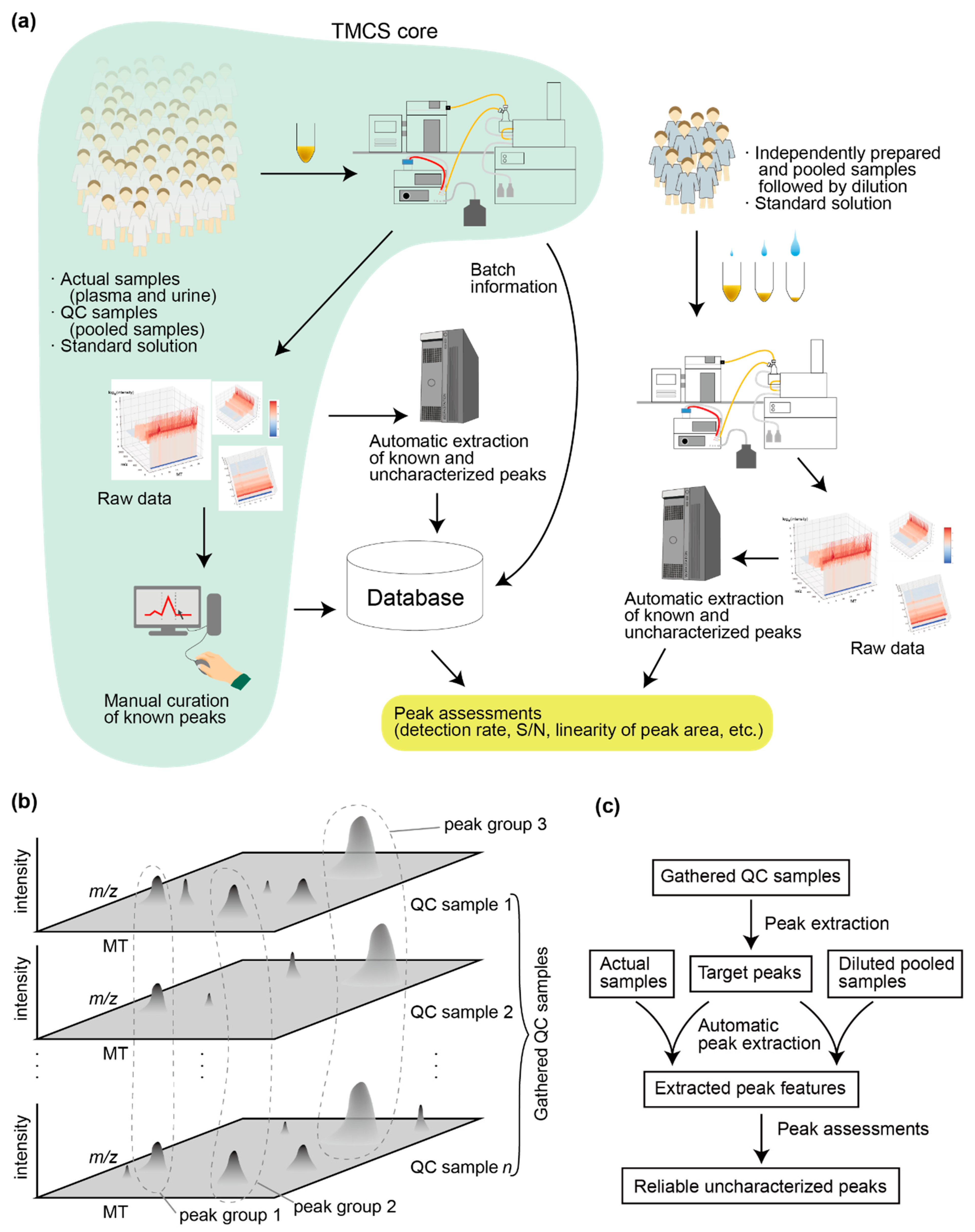
2.1. Data Collection and Computational Analyses Pipeline
2.2. Determining the Target Peaks
2.3. Automatic Peak Extraction
2.4. Dilution of Independently Prepared and Pooled Samples
2.5. Clustering Peak Fragments
3. Results
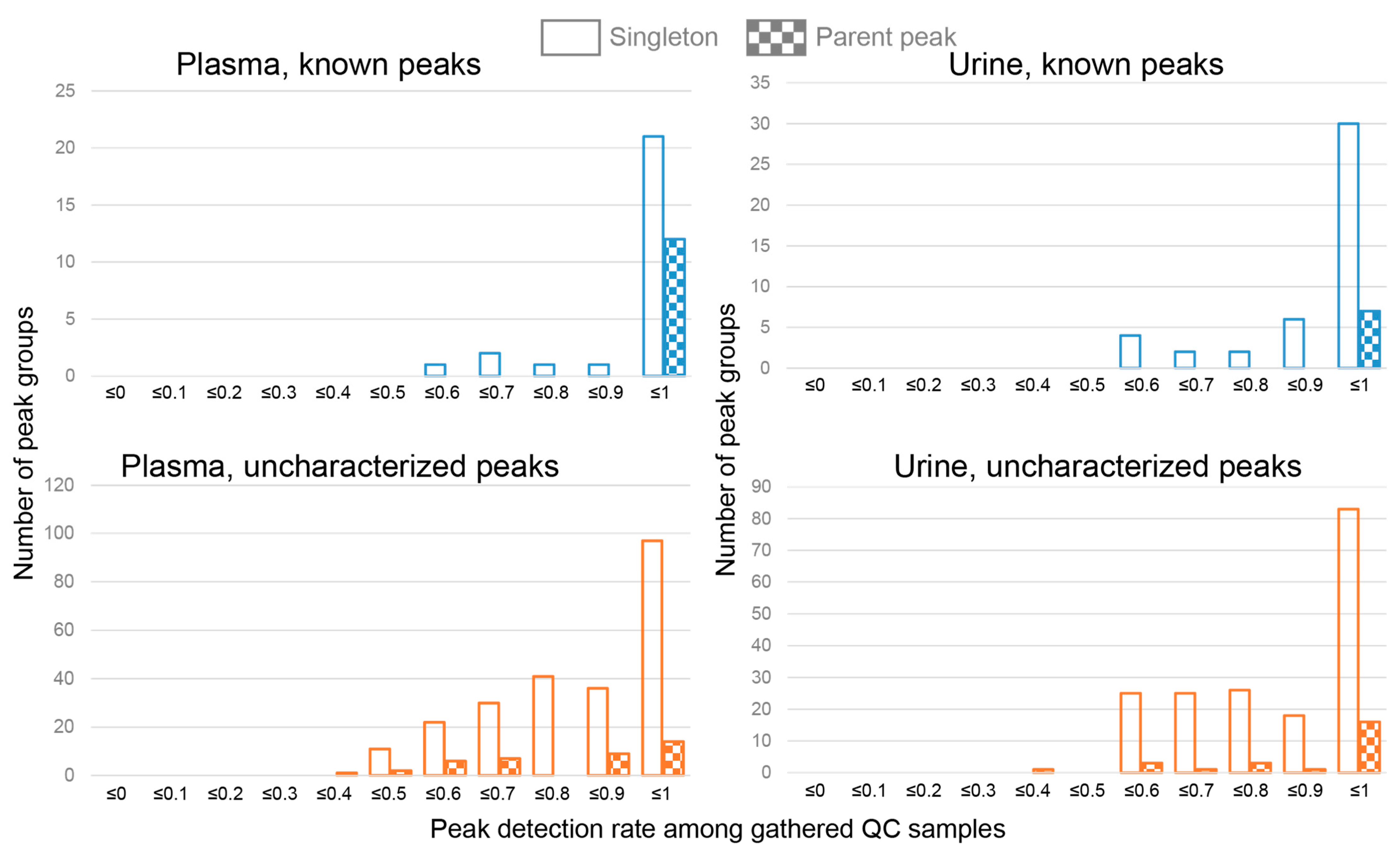
3.1. Defining Target Peaks Using the Gathered QC Samples
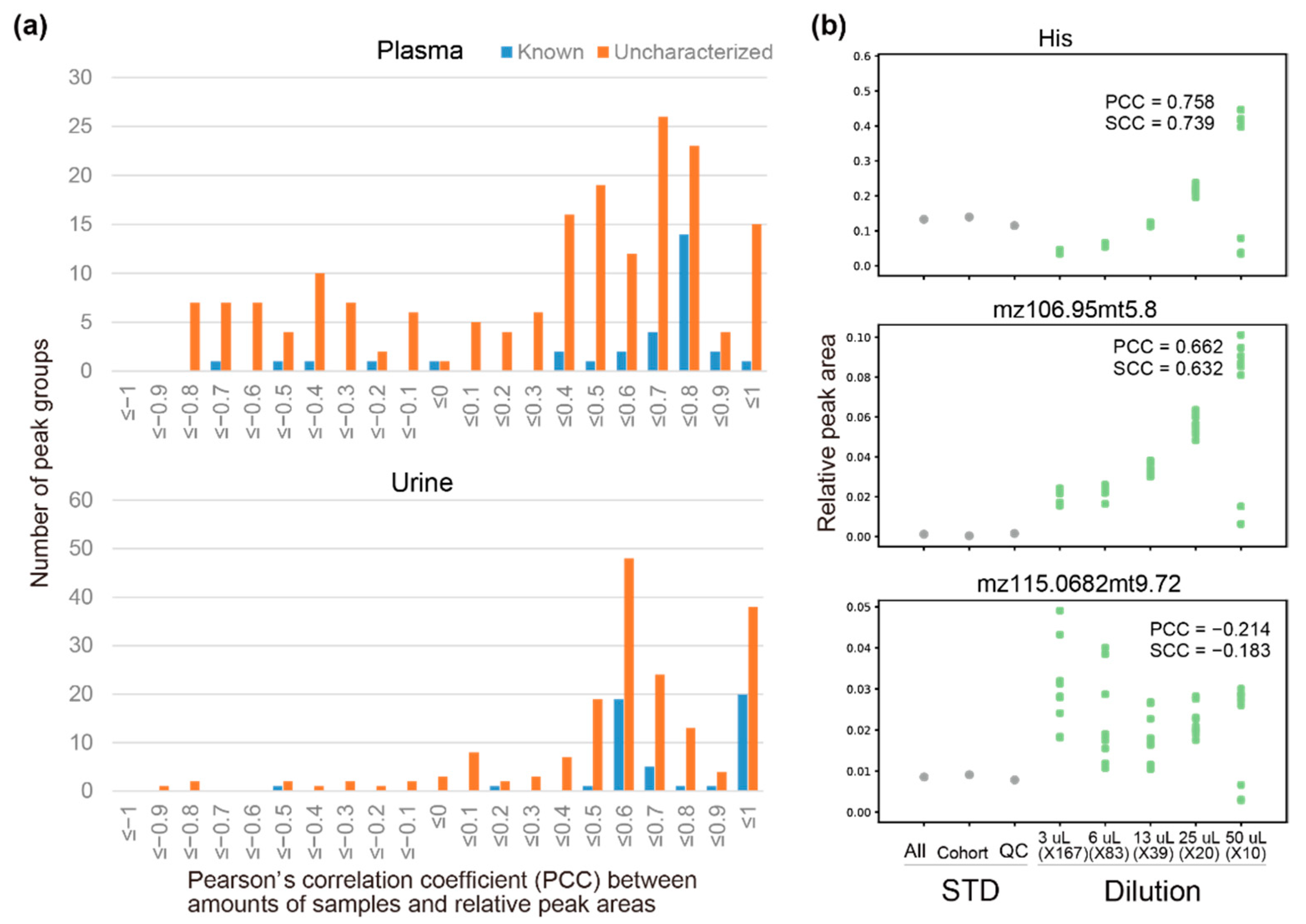
3.2. Dilution of Independently Prepared Pooled Samples
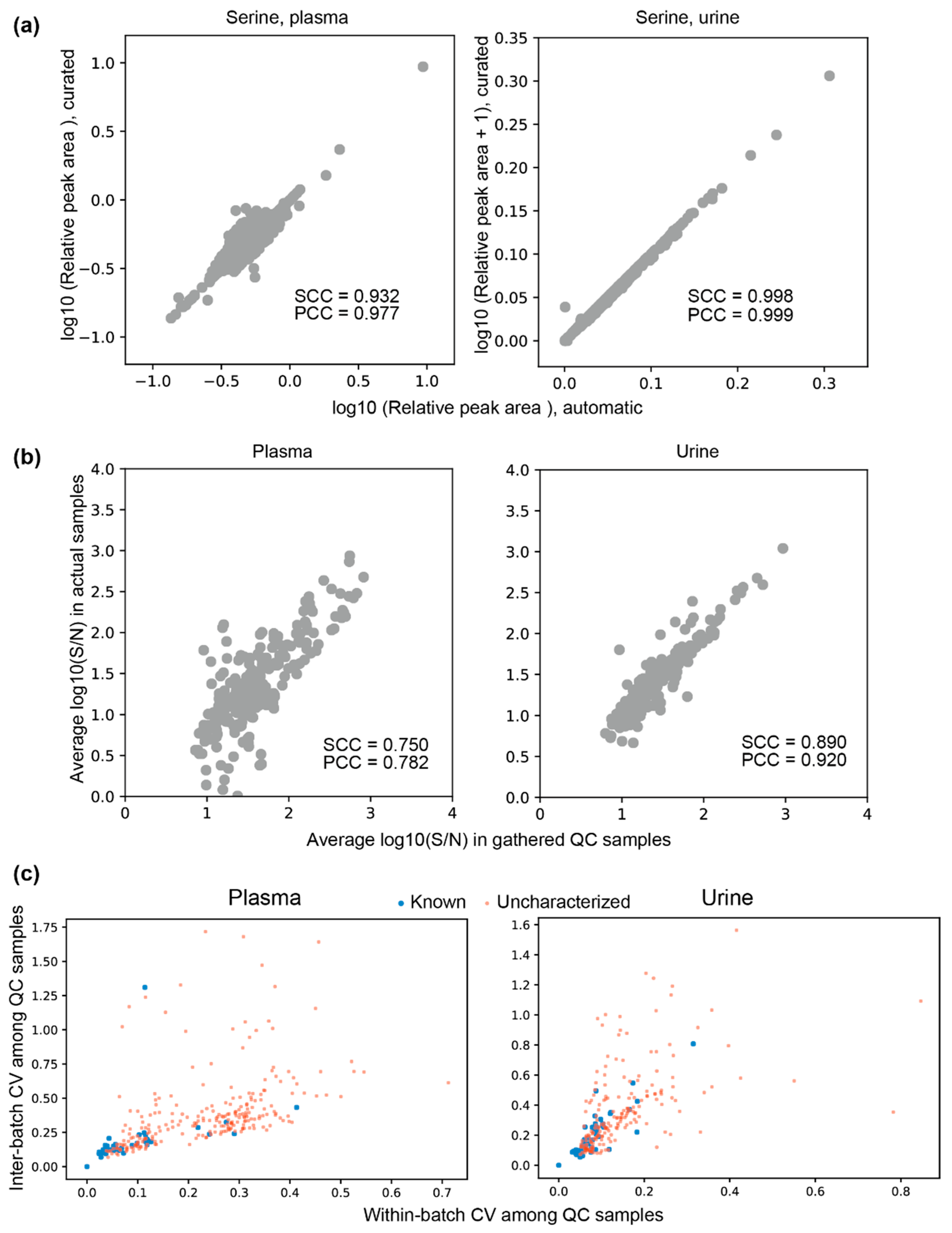
3.3. Automatic Peak Extraction from Actual Samples
3.4. Clustering Peaks Based on Inferred Origin Metabolites
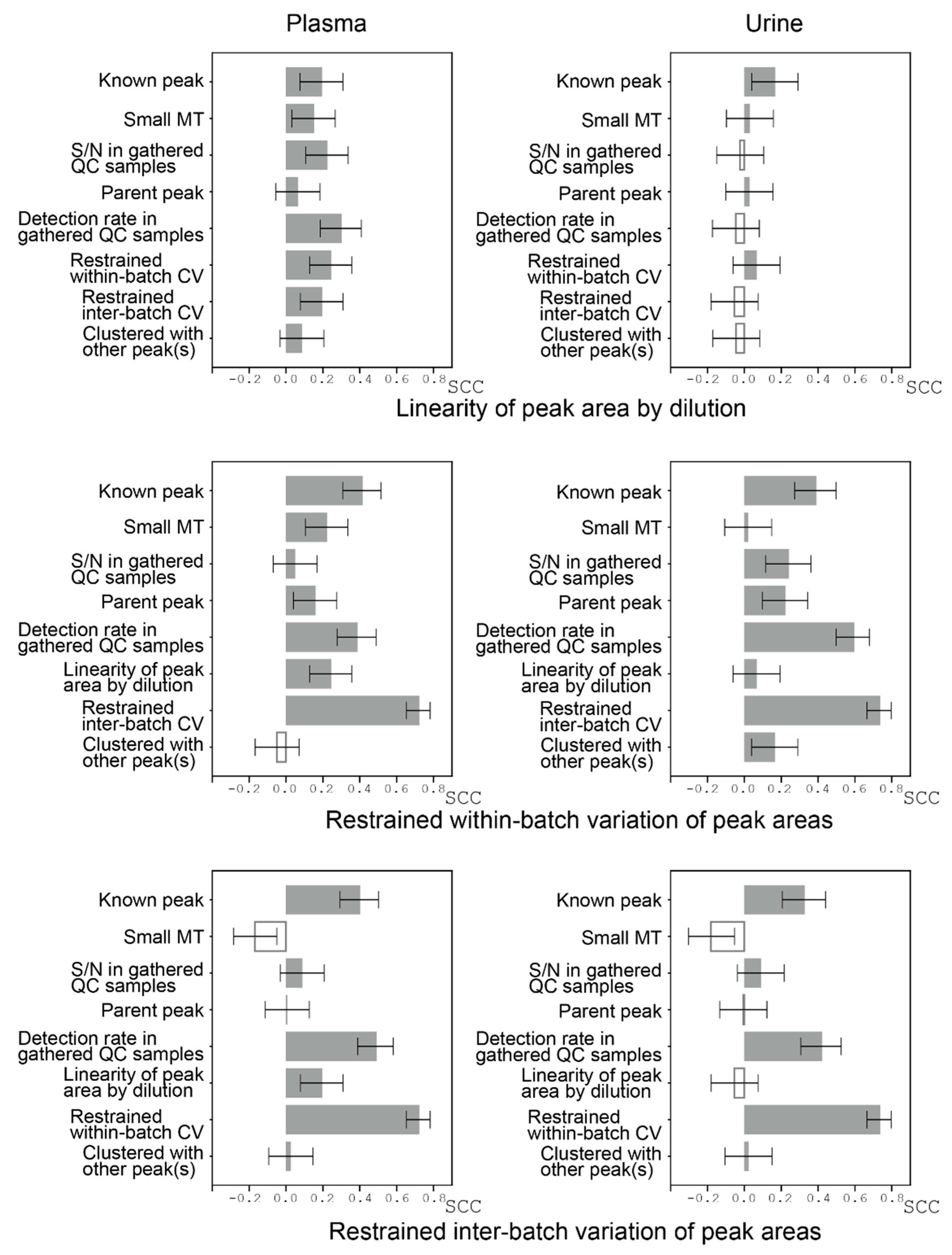
3.5. Peak Reliabilities and Their Relevant Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Yu, B.; Zanetti, K.A.; Temprosa, M.; Albanes, D.; Appel, N.; Barrera, C.B.; Ben-Shlomo, Y.; Boerwinkle, E.; Casas, J.P.; Clish, C.; et al. The Consortium of Metabolomics Studies (COMETS): Metabolomics in 47 Prospective Cohort Studies. Am. J. Epidemiol. 2019, 188, 991–1012. [Google Scholar] [CrossRef]
- Harada, S.; Hirayama, A.; Chan, Q.; Kurihara, A.; Fukai, K.; Iida, M.; Kato, S.; Sugiyama, D.; Kuwabara, K.; Takeuchi, A.; et al. Reliability of plasma polar metabolite concentrations in a large-scale cohort study using capillary electrophoresis-mass spectrometry. PLoS ONE 2018, 13, e0191230. [Google Scholar] [CrossRef]
- Saigusa, D.; Okamura, Y.; Motoike, I.N.; Katoh, Y.; Kurosawa, Y.; Saijyo, R.; Koshiba, S.; Yasuda, J.; Motohashi, H.; Sugawara, J.; et al. Establishment of Protocols for Global Metabolomics by LC-MS for Biomarker Discovery. PLoS ONE 2016, 11, e0160555. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Hirayama, A.; Nakashima, E.; Sugimoto, M.; Akiyama, S.; Sato, W.; Maruyama, S.; Matsuo, S.; Tomita, M.; Yuzawa, Y.; Soga, T. Metabolic profiling reveals new serum biomarkers for differentiating diabetic nephropathy. Anal. Bioanal. Chem. 2012, 404, 3101–3109. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Muelas, M.W.; Roberts, I.; Mughal, F.; O’Hagan, S.; Day, P.J.; Kell, D.B. An untargeted metabolomics strategy to measure differences in metabolite uptake and excretion by mammalian cell lines. Metabolomics 2020, 16, 107. [Google Scholar] [CrossRef]
- Kawamura, N.; Shinoda, K.; Sato, H.; Sasaki, K.; Suzuki, M.; Yamaki, K.; Fujimori, T.; Yamamoto, H.; Osei-Hyiaman, D.; Ohashi, Y. Plasma metabolome analysis of patients with major depressive disorder. Psychiatry Clin. Neurosci. 2018, 72, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Chong, E.Y.; Huang, Y.; Wu, H.; Ghasemzadeh, N.; Uppal, K.; Quyyumi, A.A.; Jones, D.P.; Yu, T. Local false discovery rate estimation using feature reliability in LC/MS metabolomics data. Sci. Rep. 2015, 5, 17221. [Google Scholar] [CrossRef]
- Katajamaa, M.; Oresic, M. Data processing for mass spectrometry-based metabolomics. J. Chromatogr. A 2007, 1158, 318–328. [Google Scholar] [CrossRef]
- Han, W.; Li, L. Evaluating and minimizing batch effects in metabolomics. Mass Spectrom. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wehrens, R.; Hageman, J.A.; van Eeuwijk, F.; Kooke, R.; Flood, P.J.; Wijnker, E.; Keurentjes, J.J.; Lommen, A.; van Eekelen, H.D.; Hall, R.D.; et al. Improved batch correction in untargeted MS-based metabolomics. Metabolomics 2016, 12, 88. [Google Scholar] [CrossRef]
- Phapale, P.; Rai, V.; Mohanty, A.K.; Srivastava, S. Untargeted Metabolomics Workshop Report: Quality Control Considerations from Sample Preparation to Data Analysis. J. Am. Soc. Mass Spectrom. 2020, 31, 2006–2010. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.; Ventura, D.; Prince, J.T. LC-MS alignment in theory and practice: A comprehensive algorithmic review. Brief. Bioinform. 2015, 16, 104–117. [Google Scholar] [CrossRef]
- Zhang, J.; Gonzalez, E.; Hestilow, T.; Haskins, W.; Huang, Y. Review of peak detection algorithms in liquid-chromatography-mass spectrometry. Curr. Genom. 2009, 10, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Almenara, X.; Montenegro-Burke, J.R.; Benton, H.P.; Siuzdak, G. Annotation: A Computational Solution for Streamlining Metabolomics Analysis. Anal. Chem. 2018, 90, 480–489. [Google Scholar] [CrossRef]
- Vaca Jacome, A.S.; Peckner, R.; Shulman, N.; Krug, K.; DeRuff, K.C.; Officer, A.; Christianson, K.E.; MacLean, B.; MacCoss, M.J.; Carr, S.A.; et al. Avant-garde: An automated data-driven DIA data curation tool. Nat. Methods 2020, 17, 1237–1244. [Google Scholar] [CrossRef]
- Dunn, W.B.; Wilson, I.D.; Nicholls, A.W.; Broadhurst, D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis 2012, 4, 2249–2264. [Google Scholar] [CrossRef]
- Harada, S.; Takebayashi, T.; Kurihara, A.; Akiyama, M.; Suzuki, A.; Hatakeyama, Y.; Sugiyama, D.; Kuwabara, K.; Takeuchi, A.; Okamura, T.; et al. Metabolomic profiling reveals novel biomarkers of alcohol intake and alcohol-induced liver injury in community-dwelling men. Environ. Health Prev. Med. 2016, 21, 18–26. [Google Scholar] [CrossRef]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef]
- Soga, T.; Igarashi, K.; Ito, C.; Mizobuchi, K.; Zimmermann, H.P.; Tomita, M. Metabolomic profiling of anionic metabolites by capillary electrophoresis mass spectrometry. Anal. Chem. 2009, 81, 6165–6174. [Google Scholar] [CrossRef]
- Udo, R.; Katsumata, K.; Kuwabara, H.; Enomoto, M.; Ishizaki, T.; Sunamura, M.; Nagakawa, Y.; Soya, R.; Sugimoto, M.; Tsuchida, A. Urinary charged metabolite profiling of colorectal cancer using capillary electrophoresis-mass spectrometry. Sci. Rep. 2020, 10, 21057. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Ryabova, A.; Cornette, R.; Cherkasov, A.; Watanabe, M.; Okuda, T.; Shagimardanova, E.; Kikawada, T.; Gusev, O. Combined metabolome and transcriptome analysis reveals key components of complete desiccation tolerance in an anhydrobiotic insect. Proc. Natl. Acad. Sci. USA 2020, 117, 19209–19220. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.Z.; Fung, W.W.; Ng, J.K.; Lai, K.B.; Luk, C.C.; Chow, K.M.; Li, P.K.; Szeto, C.C. Metabolomic Changes of Human Proximal Tubular Cell Line in High Glucose Environment. Sci. Rep. 2019, 9, 16617. [Google Scholar] [CrossRef] [PubMed]
- Reijeng, J.C.; Martens, J.H.; Giuliani, A.; Chiari, M. Pherogram normalization in capillary electrophoresis and micellar electrokinetic chromatography analyses in cases of sample matrix-induced migration time shifts. J. Chromatogr. B 2002, 770, 45–51. [Google Scholar] [CrossRef]
- Baran, R.; Kochi, H.; Saito, N.; Suematsu, M.; Soga, T.; Nishioka, T.; Robert, M.; Tomita, M. MathDAMP: A package for differential analysis of metabolite profiles. BMC Bioinform. 2006, 7, 530. [Google Scholar] [CrossRef]
- Soga, T.; Baran, R.; Suematsu, M.; Ueno, Y.; Ikeda, S.; Sakurakawa, T.; Kakazu, Y.; Ishikawa, T.; Robert, M.; Nishioka, T.; et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 2006, 281, 16768–16776. [Google Scholar] [CrossRef]
- Sugimoto, M.; Hirayama, A.; Ishikawa, T.; Robert, M.; Baran, R.; Uehara, K.; Kawai, K.; Soga, T.; Tomita, M. Differential metabolomics software for capillary electrophoresis-mass spectrometry data analysis. Metabolomics 2010, 6, 27–41. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Bottcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [PubMed]
- Beckers, J.L. Peak deformation in cationic analysis caused by system zones. Electrophoresis 2001, 22, 2684–2690. [Google Scholar] [CrossRef]
- Drouin, N.; van Mever, M.; Zhang, W.; Tobolkina, E.; Ferre, S.; Servais, A.C.; Gou, M.J.; Nyssen, L.; Fillet, M.; Lageveen-Kammeijer, G.S.M.; et al. Capillary Electrophoresis-Mass Spectrometry at Trial by Metabo-Ring: Effective Electrophoretic Mobility for Reproducible and Robust Compound Annotation. Anal. Chem. 2020, 92, 14103–14112. [Google Scholar] [CrossRef]
- Iida, M.; Harada, S.; Kurihara, A.; Fukai, K.; Kuwabara, K.; Sugiyama, D.; Takeuchi, A.; Okamura, T.; Akiyama, M.; Nishiwaki, Y.; et al. Profiling of plasma metabolites in postmenopausal women with metabolic syndrome. Menopause 2016, 23, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Fukai, K.; Harada, S.; Iida, M.; Kurihara, A.; Takeuchi, A.; Kuwabara, K.; Sugiyama, D.; Okamura, T.; Akiyama, M.; Nishiwaki, Y.; et al. Metabolic Profiling of Total Physical Activity and Sedentary Behavior in Community-Dwelling Men. PLoS ONE 2016, 11, e0164877. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, K.; Harada, S.; Takeuchi, A.; Kurihara, A.; Iida, M.; Fukai, K.; Kuwabara, K.; Kato, S.; Matsumoto, M.; Hirata, A.; et al. Association between dyslipidemia and plasma levels of branched-chain amino acids in the Japanese population without diabetes mellitus. J. Clin. Lipidol. 2019, 13, 932–939.e932. [Google Scholar] [CrossRef] [PubMed]
- Koshiba, S.; Motoike, I.N.; Saigusa, D.; Inoue, J.; Aoki, Y.; Tadaka, S.; Shirota, M.; Katsuoka, F.; Tamiya, G.; Minegishi, N.; et al. Identification of critical genetic variants associated with metabolic phenotypes of the Japanese population. Commun. Biol. 2020, 3, 662. [Google Scholar] [CrossRef] [PubMed]
- Izumi, Y.; Matsuda, F.; Hirayama, A.; Ikeda, K.; Kita, Y.; Horie, K.; Saigusa, D.; Saito, K.; Sawada, Y.; Nakanishi, H.; et al. Inter-Laboratory Comparison of Metabolite Measurements for Metabolomics Data Integration. Metabolites 2019, 9, 257. [Google Scholar] [CrossRef] [PubMed]
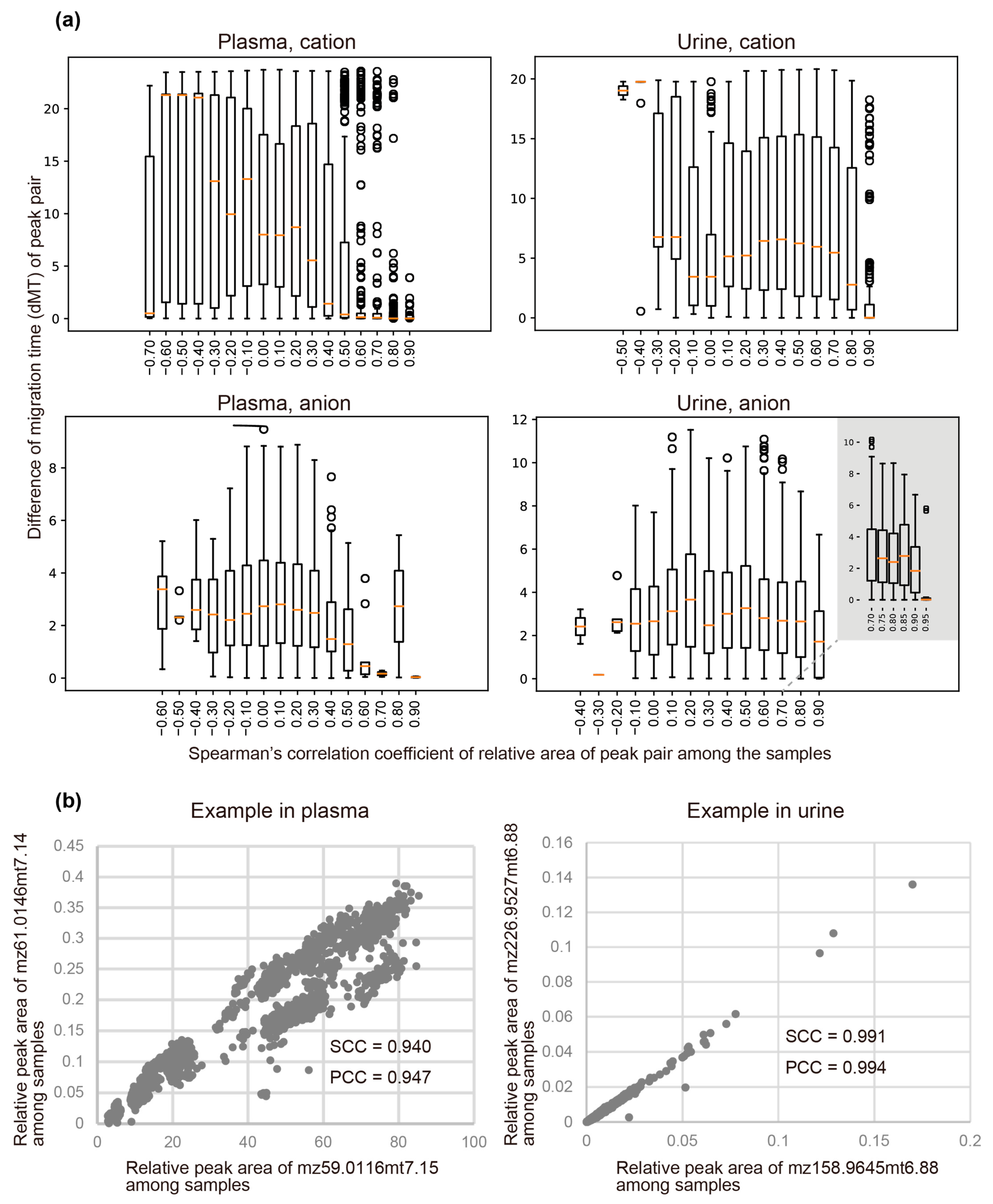
| Plasma | SCC | PCC | Urine | SCC | PCC |
|---|---|---|---|---|---|
| o-Acetylcarnitine | 0.978 | 0.978 | Carnitine | 1.000 | 1.000 |
| Tyr | 0.967 | 0.973 | Proline betaine | 0.999 | 1.000 |
| Pro | 0.966 | 0.976 | Lys | 0.999 | 0.999 |
| Trp | 0.957 | 0.963 | Gly | 0.999 | 1.000 |
| Choline | 0.949 | 0.951 | Guanidinoacetate | 0.999 | 0.999 |
| Ornithine | 0.948 | 0.980 | o-Acetylcarnitine | 0.999 | 0.999 |
| Carnitine | 0.944 | 0.953 | His | 0.999 | 0.999 |
| Citrulline | 0.944 | 0.959 | 1-Methylnicotinamide | 0.998 | 0.998 |
| Gln | 0.932 | 0.941 | Gln | 0.998 | 0.999 |
| 3-Aminopyrrolidine (*) | 0.932 | 0.905 | Ser | 0.998 | 0.999 |
| Ser | 0.932 | 0.977 | Hypoxanthine | 0.997 | 0.998 |
| Cystine | 0.924 | 0.927 | Choline | 0.997 | 0.999 |
| His | 0.920 | 0.963 | N6, N6, N6-Trimethyllysine | 0.996 | 0.998 |
| Phe | 0.915 | 0.933 | Trp | 0.996 | 0.998 |
| Gly | 0.910 | 0.908 | Tyr | 0.996 | 0.998 |
| Lys | 0.906 | 0.917 | Guanidinosuccinate | 0.995 | 0.997 |
| Malate | 0.867 | 0.103 | Hippurate | 0.994 | 0.981 |
| Met | 0.863 | 0.870 | 1-Methyladenosine | 0.994 | 0.995 |
| 5-Oxoproline | 0.849 | 0.145 | Cystine | 0.992 | 0.999 |
| Succinate | 0.849 | 0.108 | Homovanillate | 0.986 | 0.983 |
| Guanidinoacetate | 0.712 | 0.267 | 3-Indoxyl sulfate | 0.979 | 0.972 |
| Taurine | 0.585 | 0.364 | Quinate | 0.977 | 0.985 |
| Asp | 0.533 | 0.972 | Taurine | 0.976 | 0.986 |
| Proline betaine | 0.234 | 0.634 | Pimelate | 0.971 | 0.977 |
| Phe–Phe | 0.021 | 0.121 | 3-Aminopyrrolidine (*) | 0.969 | 0.989 |
| 2-Hydroxypentanoate | −0.099 | 0.023 | 2-Hydroxypentanoate | 0.963 | 0.970 |
| Hypoxanthine | −0.194 | −0.064 | Succinate | 0.956 | 0.971 |
| N-Acetylaspartate | 0.956 | 0.958 | |||
| Uridine | 0.939 | 0.905 | |||
| 4-Pyridoxate | 0.930 | 0.983 | |||
| 5-Oxoproline | 0.916 | 0.934 | |||
| N-Acetylneuraminate | 0.913 | 0.917 | |||
| Isethionate | 0.911 | 0.922 | |||
| Threonate | 0.896 | 0.899 | |||
| 3-Hydroxy-3-methylglutarate | 0.856 | 0.866 | |||
| Gluconate | 0.772 | 0.729 | |||
| Pro | 0.062 | −0.008 |
| Filtering Order | Filtering Condition | Plasma | Urine |
|---|---|---|---|
| 1 | Number of peaks with S/N > 5 in the gathered QC samples | 340 | 282 |
| 2 | Number of peaks whose MTs and m/z’s do not match with those of known metabolites | 276 | 202 |
| 3 | Peaks that are detected in independently prepared QC samples (S/N > 5) | 181 | 180 |
| 4 | Pearson’s correlation coefficient (PCC) between amounts of samples and relative peak areas is above 0.3 | 115 | 153 |
| 5 | Peaks that are not clustered with known peaks based on the differences in the migration times (dMTs) and the correlation between peak areas among the samples | 99 | 145 |
| 6 | Inferred fragment clusters based on the differences in the migration times (dMTs) and the correlation between peak areas among the samples | 53 | 114 |
| 7 | S/N of peak < 5 in the standard solution (labeled as All STD) which contains 278 and 240 standard compounds for cation and anion, respectively | 35 | 74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito, R.; Sugimoto, M.; Hirayama, A.; Soga, T.; Tomita, M.; Takebayashi, T. Quality Assessment of Untargeted Analytical Data in a Large-Scale Metabolomic Study. J. Clin. Med. 2021, 10, 1826. https://doi.org/10.3390/jcm10091826
Saito R, Sugimoto M, Hirayama A, Soga T, Tomita M, Takebayashi T. Quality Assessment of Untargeted Analytical Data in a Large-Scale Metabolomic Study. Journal of Clinical Medicine. 2021; 10(9):1826. https://doi.org/10.3390/jcm10091826
Chicago/Turabian StyleSaito, Rintaro, Masahiro Sugimoto, Akiyoshi Hirayama, Tomoyoshi Soga, Masaru Tomita, and Toru Takebayashi. 2021. "Quality Assessment of Untargeted Analytical Data in a Large-Scale Metabolomic Study" Journal of Clinical Medicine 10, no. 9: 1826. https://doi.org/10.3390/jcm10091826
APA StyleSaito, R., Sugimoto, M., Hirayama, A., Soga, T., Tomita, M., & Takebayashi, T. (2021). Quality Assessment of Untargeted Analytical Data in a Large-Scale Metabolomic Study. Journal of Clinical Medicine, 10(9), 1826. https://doi.org/10.3390/jcm10091826









