Fluorescence Approaches for Characterizing Ion Channels in Synthetic Bilayers
Abstract
:1. Introduction
2. Lipid and Membrane Bilayers
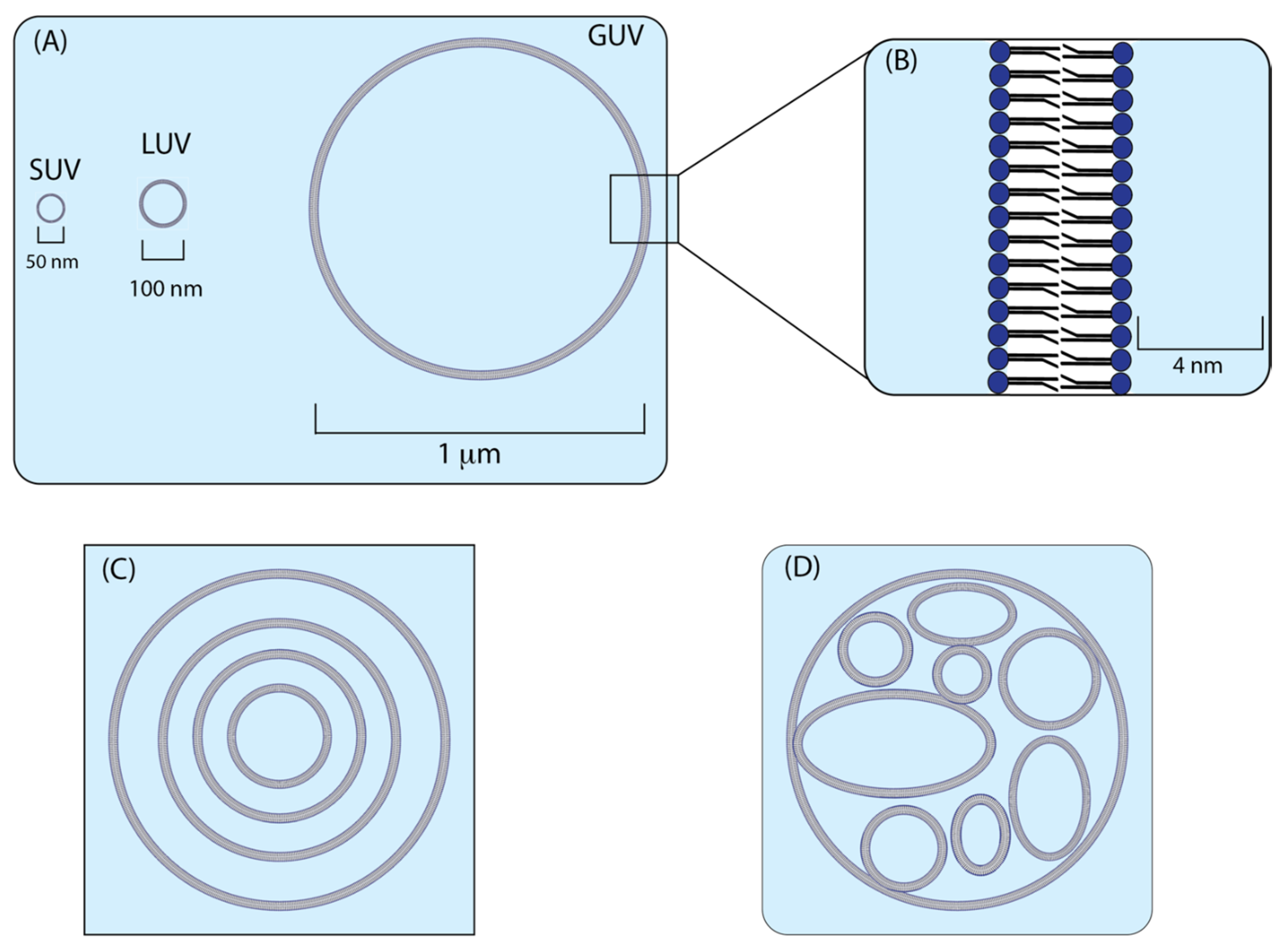
2.1. Liposomes
2.2. Synthesis of Liposomes
2.3. Microscopy
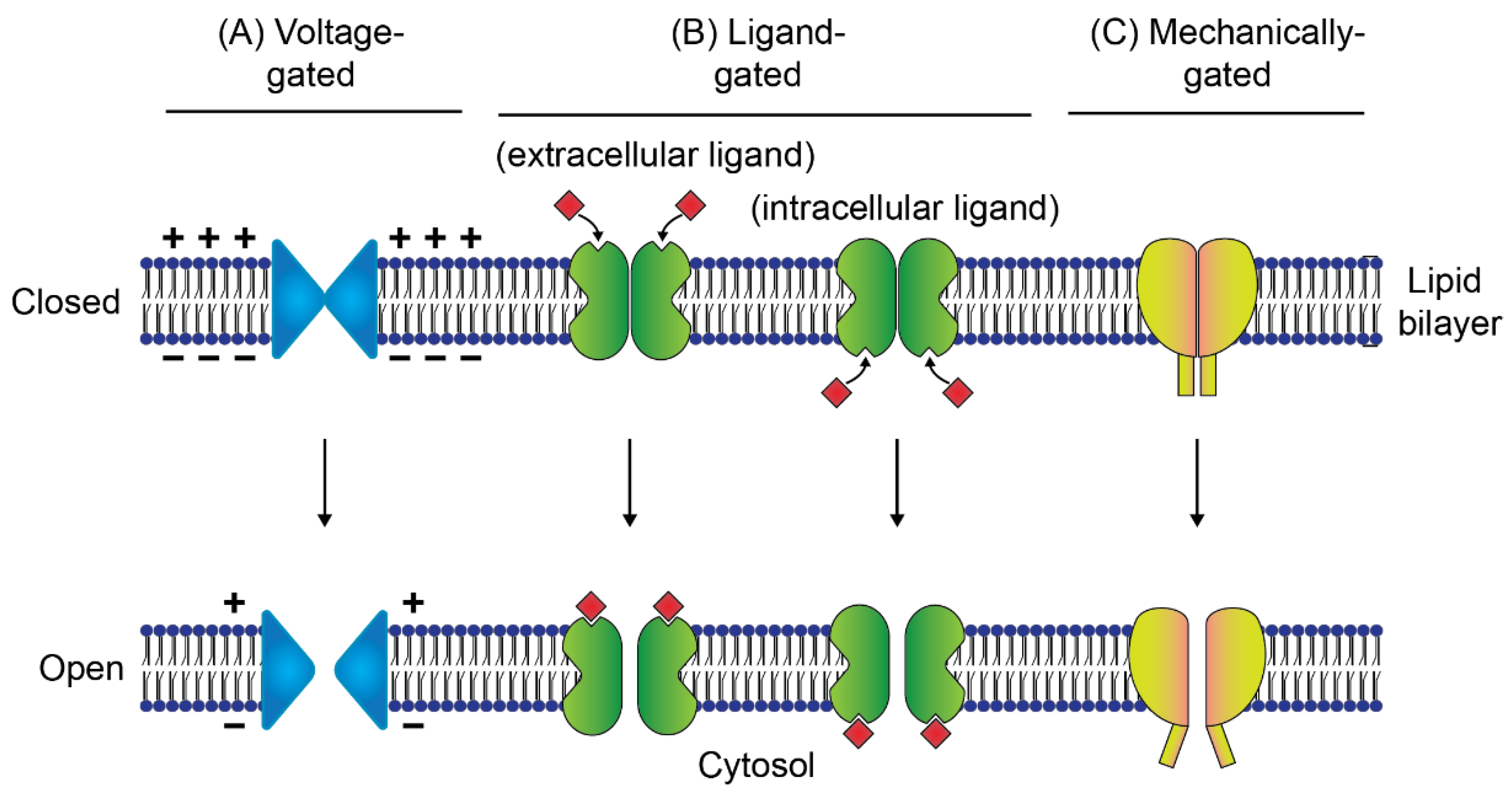
3. Ion Channels
4. Study of Ion Channels in Synthetic Liposomes
4.1. Integrating Membrane Proteins into Liposomes
4.1.1. Direct Reconstitution
4.1.2. Dehydration–Rehydration
4.1.3. Induced Fusion
4.1.4. Microfluidic Jetting
4.2. Integrating Membrane Proteins into Nanodiscs
4.3. Integrating Membrane Proteins in Cell-Free Systems
4.4. Integrating Membrane Proteins with Detergent Alternatives
5. Techniques to Characterize Ion Channels in Liposomes
5.1. Fluorescence Assays for Characterizing Ion Channels
5.1.1. Membrane-Permeable Dye-Based Assays
5.1.2. Membrane-Impermeable Dye-Based Assays
5.1.3. Lipid-Coupled Dye-Based Assays
5.2. Non-Fluorescent Assays for Characterizing Rhodopsins
6. Limitation of Fluorescent Assays for Characterizing Ion Channels
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gadsby, D.C. Ion Channels versus Ion Pumps: The Principal Difference, in Principle. Nat. Rev. Mol. Cell Biol. 2009, 10, 344–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolphin, A.C.; Insel, P.A.; Blaschke, T.F.; Meyer, U.A. Introduction to the Theme “Ion Channels and Neuropharmacology: From the Past to the Future”. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Ion Channels and the Electrical Properties of Membranes. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Lee, A.; Fakler, B.; Kaczmarek, L.K.; Isom, L.L. More Than a Pore: Ion Channel Signaling Complexes. J. Neurosci. 2014, 34, 15159–15169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, D. Ion Channels in Nerve and Muscle Cells. Curr. Opin. Cell Biol. 1990, 2, 695–707. [Google Scholar] [CrossRef]
- Littleton, J.T.; Ganetzky, B. Ion Channels and Synaptic Organization: Analysis of the Drosophila Genome. Neuron 2000, 26, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A Comprehensive Map of Molecular Drug Targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, C.J.; Colussi, P.; Clark, T.G. Ion Channels as Therapeutic Antibody Targets. In MAbs; Taylor & Francis: Abingdon, UK, 2019; Volume 11, pp. 265–296. [Google Scholar] [CrossRef]
- Charlton, F.W.; Pearson, H.M.; Hover, S.; Lippiat, J.D.; Fontana, J.; Barr, J.N.; Mankouri, J. Ion Channels as Therapeutic Targets for Viral Infections: Further Discoveries and Future Perspectives. Viruses 2020, 12, 844. [Google Scholar] [CrossRef] [PubMed]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane Lipids: Where They Are and How They Behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- van Meer, G.; de Kroon, A.I.P.M. Lipid Map of the Mammalian Cell. J. Cell Sci. 2011, 124, 5–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, A.L.; Reddy, T.; Koldsø, H.; Hélie, J.; Fowler, P.W.; Chavent, M.; Sansom, M.S.P. Protein Crowding and Lipid Complexity Influence the Nanoscale Dynamic Organization of Ion Channels in Cell Membranes. Sci. Rep. 2017, 7, 16647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booth, P.J.; Templer, R.H.; Meijberg, W.; Allen, S.J.; Curran, A.R.; Lorch, M. In Vitro Studies of Membrane Protein Folding. Crit. Rev. Biochem. Mol. Biol. 2001, 36, 501–603. [Google Scholar] [CrossRef]
- Yu, H.; Li, M.; Wang, W.; Wang, X. High Throughput Screening Technologies for Ion Channels. Acta Pharmacol. Sin. 2016, 37, 34–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, N.J.; Booth, P.J. Folding and Stability of Membrane Transport Proteins In Vitro. Biochim. Biophys. Acta (BBA)-Biomembr. 2012, 1818, 1055–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane Proteins, Lipids and Detergents: Not Just a Soap Opera. Biochim. Biophys. Acta (BBA)-Biomembr. 2004, 1666, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsia, C.-Y.; Richards, M.J.; Daniel, S. A Review of Traditional and Emerging Methods to Characterize Lipid–Protein Interactions in Biological Membranes. Anal. Methods 2015, 7, 7076–7094. [Google Scholar] [CrossRef]
- Kyrychenko, A. Using Fluorescence for Studies of Biological Membranes: A Review. Methods Appl. Fluoresc. 2015, 3, 042003. [Google Scholar] [CrossRef] [PubMed]
- Terai, T.; Nagano, T. Small-Molecule Fluorophores and Fluorescent Probes for Bioimaging. Pflügers Arch.-Eur. J. Physiol. 2013, 465, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Hu, Y.; Yoon, J. Fluorescent Probes and Bioimaging: Alkali Metals, Alkaline Earth Metals and PH. Chem. Soc. Rev. 2015, 44, 4619–4644. [Google Scholar] [CrossRef]
- Diekmann, Y.; Pereira-Leal, J.B. Evolution of Intracellular Compartmentalization. Biochem. J. 2013, 449, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A Comprehensive Classification System for Lipids. Eur. J. Lipid Sci. Technol. 2005, 107, 337–364. [Google Scholar] [CrossRef]
- Ulrich, A.S. Biophysical Aspects of Using Liposomes as Delivery Vehicles. Biosci. Rep. 2002, 22, 129–150. [Google Scholar] [CrossRef] [PubMed]
- Gruner, S.M.; Lenk, R.P.; Janoff, A.S.; Ostro, N.J. Novel Multilayered Lipid Vesicles: Comparison of Physical Characteristics of Multilamellar Liposomes and Stable Plurilamellar Vesicles. Biochemistry 1985, 24, 2833–2842. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, Preparation, and Applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. MRNA Vaccines—A New Era in Vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Swaay, D.; DeMello, A. Microfluidic Methods for Forming Liposomes. Lab Chip 2013, 13, 752–767. [Google Scholar] [CrossRef]
- Alavi, M.; Karimi, N.; Safaei, M. Application of Various Types of Liposomes in Drug Delivery Systems. Adv. Pharm. Bull. 2017, 7, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jacobs, R.E.; White, S.H. Preparation of Multilamellar Vesicles of Defined Size-Distribution by Solvent-Spherule Evaporation. Biochim. Biophys. Acta 1985, 812, 793–801. [Google Scholar] [CrossRef]
- Payne, N.I.; Timmins, P.; Ambrose, C.V.; Ward, M.D.; Ridgway, F. Proliposomes: A Novel Solution to an Old Problem. J. Pharm. Sci. 1986, 75, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Angelova, M.I.; Dimitrov, D.S. Liposome Electroformation. Faraday Discuss. Chem. Soc. 1986, 81, 303–311. [Google Scholar] [CrossRef]
- Peruzzi, J.; Gutierrez, M.G.; Mansfield, K.; Malmstadt, N. Dynamics of Hydrogel-Assisted Giant Unilamellar Vesicle Formation from Unsaturated Lipid Systems. Langmuir 2016, 32, 12702–12709. [Google Scholar] [CrossRef]
- Hotani, H.; Nomura, F.; Suzuki, Y. Giant Liposomes: From Membrane Dynamics to Cell Morphogenesis. Curr. Opin. Colloid Interface Sci. 1999, 4, 358–368. [Google Scholar] [CrossRef]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Production of Unilamellar Vesicles Using an Inverted Emulsion. Langmuir 2003, 19, 2870–2879. [Google Scholar] [CrossRef]
- Hargreaves, W.R.; Deamer, D.W. Liposomes from Ionic, Single-Chain Amphiphiles. Biochemistry 1978, 17, 3759–3768. [Google Scholar] [CrossRef]
- Bibi, S.; Kaur, R.; Henriksen-Lacey, M.; McNeil, S.E.; Wilkhu, J.; Lattmann, E.; Christensen, D.; Mohammed, A.R.; Perrie, Y. Microscopy Imaging of Liposomes: From Coverslips to Environmental SEM. Int. J. Pharm. 2011, 417, 138–150. [Google Scholar] [CrossRef]
- Baxa, U. Imaging of Liposomes by Transmission Electron Microscopy. In Characterization of Nanoparticles Intended for Drug Delivery; McNeil, S.E., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 73–88. ISBN 978-1-4939-7352-1. [Google Scholar]
- Talmon, Y. Staining and Drying-Induced Artifacts in Electron Microscopy of Surfactant Dispersions. J. Colloid Interface Sci. 1983, 93, 366–382. [Google Scholar] [CrossRef]
- Tonggu, L.; Wang, L. Cryo-EM Sample Preparation Method for Extremely Low Concentration Liposomes. Ultramicroscopy 2020, 208, 112849. [Google Scholar] [CrossRef] [PubMed]
- Bouvrais, H.; Pott, T.; Bagatolli, L.A.; Ipsen, J.H.; Méléard, P. Impact of Membrane-Anchored Fluorescent Probes on the Mechanical Properties of Lipid Bilayers. Biochim. Biophys. Acta (BBA)-Biomembr. 2010, 1798, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, D. Carbocyanine Dye Orientation in Red Cell Membrane Studied by Microscopic Fluorescence Polarization. Biophys. J. 1979, 26, 557–573. [Google Scholar] [CrossRef] [Green Version]
- Maier, O.; Oberle, V.; Hoekstra, D. Fluorescent Lipid Probes: Some Properties and Applications (a Review). Chem. Phys. Lipids 2002, 116, 3–18. [Google Scholar] [CrossRef]
- Mizuno, H.; Abe, M.; Dedecker, P.; Makino, A.; Rocha, S.; Ohno-Iwashita, Y.; Hofkens, J.; Kobayashi, T.; Miyawaki, A. Fluorescent Probes for Superresolution Imaging of Lipid Domains on the Plasma Membrane. Chem. Sci. 2011, 2, 1548–1553. [Google Scholar] [CrossRef]
- Goudsmits, J.M.H.; van Oijen, A.M.; Slotboom, D.J. Chapter Four-Single-Molecule Fluorescence Studies of Membrane Transporters Using Total Internal Reflection Microscopy. In Methods in Enzymology; Ziegler, C., Ed.; A Structure-Function Toolbox for Membrane Transporter and Channels; Academic Press: Cambridge, MA, USA, 2017; Volume 594, pp. 101–121. [Google Scholar]
- Nishimura, K.; Matsuura, T.; Nishimura, K.; Sunami, T.; Suzuki, H.; Yomo, T. Cell-Free Protein Synthesis inside Giant Unilamellar Vesicles Analyzed by Flow Cytometry. Langmuir 2012, 28, 8426–8432. [Google Scholar] [CrossRef]
- Korlach, J.; Schwille, P.; Webb, W.W.; Feigenson, G.W. Characterization of Lipid Bilayer Phases by Confocal Microscopy and Fluorescence Correlation Spectroscopy. Proc. Natl. Acad. Sci. USA 1999, 96, 8461–8466. [Google Scholar] [CrossRef] [Green Version]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Klymchenko, A.S. Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications. Acc. Chem. Res. 2017, 50, 366–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Cao, J.; He, Y.; Yang, J.H.; Kim, T.; Peng, X.; Kim, J.S. Macro-/Micro-Environment-Sensitive Chemosensing and Biological Imaging. Chem. Soc. Rev. 2014, 43, 4563–4601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sezgin, E.; Sadowski, T.; Simons, K. Measuring Lipid Packing of Model and Cellular Membranes with Environment Sensitive Probes. Langmuir 2014, 30, 8160–8166. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, A.P.; Mély, Y.; Duportail, G.; Klymchenko, A.S. Monitoring Biophysical Properties of Lipid Membranes by Environment-Sensitive Fluorescent Probes. Biophys. J. 2009, 96, 3461–3470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.S.; Kabir, A.M.R.; Inoue, D.; Sada, K.; Kakugo, A. Enhanced Dynamic Instability of Microtubules in a ROS Free Inert Environment. Biophys. Chem. 2016, 211, 1–8. [Google Scholar] [CrossRef] [PubMed]
- VanDelinder, V.; Bachand, G.D. Photodamage and the Importance of Photoprotection in Biomolecular-Powered Device Applications. Anal. Chem. 2014, 86, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Roux, B. Ion Channels and Ion Selectivity Essays in Biochemistry. Essays Biochem. 2017, 61, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-X.; McCammon, J.A. The Gates of Ion Channels and Enzymes. Trends Biochem. Sci. 2010, 35, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Tombola, F.; Pathak, M.M.; Isacoff, E.Y. How Does Voltage Open an Ion Channel? Annu. Rev. Cell Dev. Biol. 2006, 22, 23–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemoine, D.; Jiang, R.; Taly, A.; Chataigneau, T.; Specht, A.; Grutter, T. Ligand-Gated Ion Channels: New Insights into Neurological Disorders and Ligand Recognition. Chem. Rev. 2012, 112, 6285–6318. [Google Scholar] [CrossRef] [PubMed]
- Krasowski, M.D.; Harrison, N.L. General Anaesthetic Actions on Ligand-Gated Ion Channels. Cell. Mol. Life Sci. 1999, 55, 1278–1303. [Google Scholar] [CrossRef]
- Rammes, G.; Rupprecht, R. Modulation of Ligand-Gated Ion Channels by Antidepressants and Antipsychotics. Mol. Neurobiol. 2007, 35, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Ranade, S.S.; Syeda, R.; Patapoutian, A. Mechanically Activated Ion Channels. Neuron 2015, 87, 1162–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gargalionis, A.N.; Basdra, E.K.; Papavassiliou, A.G. Tumor Mechanosensing and Its Therapeutic Potential. J. Cell. Biochem. 2018, 119, 4304–4308. [Google Scholar] [CrossRef]
- Drexler, S.; Wann, A.; Vincent, T.L. Are Cellular Mechanosensors Potential Therapeutic Targets in Osteoarthritis? Int. J. Clin. Rheumatol. 2014, 9, 155–167. [Google Scholar] [CrossRef]
- Grote, M.; Engelhard, M.; Hegemann, P. Of Ion Pumps, Sensors and Channels—Perspectives on Microbial Rhodopsins between Science and History. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 533–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanyi, J.K. Mechanism of Ion Transport across Membranes. J. Biol. Chem. 1997, 272, 31209–31212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kono, M.; Goletz, P.W.; Crouch, R.K. 11-Cis and All-Trans Retinols Can Activate Rod Opsin: Rational Design of the Visual Cycle. Biochemistry 2008, 47, 7567–7571. [Google Scholar] [CrossRef] [Green Version]
- Yan, B.; Takahashi, T.; Johnson, R.; Derguini, F.; Nakanishi, K.; Spudich, J.L. All-Trans/13-Cis Isomerization of Retinal Is Required for Phototaxis Signaling by Sensory Rhodopsins in Halobacterium Halobium. Biophys. J. 1990, 57, 807–814. [Google Scholar] [CrossRef] [Green Version]
- Nakanishi, K. Why 11-Cis-Retinal? Am. Zool. 1991, 31, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Kühlbrandt, W. Bacteriorhodopsin—The Movie. Nature 2000, 406, 569–570. [Google Scholar] [CrossRef]
- Henderson, R.; Schertler, G.F.X. The Structure of Bacteriorhodopsin and Its Relevance to the Visual Opsins and Other Seven-Helix G-Protein Coupled Receptors. Philos. Trans. R. Soc. London. B Biol. Sci. 1990, 326, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Bamann, C.; Bamberg, E.; Wachtveitl, J.; Glaubitz, C. Proteorhodopsin. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 614–625. [Google Scholar] [CrossRef] [Green Version]
- Shevchenko, V.; Mager, T.; Kovalev, K.; Polovinkin, V.; Alekseev, A.; Juettner, J.; Chizhov, I.; Bamann, C.; Vavourakis, C.; Ghai, R.; et al. Inward H+ Pump Xenorhodopsin: Mechanism and Alternative Optogenetic Approach. Sci. Adv. 2017, 3, e1603187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ugalde, J.A.; Podell, S.; Narasingarao, P.; Allen, E.E. Xenorhodopsins, an Enigmatic New Class of Microbial Rhodopsins Horizontally Transferred between Archaea and Bacteria. Biol. Direct 2011, 6, 52. [Google Scholar] [CrossRef] [Green Version]
- Mukohata, Y.; Sugiyama, Y.; Ihara, K.; Yoshida, M. An Australian Halobacterium Contains a Novel Proton Pump Retinal Protein: Archaerhodopsin. Biochem. Biophys. Res. Commun. 1988, 151, 1339–1345. [Google Scholar] [CrossRef]
- Hargrave, P.A.; McDowell, J.H.; Curtis, D.R.; Wang, J.K.; Juszczak, E.; Fong, S.-L.; Mohana Rao, J.K.; Argos, P. The Structure of Bovine Rhodopsin. Biophys. Struct. Mech. 1983, 9, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Deisseroth, K.; Hegemann, P. The Form and Function of Channelrhodopsin. Science 2017, 357, eaan5544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, G.; Szellas, T.; Huhn, W.; Kateriya, S.; Adeishvili, N.; Berthold, P.; Ollig, D.; Hegemann, P.; Bamberg, E. Channelrhodopsin-2, a Directly Light-Gated Cation-Selective Membrane Channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schobert, B.; Lanyi, J.K. Halorhodopsin Is a Light-Driven Chloride Pump. J. Biol. Chem. 1982, 257, 10306–10313. [Google Scholar] [CrossRef]
- Lutz, I.; Sieg, A.; Wegener, A.A.; Engelhard, M.; Boche, I.; Otsuka, M.; Oesterhelt, D.; Wachtveitl, J.; Zinth, W. Primary Reactions of Sensory Rhodopsins. Proc. Natl. Acad. Sci. USA 2001, 98, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Oesterhelt, D. The Structure and Mechanism of the Family of Retinal Proteins from Halophilic Archaea. Curr. Opin. Struct. Biol. 1998, 8, 489–500. [Google Scholar] [CrossRef]
- Chan, Y.-H.M.; Boxer, S.G. Model Membrane Systems and Their Applications. Curr. Opin. Chem. Biol. 2007, 11, 581–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreir, M.; Farre, C.; Beckler, M.; George, M.; Fertig, N. Rapid Screening of Membrane Protein Activity: Electrophysiological Analysis of OmpF Reconstituted in Proteoliposomes. Lab Chip 2008, 8, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Dezi, M.; Cicco, A.D.; Bassereau, P.; Lévy, D. Detergent-Mediated Incorporation of Transmembrane Proteins in Giant Unilamellar Vesicles with Controlled Physiological Contents. Proc. Natl. Acad. Sci. USA 2013, 110, 7276–7281. [Google Scholar] [CrossRef] [Green Version]
- Rigaud, J.-L.; Mosser, G.; Lacapere, J.-J.; Olofsson, A.; Levy, D.; Ranck, J.-L. Bio-Beads: An Efficient Strategy for Two-Dimensional Crystallization of Membrane Proteins. J. Struct. Biol. 1997, 118, 226–235. [Google Scholar] [CrossRef]
- Reeves, J.P.; Dowben, R.M. Formation and Properties of Thin-Walled Phospholipid Vesicles. J. Cell. Physiol. 1969, 73, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Girard, P.; Pécréaux, J.; Lenoir, G.; Falson, P.; Rigaud, J.-L.; Bassereau, P. A New Method for the Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles. Biophys. J. 2004, 87, 419–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahya, N.; Pécheur, E.-I.; de Boeij, W.P.; Wiersma, D.A.; Hoekstra, D. Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles via Peptide-Induced Fusion. Biophys. J. 2001, 81, 1464–1474. [Google Scholar] [CrossRef] [Green Version]
- Stachowiak, J.C.; Richmond, D.L.; Li, T.H.; Liu, A.P.; Parekh, S.H.; Fletcher, D.A. Unilamellar Vesicle Formation and Encapsulation by Microfluidic Jetting. Proc. Natl. Acad. Sci. USA 2008, 105, 4697–4702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richmond, D.L.; Schmid, E.M.; Martens, S.; Stachowiak, J.C.; Liska, N.; Fletcher, D.A. Forming Giant Vesicles with Controlled Membrane Composition, Asymmetry, and Contents. Proc. Natl. Acad. Sci. USA 2011, 108, 9431–9436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borch, J.; Hamann, T. The Nanodisc: A Novel Tool for Membrane Protein Studies. Biol. Chem. 2009, 390, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 2017, 117, 4669–4713. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Grinkova, Y.V.; Sligar, S.G. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar] [CrossRef]
- Chen, A.; Majdinasab, E.J.; Fiori, M.C.; Liang, H.; Altenberg, G.A. Polymer-Encased Nanodiscs and Polymer Nanodiscs: New Platforms for Membrane Protein Research and Applications. Front. Bioeng. Biotechnol. 2020, 8, 1329. [Google Scholar] [CrossRef] [PubMed]
- Ravula, T.; Hardin, N.Z.; Ramamoorthy, A. Polymer Nanodiscs: Advantages and Limitations. Chem. Phys. Lipids 2019, 219, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Midtgaard, S.R.; Pedersen, M.C.; Kirkensgaard, J.J.K.; Sørensen, K.K.; Mortensen, K.; Jensen, K.J.; Arleth, L. Self-Assembling Peptides Form Nanodiscs That Stabilize Membrane Proteins. Soft Matter 2014, 10, 738–752. [Google Scholar] [CrossRef] [PubMed]
- Sachse, R.; Dondapati, S.K.; Fenz, S.F.; Schmidt, T.; Kubick, S. Membrane Protein Synthesis in Cell-Free Systems: From Bio-Mimetic Systems to Bio-Membranes. FEBS Lett. 2014, 588, 2774–2781. [Google Scholar] [CrossRef] [PubMed]
- Underwood, K.A.; Swartz, J.R.; Puglisi, J.D. Quantitative Polysome Analysis Identifies Limitations in Bacterial Cell-Free Protein Synthesis. Biotechnol. Bioeng. 2005, 91, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Kanamori, T.; Ueda, T. Protein Synthesis by Pure Translation Systems. Methods 2005, 36, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Kalmbach, R.; Chizhov, I.; Schumacher, M.C.; Friedrich, T.; Bamberg, E.; Engelhard, M. Functional Cell-Free Synthesis of a Seven Helix Membrane Protein: In Situ Insertion of Bacteriorhodopsin into Liposomes. J. Mol. Biol. 2007, 371, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Goren, M.A.; Fox, B.G. Wheat Germ Cell-Free Translation, Purification, and Assembly of a Functional Human Stearoyl-CoA Desaturase Complex. Protein Expr. Purif. 2008, 62, 171–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneda, M.; Nomura, S.M.; Ichinose, S.; Kondo, S.; Nakahama, K.; Akiyoshi, K.; Morita, I. Direct Formation of Proteo-Liposomes by in Vitro Synthesis and Cellular Cytosolic Delivery with Connexin-Expressing Liposomes. Biomaterials 2009, 30, 3971–3977. [Google Scholar] [CrossRef] [PubMed]
- Long, A.R.; O’Brien, C.C.; Malhotra, K.; Schwall, C.T.; Albert, A.D.; Watts, A.; Alder, N.N. A Detergent-Free Strategy for the Reconstitution of Active Enzyme Complexes from Native Biological Membranes into Nanoscale Discs. BMC Biotechnol. 2013, 13, 41. [Google Scholar] [CrossRef] [Green Version]
- Popot, J.-L.; Althoff, T.; Bagnard, D.; Banères, J.-L.; Bazzacco, P.; Billon-Denis, E.; Catoire, L.J.; Champeil, P.; Charvolin, D.; Cocco, M.J.; et al. Amphipols From A to Z. Annu. Rev. Biophys. 2011, 40, 379–408. [Google Scholar] [CrossRef]
- Gorzelle, B.M.; Hoffman, A.K.; Keyes, M.H.; Gray, D.N.; Ray, D.G.; Sanders, C.R. Amphipols Can Support the Activity of a Membrane Enzyme. J. Am. Chem. Soc. 2002, 124, 11594–11595. [Google Scholar] [CrossRef] [PubMed]
- Althoff, T.; Mills, D.J.; Popot, J.-L.; Kuhlbrandt, W. Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I1III2IV1. EMBO J. 2011, 30, 4652–4664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orwick, M.C.; Judge, P.J.; Procek, J.; Lindholm, L.; Graziadei, A.; Engel, A.; Gröbner, G.; Watts, A. Detergent-Free Formation and Physicochemical Characterization of Nanosized Lipid–Polymer Complexes: Lipodisq. Angew. Chem. Int. Ed. 2012, 51, 4653–4657. [Google Scholar] [CrossRef] [PubMed]
- Orwick-Rydmark, M.; Lovett, J.E.; Graziadei, A.; Lindholm, L.; Hicks, M.R.; Watts, A. Detergent-Free Incorporation of a Seven-Transmembrane Receptor Protein into Nanosized Bilayer Lipodisq Particles for Functional and Biophysical Studies. Nano Lett. 2012, 12, 4687–4692. [Google Scholar] [CrossRef]
- Fiori, M.C.; Zheng, W.; Kamilar, E.; Simiyu, G.; Altenberg, G.A.; Liang, H. Extraction and Reconstitution of Membrane Proteins into Lipid Nanodiscs Encased by Zwitterionic Styrene-Maleic Amide Copolymers. Sci. Rep. 2020, 10, 9940. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.J.; Finka, R.; Smith, C.; Lin, Y.-P.; Dafforn, T.; Overduin, M. Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. J. Am. Chem. Soc. 2009, 131, 7484–7485. [Google Scholar] [CrossRef]
- Ernst, O.P.; Lodowski, D.T.; Elstner, M.; Hegemann, P.; Brown, L.S.; Kandori, H. Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev. 2014, 114, 126–163. [Google Scholar] [CrossRef] [PubMed]
- Hoi, H.; Qi, Z.; Zhou, H.; Montemagno, C.D. Enhanced Overexpression, Purification of a Channelrhodopsin and a Fluorescent Flux Assay for Its Functional Characterization. J. Biotechnol. 2018, 281, 99–105. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Letts, J.A.; MacKinnon, R. Functional Reconstitution of Purified Human Hv1 H+ Channels. J. Mol. Biol. 2009, 387, 1055–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Feng, Y.; Forgac, M. Proton Conduction and Bafilomycin Binding by the V0 Domain of the Coated Vesicle V-ATPase. J. Biol. Chem. 1994, 269, 23518–23523. [Google Scholar] [CrossRef]
- Justesen, B.H.; Hansen, R.W.; Martens, H.J.; Theorin, L.; Palmgren, M.G.; Martinez, K.L.; Pomorski, T.G.; Fuglsang, A.T. Active Plasma Membrane P-Type H+-ATPase Reconstituted into Nanodiscs Is a Monomer. J. Biol. Chem. 2013, 288, 26419–26429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishmukhametov, R.; Hornung, T.; Spetzler, D.; Frasch, W.D. Direct Observation of Stepped Proteolipid Ring Rotation in E. coli FoF1-ATP Synthase. EMBO J. 2010, 29, 3911–3923. [Google Scholar] [CrossRef] [Green Version]
- Berhanu, S.; Ueda, T.; Kuruma, Y. Artificial Photosynthetic Cell Producing Energy for Protein Synthesis. Nat. Commun. 2019, 10, 1325. [Google Scholar] [CrossRef] [PubMed]
- Cabanos, C.; Wang, M.; Han, X.; Hansen, S.B. A Soluble Fluorescent Binding Assay Reveals PIP 2 Antagonism of TREK-1 Channels. Cell Rep. 2017, 20, 1287–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukamoto, H.; Higashi, M.; Motoki, H.; Watanabe, H.; Ganser, C.; Nakajo, K.; Kubo, Y.; Uchihashi, T.; Furutani, Y. Structural Properties Determining Low K+ Affinity of the Selectivity Filter in the TWIK1 K+ Channel. J. Biol. Chem. 2018, 293, 6969–6984. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Campbell, E.B.; MacKinnon, R. Molecular Mechanism of Proton Transport in CLC Cl−/Hþ Exchange Transporters. Proc. Natl. Acad. Sci. USA 2012, 109, 11699–11704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avnir, Y.; Barenholz, Y. PH Determination by Pyranine: Medium-Related Artifacts and Their Correction. Anal. Biochem. 2005, 347, 34–41. [Google Scholar] [CrossRef]
- Göpfrich, K.; Haller, B.; Staufer, O.; Dreher, Y.; Mersdorf, U.; Platzman, I.; Spatz, J.P. One-Pot Assembly of Complex Giant Unilamellar Vesicle-Based Synthetic Cells. ACS Synth. Biol. 2019, 8, 937–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clement, N.R.; Gould, J.M. Pyranine (8-Hydroxy-1,3,6-Pyrenetrisulfonate) as a Probe of Internal Aqueous Hydrogen Ion Concentration in Phospholipid Vesicles. Biochemistry 1981, 20, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
- Kano, K.; Fendler, J.H. Pyranine as a Sensitive PH Probe for Liposome Interiors and Surfaces. PH Gradients across Phospholipid Vesicles. Biochim. Biophys. Acta (BBA)-Biomembr. 1978, 509, 289–299. [Google Scholar] [CrossRef]
- Heberle, J. Proton Transfer Reactions across Bacteriorhodopsin and along the Membrane. Biochim. Biophys. Acta (BBA)-Bioenerg. 2000, 1458, 135–147. [Google Scholar] [CrossRef] [Green Version]
- Verchère, A.; Dezi, M.; Broutin, I.; Picard, M. In Vitro Investigation of the MexAB Efflux Pump from Pseudomonas Aeruginosa. J. Vis. Exp. 2014, 50894. [Google Scholar] [CrossRef] [Green Version]
- Verchère, A.; Broutin, I.; Picard, M. Photo-Induced Proton Gradients for the in Vitro Investigation of Bacterial Efflux Pumps. Sci. Rep. 2012, 2, 306. [Google Scholar] [CrossRef] [Green Version]
- Seigneuret, M.; Rigaud, J.-L. Use of the Fluorescent PH Probe Pyranine to Detect Heterogeneous Directions of Proton Movement in Bacteriorhodopsin Reconstituted Large Liposomes. FEBS Lett. 1985, 188, 101–106. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Ho, D.; Kuo, K.; Montemagno, C.D. Vectorial Insertion of Bacteriorhodopsin for Directed Orientation Assays in Various Polymeric Biomembranes. Polymer 2006, 47, 2935–2941. [Google Scholar] [CrossRef]
- Olson, K.D.; Deval, P.; Spudich, J.L. Absorption and Photochemistry of Sensory Rhodopsin—I: PH Effects. Photochem. Photobiol. 1992, 56, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, J.; Spudich, J.L. Proton Circulation During the Photocycle of Sensory Rhodopsin II. Biophys. J. 1999, 77, 2145–2152. [Google Scholar] [CrossRef] [Green Version]
- Inoue, K.; Tahara, S.; Kato, Y.; Takeuchi, S.; Tahara, T.; Kandori, H. Spectroscopic Study of Proton-Transfer Mechanism of Inward Proton-Pump Rhodopsin, Parvularcula Oceani Xenorhodopsin. J. Phys. Chem. B 2018, 122, 6453–6461. [Google Scholar] [CrossRef]
- Li, H.; Sineshchekov, O.A.; Wu, G.; Spudich, J.L. In Vitro Activity of a Purified Natural Anion Channelrhodopsin. J. Biol. Chem. 2016, 291, 25319–25325. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Ding, X.; Peng, B.; Zhao, Y.; Ding, J.; Watts, A.; Zhao, X. Novel Expression and Characterization of a Light Driven Proton Pump Archaerhodopsin 4 in a Halobacterium Salinarum Strain. Biochim. Biophys. Acta (BBA)-Bioenerg. 2015, 1847, 390–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dioumaev, A.K.; Brown, L.S.; Shih, J.; Spudich, E.N.; Spudich, J.L.; Lanyi, J.K. Proton Transfers in the Photochemical Reaction Cycle of Proteorhodopsin. Biochemistry 2002, 41, 5348–5358. [Google Scholar] [CrossRef] [PubMed]
- Lanyi, J.K.; Balashov, S.P. Xanthorhodopsin: A Bacteriorhodopsin-like Proton Pump with a Carotenoid Antenna. Biochim. Biophys. Acta (BBA)-Bioenerg. 2008, 1777, 684–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balashov, S.P.; Imasheva, E.S.; Boichenko, V.A.; Antón, J.; Wang, J.M.; Lanyi, J.K. Xanthorhodopsin: A Proton Pump with a Light-Harvesting Carotenoid Antenna. Science 2005, 309, 2061–2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghilarov, D.; Inaba-Inoue, S.; Stepien, P.; Qu, F.; Michalczyk, E.; Pakosz, Z.; Nomura, N.; Ogasawara, S.; Walker, G.C.; Rebuffat, S.; et al. Molecular Mechanism of SbmA, a Promiscuous Transporter Exploited by Antimicrobial Peptides. Sci. Adv. 2021, 7, eabj5363. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, H.; Xiang, Y.; Pang, S.; Bao, C.; Zhu, L. A Synthetic Phospholipid Derivative Mediates Ion Transport Across Lipid Bilayers. Front. Chem. 2021, 9, 267. [Google Scholar] [CrossRef]
- Kemmer, G.C.; Bogh, S.A.; Urban, M.; Palmgren, M.G.; Vosch, T.; Schiller, J.; Pomorski, T.G. Lipid-Conjugated Fluorescent PH Sensors for Monitoring PH Changes in Reconstituted Membrane Systems. Analyst 2015, 140, 6313–6320. [Google Scholar] [CrossRef] [Green Version]
- Bolli, R.; Müller, M.; Nalȩcs, K.; Azzi, A. Cytochrome c Oxidase: Example of a Redox-Coupled Proton Pump. Mol. Asp. Med. 1988, 10, 249–255. [Google Scholar] [CrossRef]
- Veshaguri, S.; Christensen, S.M.; Kemmer, G.C.; Ghale, G.; Moller, M.P.; Lohr, C.; Christensen, A.L.; Justesen, B.H.; Jorgensen, I.L.; Schiller, J.; et al. Direct Observation of Proton Pumping by a Eukaryotic P-Type ATPase. Science 2016, 351, 1469–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwamborn, M.; Schumacher, J.; Sibold, J.; Teiwes, N.K.; Steinem, C. Monitoring ATPase Induced PH Changes in Single Proteoliposomes with the Lipid-Coupled Fluorophore Oregon Green 488. Analyst 2017, 142, 2670–2677. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, B.; Rixen, R.M.; Kramer, K.; Forbrig, E.; Hildebrandt, P.; Steinem, C. Quantification of Hv1-Induced Proton Translocation by a Lipid-Coupled Oregon Green 488-Based Assay. Anal. Bioanal. Chem. 2018, 410, 6497–6505. [Google Scholar] [CrossRef]
- Zoccarato, F.; Cavallini, L.; Alexandre, A. The PH-Sensitive Dye Acridine Orange as a Tool to MonitorExocytosis/Endocytosis in Synaptosomes. J. Neurochem. 1999, 72, 625–633. [Google Scholar] [CrossRef]
- Byvaltsev, V.A.; Bardonova, L.A.; Onaka, N.R.; Polkin, R.A.; Ochkal, S.V.; Shepelev, V.V.; Aliyev, M.A.; Potapov, A.A. Acridine Orange: A Review of Novel Applications for Surgical Cancer Imaging and Therapy. Front. Oncol. 2019, 9, 925. [Google Scholar] [CrossRef] [PubMed]
- Reifenrath, M.; Boles, E. A Superfolder Variant of PH-Sensitive PHluorin for In Vivo PH Measurements in the Endoplasmic Reticulum. Sci. Rep. 2018, 8, 11985. [Google Scholar] [CrossRef] [Green Version]
- Nekrasova, O.V.; Wulfson, A.N.; Tikhonov, R.V.; Yakimov, S.A.; Simonova, T.N.; Tagvey, A.I.; Dolgikh, D.A.; Ostrovsky, M.A.; Kirpichnikov, M.P. A New Hybrid Protein for Production of Recombinant Bacteriorhodopsin in Escherichia Coli. J. Biotechnol. 2010, 147, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Ritzmann, N.; Thoma, J.; Hirschi, S.; Kalbermatter, D.; Fotiadis, D.; Müller, D.J. Fusion Domains Guide the Oriented Insertion of Light-Driven Proton Pumps into Liposomes. Biophys. J. 2017, 113, 1181–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramaniam, V.; Alves, I.D.; Salgado, G.F.J.; Lau, P.; Wysocki, R.J.; Salamon, Z.; Tollin, G.; Hruby, V.J.; Brown, M.F.; Saavedra, S.S. Rhodopsin Reconstituted into a Planar-Supported Lipid Bilayer Retains Photoactivity after Cross-Linking Polymerization of Lipid Monomers. J. Am. Chem. Soc. 2005, 127, 5320–5321. [Google Scholar] [CrossRef] [PubMed]
- Bieri, C.; Ernst, O.P.; Heyse, S.; Hofmann, K.P.; Vogel, H. Micropatterned Immobilization of a G Protein–Coupled Receptor and Direct Detection of G Protein Activation. Nat. Biotechnol. 1999, 17, 1105–1108. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, O.P.; Lofas, S. Flow-Mediated On-Surface Reconstitution of G-Protein Coupled Receptors for Applications in Surface Plasmon Resonance Biosensors. Anal. Biochem. 2002, 300, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lu, G.; Chiang, E.Y.; Chernov-Rogan, T.; Grogan, J.L.; Chen, J. High-Throughput Electrophysiological Assays for Voltage Gated Ion Channels Using SyncroPatch 768PE. PLoS ONE 2017, 12, e0180154. [Google Scholar] [CrossRef] [Green Version]
- Jaggers, O.B.; Ridone, P.; Martinac, B.; Baker, M.A.B. Fluorescence Microscopy of Piezo1 in Droplet Hydrogel Bilayers. Channels 2019, 13, 102–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, M.D.; Gordon, S.E. Giant Liposome Preparation for Imaging and Patch-Clamp Electrophysiology. J. Vis. Exp. 2013, 50227. [Google Scholar] [CrossRef] [PubMed]
- Battle, A.R.; Ridone, P.; Bavi, N.; Nakayama, Y.; Nikolaev, Y.A.; Martinac, B. Lipid–Protein Interactions: Lessons Learned from Stress. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 1744–1756. [Google Scholar] [CrossRef]
- Cho, H.-Y.; Chuang, T.-H.; Wu, S.-N. Effective Perturbations on the Amplitude and Hysteresis of Erg-Mediated Potassium Current Caused by 1-Octylnonyl 8-[(2-Hydroxyethyl)[6-Oxo-6(Undecyloxy)Hexyl]Amino]-Octanoate (SM-102), a Cationic Lipid. Biomedicines 2021, 9, 1367. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.-H.; Liu, P.-Y.; Wu, S.-N. Characterization of Perturbing Actions by Verteporfin, a Benzoporphyrin Photosensitizer, on Membrane Ionic Currents. Front. Chem. 2019, 7, 566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.-N.; Lin, M.-W.; Wang, Y.-J. Stimulatory Actions of Di-8-Butyl-Amino-Naphthyl-Ethylene-Pyridinium-Propyl-Sulfonate (Di-8-ANEPPS), Voltage-Sensitive Dye, on the BKCa Channel in Pituitary Tumor (GH3) Cells. Pflügers Arch.-Eur. J. Physiol. 2008, 455, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Booth, M.J.; Restrepo Schild, V.; Box, S.J.; Bayley, H. Light-Patterning of Synthetic Tissues with Single Droplet Resolution. Sci. Rep. 2017, 7, 9315. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Dye | Type | λex/em, nm | Used to Monitor | References |
|---|---|---|---|---|
| ACMA | Membrane-impermeable, hydrophobic | 410/490 | bR | [116] |
| ChIEF | [111] | |||
| Hv1 | [112] | |||
| V-ATPases | [113,114] | |||
| Pyranine | Membrane-permeable, hydrophilic | 454/520 | bR | [83,86,125,126,127,128] |
| pR | [134] | |||
| sR | [129,130] | |||
| xR | [131] | |||
| ChR | [132] | |||
| aR | [133] | |||
| Xanthorhodopsin | [135] | |||
| SbmA | [137] | |||
| Oregon Green 488-DHPE | Lipid-coupled | 508/534 | ATPase, Hv1 | [142,143] |
| Fluorescein-PE | Lipid-coupled | 498/517 | Cytochrome c oxidase | [140] |
| pHrodo-DOPE | Lipid-coupled | 532/585 | ATPase | [139,141] |
| Acridine orange | Hydrophobic | 490/520 | Synaptosome, acidic vesicular organelles | [144,145] |
| pHluorin | GFP variant | 470/512 | Cytosol and endoplasmic reticulum | [146] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.S.; Gaston, J.P.; Baker, M.A.B. Fluorescence Approaches for Characterizing Ion Channels in Synthetic Bilayers. Membranes 2021, 11, 857. https://doi.org/10.3390/membranes11110857
Islam MS, Gaston JP, Baker MAB. Fluorescence Approaches for Characterizing Ion Channels in Synthetic Bilayers. Membranes. 2021; 11(11):857. https://doi.org/10.3390/membranes11110857
Chicago/Turabian StyleIslam, Md. Sirajul, James P. Gaston, and Matthew A. B. Baker. 2021. "Fluorescence Approaches for Characterizing Ion Channels in Synthetic Bilayers" Membranes 11, no. 11: 857. https://doi.org/10.3390/membranes11110857
APA StyleIslam, M. S., Gaston, J. P., & Baker, M. A. B. (2021). Fluorescence Approaches for Characterizing Ion Channels in Synthetic Bilayers. Membranes, 11(11), 857. https://doi.org/10.3390/membranes11110857






