The Mechanism of Metal Homeostasis in Plants: A New View on the Synergistic Regulation Pathway of Membrane Proteins, Lipids and Metal Ions
Abstract
1. Introduction
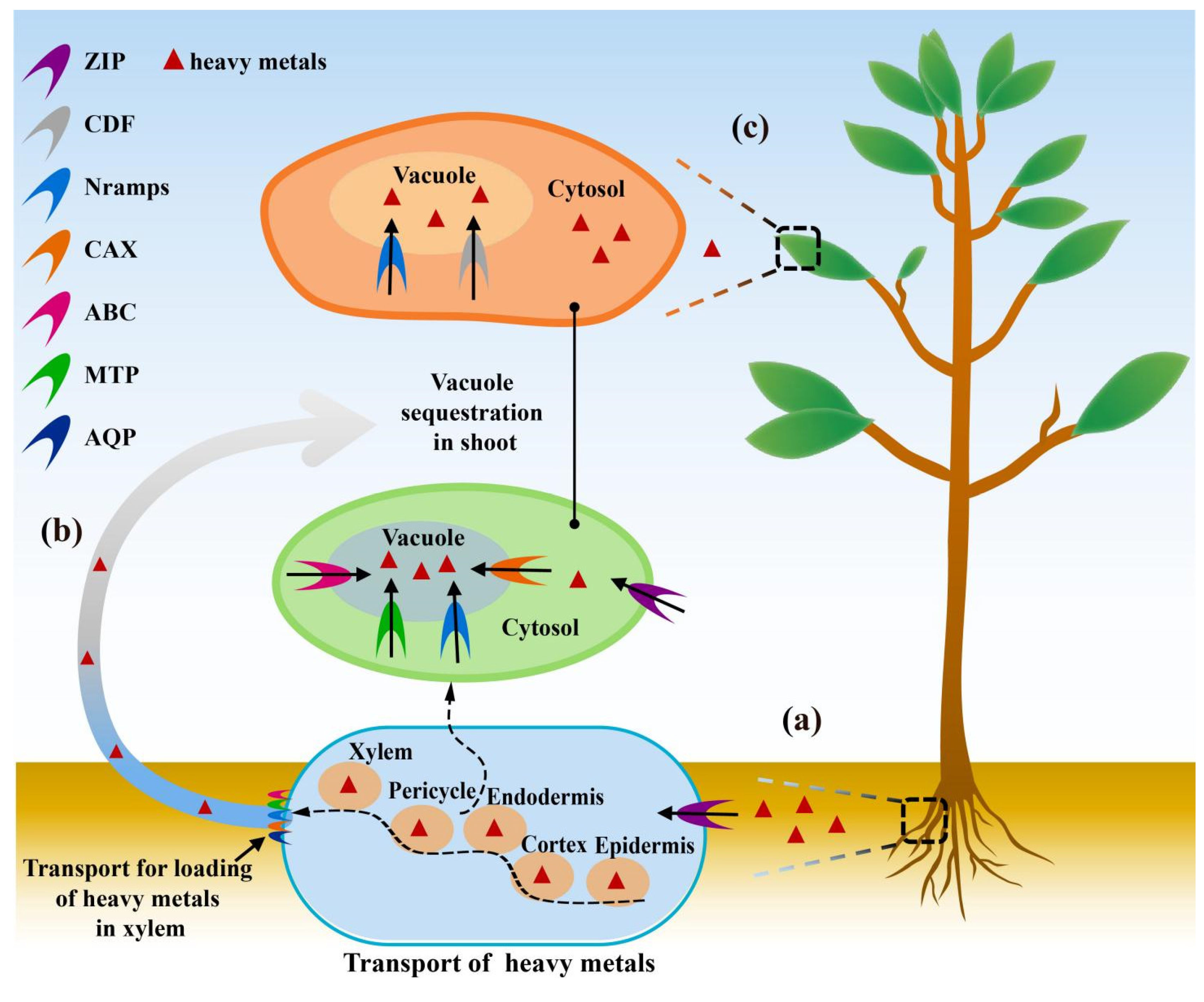
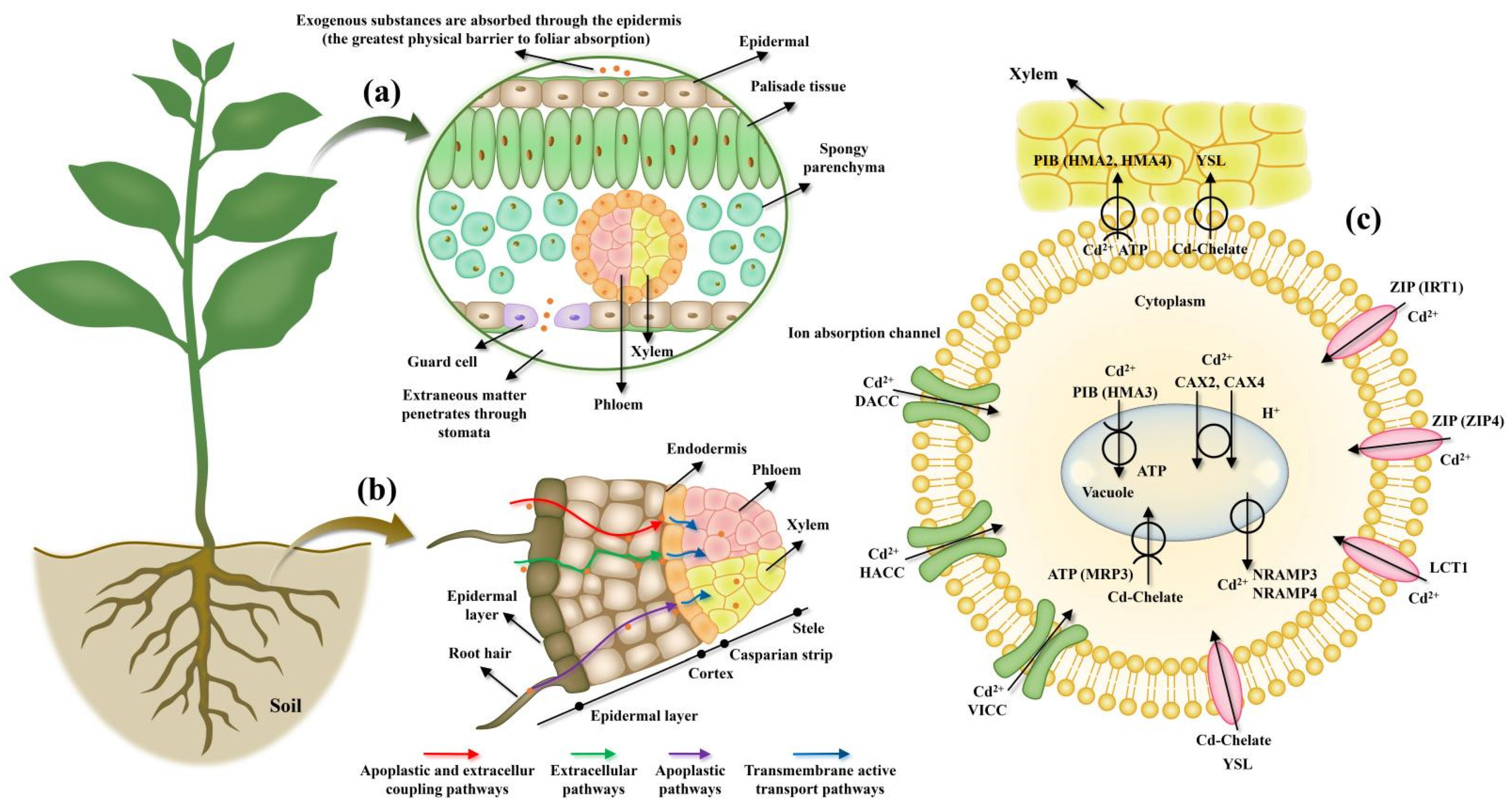
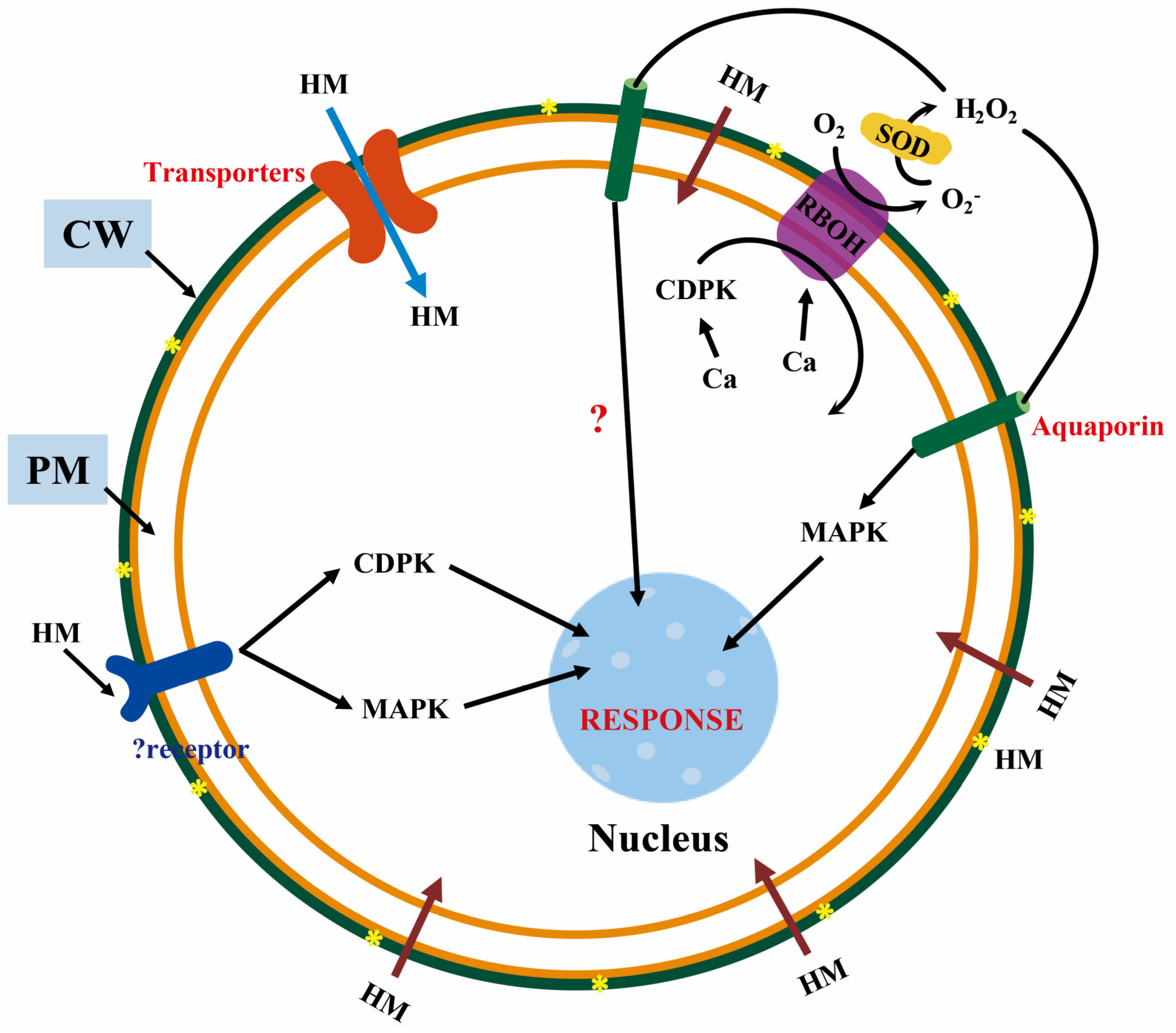
2. Role Cell Membrane Proteins in HMS Tolerance in Plants
2.1. The Reorganization of Endomembranes
2.2. Detoxification Mechanism
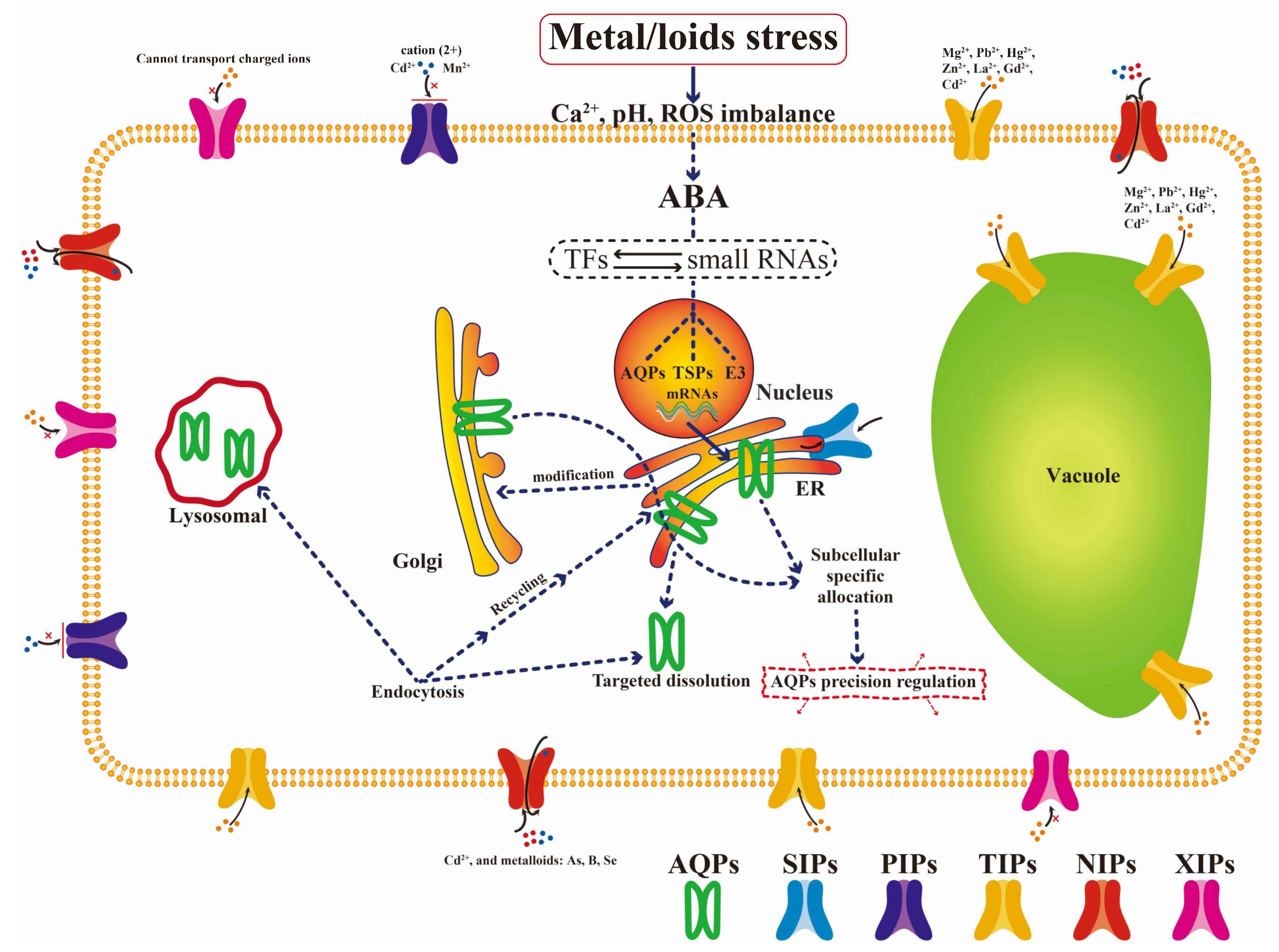
2.3. Important Aquaporin Family
3. The Role of Lipid Regulation in Plant Responses to HMS
3.1. Cascade Signal Transduction
3.2. Abscisic Acid
3.3. Plant TFs and miRNAs Play Important Roles in Regulating Metal Homeostasis
4. Lipid Regulation in Plants under HMS
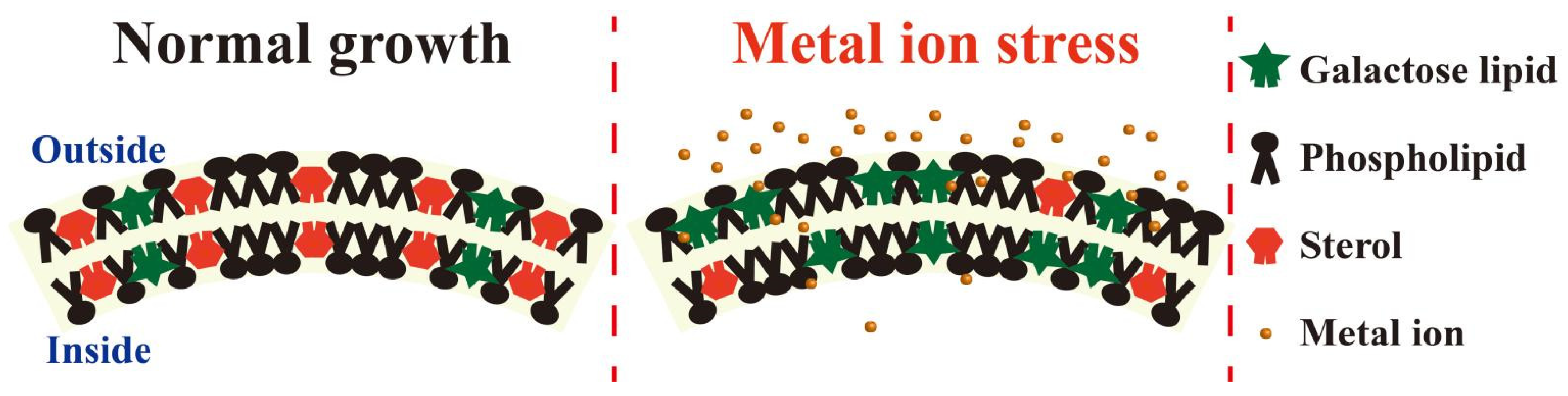
4.1. Membrane Fluidity
4.2. Sphingolipids
5. Interactions between Membrane Proteins and Lipids under HMS
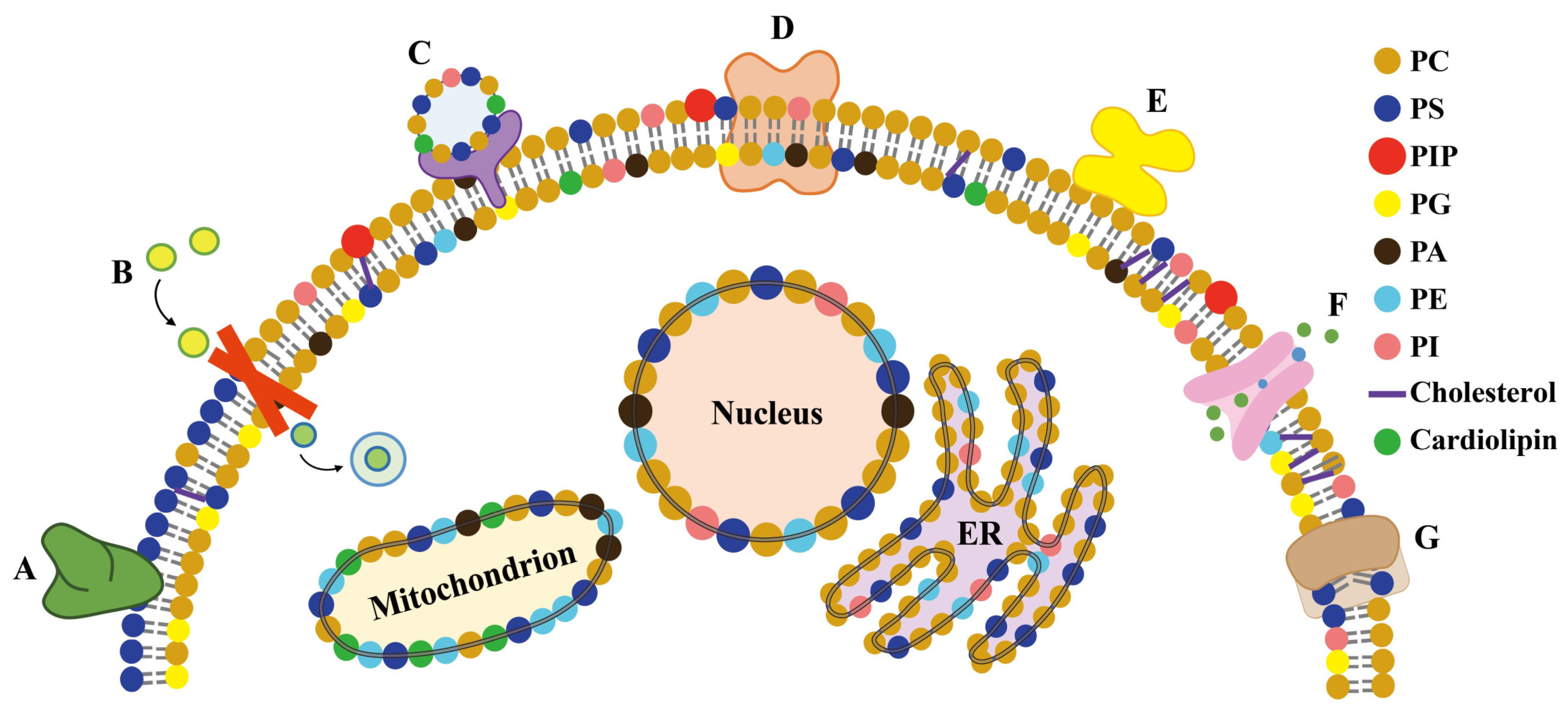
5.1. Membrane Proteins and Lipid Remodeling
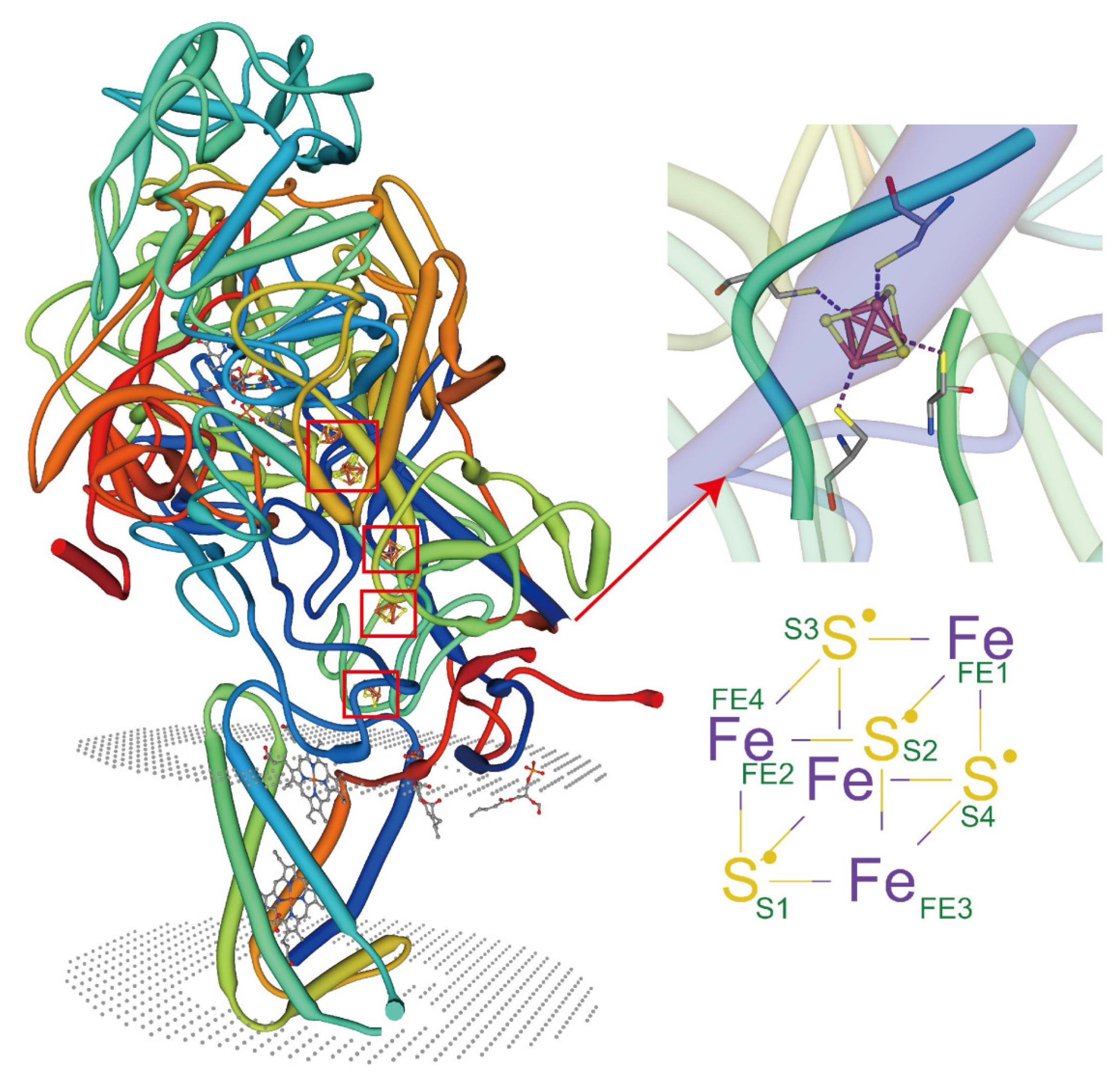
5.2. Metal Coordination of Lipids with Membrane Proteins
5.3. Special Ion Channels as Lipid Sensors
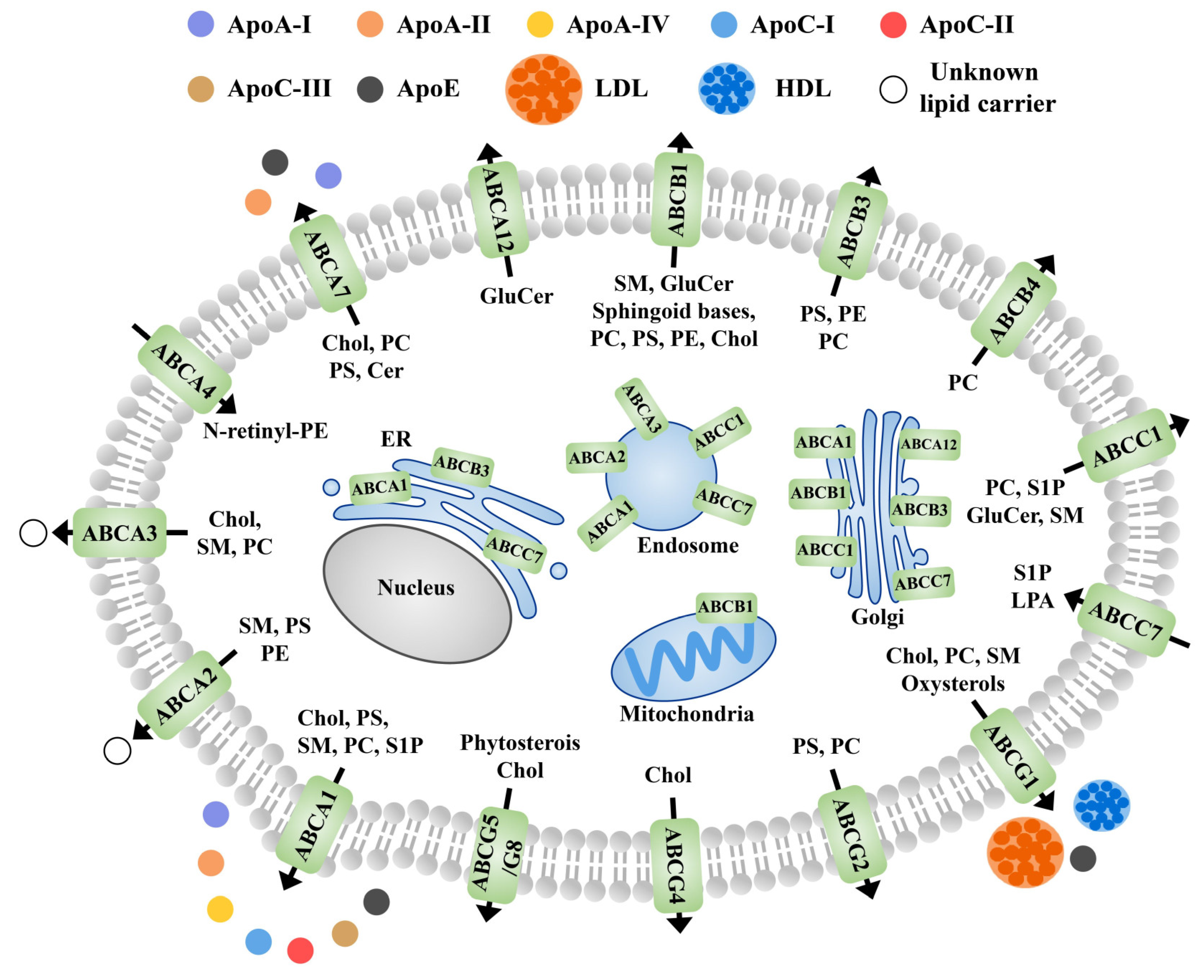
5.4. Role of Membrane Proteins in the Lipid Outflow
5.5. Synergistic Participation in Specific Binding of Metal Ions
6. Analysis of Membrane Protein-Metal Binding Domains and Interaction Based on the Novel Metallomics Database
7. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jalmi, S.; Bhagat, P.K.; Verma, D.; Noryang, S.; Tayyeba, S.; Singh, K.; Sharma, D.; Sinha, A.K. Traversing the Links between Heavy Metal Stress and Plant Signaling. Front. Plant Sci. 2018, 9, 12. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Shinozaki, K. Long-distance signaling in plant stress response. Curr. Opin. Plant Biol. 2019, 47, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Deason-Towne, F.; Perraud, A.-L.; Schmitz, C. Identification of Ser/Thr phosphorylation sites in the C2-domain of phospholipase C γ2 (PLCγ2) using TRPM7-kinase. Cell. Signal. 2012, 24, 2070–2075. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Ci, X.; Huang, J.; Liu, Q.; Yu, Q.; Zhou, J.; Deng, X. Asiatic acid enhances Nrf2 signaling to protect HepG2 cells from oxidative damage through Akt and ERK activation. Biomed. Pharmacother. 2017, 88, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xu, F.; Chen, Z.; Teng, Z.; Sun, K.; Li, X.; Yu, J.; Zhang, G.; Liang, Y.; Huang, X.; et al. Synergistic interplay of ABA and BR signal in regulating plant growth and adaptation. Nat. Plants 2021, 7, 1108–1118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Rankenberg, T.; Geldhof, B.; van Veen, H.; Holsteens, K.; Van de Poel, B.; Sasidharan, R. Age-Dependent Abiotic Stress Resilience in Plants. Trends Plant Sci. 2021, 26, 692–705. [Google Scholar] [CrossRef]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Krüler, V.; Winkelmüller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef]
- Savvides, A.; Ali, S.; Tester, M.; Fotopoulos, V. Chemical Priming of Plants Against Multiple Abiotic Stresses: Mission Possible? Trends Plant Sci. 2016, 21, 329–340. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of Jasmonic Acid in Plant Regulation and Response to Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef] [PubMed]
- Sako, K.; Nguyen, H.M.; Seki, M. Advances in Chemical Priming to Enhance Abiotic Stress Tolerance in Plants. Plant Cell Physiol. 2021, 61, 1995–2003. [Google Scholar] [CrossRef] [PubMed]
- Emamverdian, A.; Ding, Y.; Mokhberdoran, F. The role of salicylic acid and gibberellin signaling in plant responses to abiotic stress with an emphasis on heavy metals. Plant Signal. Behav. 2020, 15, 1777372. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Das, S.; Bansal, S.; Singh, G.; Sardar, S.; Dhar, H.; Ram, H. Heavy metal stress in rice: Uptake, transport, signaling, and tolerance mechanisms. Physiol. Plant. 2021, 173, 430–448. [Google Scholar] [CrossRef]
- Sinha, A.K.; Jaggi, M.; Raghuram, B.; Tuteja, N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 2011, 6, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and Calcineurin B–like Proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14, S389–S400. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541. [Google Scholar] [CrossRef]
- Mallhi, Z.I.; Rizwan, M.; Mansha, A.; Ali, Q.; Asim, S.; Ali, S.; Hussain, A.; Alrokayan, S.H.; Khan, H.A.; Alam, P.; et al. Citric Acid Enhances Plant Growth, Photosynthesis, and Phytoextraction of Lead by Alleviating the Oxidative Stress in Castor Beans. Plants 2019, 8, 525. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ding, L.; Xia, Y.; Wang, F.; Zhu, C. Emerging Roles of microRNAs in Plant Heavy Metal Tolerance and Homeostasis. J. Agric. Food Chem. 2020, 68, 1958–1965. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Mishra, M.; Patil, G.; Mulkey, S.; Ramawat, N.; Pratap Singh, V.; Deshmukh, R.; Kumar Tripathi, D.; Nguyen, H.T.; Sharma, S. Avenues of the membrane transport system in adaptation of plants to abiotic stresses. Crit. Rev. Biotechnol. 2019, 39, 861–883. [Google Scholar] [CrossRef] [PubMed]
- Laganowsky, A.; Reading, E.; Allison, T.M.; Ulmschneider, M.B.; Degiacomi, M.; Baldwin, A.J.; Robinson, C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nat. Cell Biol. 2014, 510, 172–175. [Google Scholar] [CrossRef]
- Hilgemann, D.W.; Dai, G.; Collins, A.; Larricia, V.; Magi, S.; Deisl, C.; Fine, M. Lipid signaling to membrane proteins: From second messengers to membrane domains and adapter-free endocytosis. J. Gen. Physiol. 2018, 150, 211–224. [Google Scholar] [CrossRef]
- Minami, A.; Takahashi, D.; Kawamura, Y.; Uemura, M. Isolation of Plasma Membrane and Plasma Membrane Microdomains. Adv. Struct. Saf. Stud. 2017, 1511, 199–212. [Google Scholar] [CrossRef]
- Cassim, A.M.; Gouguet, P.; Gronnier, J.; Laurent, N.; Germain, V.; Grison, M.S.; Boutté, Y.; Gerbeau-Pissot, P.; Simon-Plas, F.; Mongrand, S. Plant lipids: Key players of plasma membrane organization and function. Prog. Lipid Res. 2019, 73, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Sarabia, L.D.; Boughton, B.A.; Rupasinghe, T.; Callahan, D.; Hill, C.; Roessner, U. Comparative spatial lipidomics analysis reveals cellular lipid remodelling in different developmental zones of barley roots in response to salinity. Plant Cell Environ. 2019, 43, 327–343. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Singla-Pareek, S.L.; Pareek, A. Membrane dynamics during individual and combined abiotic stresses in plants and tools to study the same. Physiol. Plant. 2021, 171, 653–676. [Google Scholar] [CrossRef] [PubMed]
- De Caroli, M.; Furini, A.; DalCorso, G.; Rojas, M.; Di Sansebastiano, G.-P. Endomembrane Reorganization Induced by Heavy Metals. Plants 2020, 9, 482. [Google Scholar] [CrossRef]
- Bellini, E.; Maresca, V.; Betti, C.; Castiglione, M.R.; Fontanini, D.; Capocchi, A.; Sorce, C.; Borsò, M.; Bruno, L.; Sorbo, S.; et al. The Moss Leptodictyum riparium Counteracts Severe Cadmium Stress by Activation of Glutathione Transferase and Phytochelatin Synthase, but Slightly by Phytochelatins. Int. J. Mol. Sci. 2020, 21, 1583. [Google Scholar] [CrossRef] [PubMed]
- Barozzi, F.; Papadia, P.; Stefano, G.; Renna, L.; Brandizzi, F.; Migoni, D.; Fanizzi, F.P.; Piro, G.; Di Sansebastiano, G.-P. Variation in Membrane Trafficking Linked to SNARE AtSYP51 Interaction With Aquaporin NIP1;1. Front. Plant Sci. 2019, 9, 1949. [Google Scholar] [CrossRef]
- Emamverdian, A.; Ding, Y.; Mokhberdoran, F.; Xie, Y. Heavy Metal Stress and Some Mechanisms of Plant Defense Response. Sci. World J. 2015, 2015, 1–18. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.-J.; Mimura, T. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant Cell Environ. 2016, 39, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Kambe, T. The Functions of Metallothionein and ZIP and ZnT Transporters: An Overview and Perspective. Int. J. Mol. Sci. 2016, 17, 336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Zhao, K.X.; Yang, Z.M. Identification of genomic ATP binding cassette (ABC) transporter genes and Cd-responsive ABCs in Brassica napus. Gene 2018, 664, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Barber-Zucker, S.; Hall, J.; Froes, A.; Kolusheva, S.; MacMillan, F.; Zarivach, R. The cation diffusion facilitator protein MamM’s cytoplasmic domain exhibits metal-type dependent binding modes and discriminates against Mn2+. J. Biol. Chem. 2020, 295, 16614–16629. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, F.; Liu, J.; Xie, W.; Zhang, L.; Chen, Z.; Peng, Z.; Ou, Y.; Yao, Y. Genome-Wide Identification of Metal Tolerance Protein Genes in Populus trichocarpa and Their Roles in Response to Various Heavy Metal Stresses. Int. J. Mol. Sci. 2020, 21, 1680. [Google Scholar] [CrossRef] [PubMed]
- Yokosho, K.; Yamaji, N.; Ma, J.F. Buckwheat FeNramp5 Mediates High Manganese Uptake in Roots. Plant Cell Physiol. 2021, 62, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Modareszadeh, M.; Bahmani, R.; Kim, D.; Hwang, S. CAX3 (cation/proton exchanger) mediates a Cd tolerance by decreasing ROS through Ca elevation in Arabidopsis. Plant Mol. Biol. 2021, 105, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Vats, S.; Sudhakaran, S.; Bhardwaj, A.; Mandlik, R.; Sharma, Y.; Kumar, S.; Tripathi, D.K.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Targeting aquaporins to alleviate hazardous metal(loid)s imposed stress in plants. J. Hazard. Mater. 2021, 408, 124910. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, Y.; Liu, Z.; Tian, J.; Liang, L.; Qiu, Y.; Wang, G.; Du, Q.; Cheng, D.; Cai, H.; et al. A high activity zinc transporter OsZIP9 mediates zinc uptake in rice. Plant J. 2020, 103, 1695–1709. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Tang, L.; Peng, Y.; Qian, M.; Guo, Y.; Rui, H.; Zhang, F.; Hu, Z.; Chen, Y.; et al. The root iron transporter 1 governs cadmium uptake in Vicia sativa roots. J. Hazard. Mater. 2020, 398, 122873. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, T.; Burla, B.; Lee, Y.; Martinoia, E.; Nagy, R. Functions of ABC transporters in plants. Essays Biochem. 2011, 50, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Dahuja, A.; Kumar, R.R.; Sakhare, A.; Watts, A.; Singh, B.; Goswami, S.; Sachdev, A.; Praveen, S. Role of ATP-binding cassette transporters in maintaining plant homeostasis under abiotic and biotic stresses. Physiol. Plant. 2021, 171, 785–801. [Google Scholar] [CrossRef]
- Song, W.-Y.; Park, J.; Cozatl, D.M.; Suter-Grotemeyer, M.; Shim, D.; Hörtensteiner, S.; Geisler, M.; Weder, B.; Rea, P.A.; Rentsch, D.; et al. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 21187–21192. [Google Scholar] [CrossRef]
- Ullah, I.; Wang, Y.; Eide, D.J.; Dunwell, J.M. Evolution, and functional analysis of Natural Resistance-Associated Macrophage Proteins (NRAMPs) from Theobroma cacao and their role in cadmium accumulation. Sci. Rep. 2018, 8, 14412. [Google Scholar] [CrossRef]
- Lanquar, V.; Ramos, M.S.; Lelièvre, F.; Barbier-Brygoo, H.; Krieger-Liszkay, A.; Kraemer, U.; Thomine, S. Export of Vacuolar Manganese by AtNRAMP3 and AtNRAMP4 Is Required for Optimal Photosynthesis and Growth under Manganese Deficiency. Plant Physiol. 2010, 152, 1986–1999. [Google Scholar] [CrossRef]
- Wang, N.; Qiu, W.; Dai, J.; Guo, X.; Lu, Q.; Wang, T.; Li, S.; Liu, T.; Zuo, Y. AhNRAMP1 Enhances Manganese and Zinc Uptake in Plants. Front. Plant Sci. 2019, 10, 415. [Google Scholar] [CrossRef]
- Barberon, M.; Geldner, N. Radial Transport of Nutrients: The Plant Root as a Polarized Epithelium. Plant Physiol. 2014, 166, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Schenk, H.J.; Jansen, S.; Hölttä, T. Positive pressure in xylem and its role in hydraulic function. New Phytol. 2020, 230, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Ovečka, M.; Takáč, T. Managing heavy metal toxicity stress in plants: Biological and biotechnological tools. Biotechnol. Adv. 2014, 32, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Zhang, R.; Zhou, F.; Li, P. Foliar uptake and transport of atmospheric trace metals bounded on particulate matters in epiphytic Tillandsia brachycaulos. Int. J. Phytoremediation 2021, 23, 400–406. [Google Scholar] [CrossRef]
- Shahid, M.; Dumat, C.; Khalid, S.; Schreck, E.; Xiong, T.; Niazi, N.K. Foliar heavy metal uptake, toxicity and detoxification in plants: A comparison of foliar and root metal uptake. J. Hazard. Mater. 2017, 325, 36–58. [Google Scholar] [CrossRef] [PubMed]
- Kapilan, R.; Vaziri, M.; Zwiazek, J.J. Regulation of aquaporins in plants under stress. Biol. Res. 2018, 51, 4. [Google Scholar] [CrossRef]
- Kaldenhoff, R.; Fischer, M. Aquaporins in plants. Acta Physiol. 2006, 187, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ballesta, M.D.C.; Carvajal, M. Mutual Interactions between Aquaporins and Membrane Components. Front. Plant Sci. 2016, 7, 1322. [Google Scholar] [CrossRef]
- Quiroga, G.; Erice, G.; Aroca, R.; Ruiz-Lozano, J.M. Elucidating the Possible Involvement of Maize Aquaporins in the Plant Boron Transport and Homeostasis Mediated by Rhizophagus irregularis under Drought Stress Conditions. Int. J. Mol. Sci. 2020, 21, 1748. [Google Scholar] [CrossRef]
- Rios, J.J.; Martínez-Ballesta, M.C.; Ruiz, J.M.; Blasco, B.; Carvajal, M. Silicon-mediated Improvement in Plant Salinity Tolerance: The Role of Aquaporins. Front. Plant Sci. 2017, 8, 948. [Google Scholar] [CrossRef]
- Feng, Z.-J.; Liu, N.; Zhang, G.-W.; Niu, F.-G.; Xu, S.-C.; Gong, Y.-M. Investigation of the AQP Family in Soybean and the Promoter Activity of TIP2;6 in Heat Stress and Hormone Responses. Int. J. Mol. Sci. 2019, 20, 262. [Google Scholar] [CrossRef]
- Neri, A.; Traversari, S.; Andreucci, A.; Francini, A.; Sebastiani, L. The Role of Aquaporin Overexpression in the Modulation of Transcription of Heavy Metal Transporters under Cadmium Treatment in Poplar. Plants 2020, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Chaumont, F.; Moshelion, M.; Daniels, M.J. Regulation of plant aquaporin activity. Biol. Cell 2005, 97, 749–764. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Briggs, M.M.; McIntosh, T.J. Water Permeability of Aquaporin-4 Channel Depends on Bilayer Composition, Thickness, and Elasticity. Biophys. J. 2012, 103, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Bermúdez, A.; Recuero, S.; Llavanera, M.; Mateo-Otero, Y.; Sandu, A.; Barranco, I.; Ribas-Maynou, J.; Yeste, M. Aquaporins Are Essential to Maintain Motility and Membrane Lipid Architecture During Mammalian Sperm Capacitation. Front. Cell Dev. Biol. 2021, 9, 2451. [Google Scholar] [CrossRef]
- Hachez, C.; Laloux, T.; Reinhardt, H.; Cavez, D.; Degand, H.; Grefen, C.; De Rycke, R.; Inzé, D.; Blatt, M.R.; Russinova, E.; et al. Arabidopsis SNAREs SYP61 and SYP121 Coordinate the Trafficking of Plasma Membrane Aquaporin PIP2;7 to Modulate the Cell Membrane Water Permeability. Plant Cell 2014, 26, 3132–3147. [Google Scholar] [CrossRef] [PubMed]
- Besserer, A.; Burnotte, E.; Bienert, G.; Chevalier, A.S.; Errachid, A.; Grefen, C.; Blatt, M.R.; Chaumont, F. Selective Regulation of Maize Plasma Membrane Aquaporin Trafficking and Activity by the SNARE SYP121. Plant Cell 2012, 24, 3463–3481. [Google Scholar] [CrossRef] [PubMed]
- Sharipova, G.; Veselov, D.; Kudoyarova, G.; Fricke, W.; Dodd, I.C.; Katsuhara, M.; Furuichi, T.; Ivanov, I.; Veselov, S. Exogenous application of abscisic acid (ABA) increases root and cell hydraulic conductivity and abundance of some aquaporin isoforms in the ABA-deficient barley mutant Az34. Ann. Bot. 2016, 118, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Maurel, C.; Santoni, V.; Luu, D.-T.; Wudick, M.M.; Verdoucq, L. The cellular dynamics of plant aquaporin expression and functions. Curr. Opin. Plant Biol. 2009, 12, 690–698. [Google Scholar] [CrossRef]
- Rusinova, R.; Hobart, E.A.; Koeppe, R.; Andersen, O.S. Phosphoinositides alter lipid bilayer properties. J. Gen. Physiol. 2013, 141, 673–690. [Google Scholar] [CrossRef]
- Ishikawa, F.; Suga, S.; Uemura, T.; Sato, M.H.; Maeshima, M. Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression inArabidopsis thaliana. FEBS Lett. 2005, 579, 5814–5820. [Google Scholar] [CrossRef]
- Verdoucq, L.; Grondin, A.; Maurel, C. Structure–function analysis of plant aquaporin AtPIP2;1 gating by divalent cations and protons. Biochem. J. 2008, 415, 409–416. [Google Scholar] [CrossRef]
- Gattolin, S.; Sorieul, M.; Frigerio, L. Mapping of Tonoplast Intrinsic Proteins in Maturing and Germinating Arabidopsis Seeds Reveals Dual Localization of Embryonic TIPs to the Tonoplast and Plasma Membrane. Mol. Plant 2011, 4, 180–189. [Google Scholar] [CrossRef]
- Sabir, F.; Gomes, S.; Loureiro-Dias, M.C.; Soveral, G.; Prista, C. Molecular and Functional Characterization of Grapevine NIPs through Heterologous Expression in aqy-Null Saccharomyces cerevisiae. Int. J. Mol. Sci. 2020, 21, 663. [Google Scholar] [CrossRef]
- Bienert, G.; Bienert, M.D.; Jahn, T.P.; Boutry, M.; Chaumont, F. Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 2011, 66, 306–317. [Google Scholar] [CrossRef]
- Simons, K.; Sampaio, J. Membrane Organization and Lipid Rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef] [PubMed]
- Gromiha, M.M.; Ou, Y.-Y. Bioinformatics approaches for functional annotation of membrane proteins. Briefings Bioinform. 2013, 15, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Lebrun-Garcia, A.; Bourque, S.; Binet, M.-N.; Ouaked, F.; Wendehenne, D.; Chiltz, A.; Schäffner, A.; Pugin, A. Involvement of plasma membrane proteins in plant defense responses. Analysis of the cryptogein signal transduction in tobacco. Biochim. 1999, 81, 663–668. [Google Scholar] [CrossRef]
- Cheng, F.; Brewer, C. Conversion of protein-rich lignocellulosic wastes to bio-energy: Review and recommendations for hydrolysis + fermentation and anaerobic digestion. Renew. Sustain. Energy Rev. 2021, 146, 111167. [Google Scholar] [CrossRef]
- Kosakivska, I.V.; Babenko, L.M.; Romanenko, K.O.; Korotka, I.Y.; Potters, G. Molecular mechanisms of plant adaptive responses to heavy metals stress. Cell Biol. Int. 2021, 45, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Pourrut, B.; Dumat, C.; Nadeem, M.; Aslam, M.; Pinelli, E. Heavy-metal-induced reactive oxygen species: Phytotoxicity and physicochemical changes in plants. Rev. Environ. Contam Toxicol 2014, 232, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Keyster, M.; Niekerk, L.-A.; Basson, G.; Carelse, M.; Bakare, O.; Ludidi, N.; Klein, A.; Mekuto, L.; Gokul, A. Decoding Heavy Metal Stress Signalling in Plants: Towards Improved Food Security and Safety. Plants 2020, 9, 1781. [Google Scholar] [CrossRef]
- Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular Mechanism for the Regulation of ABA Homeostasis During Plant Development and Stress Responses. Int. J. Mol. Sci. 2018, 19, 3643. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Li, Z.; Waadt, R.; Schroeder, J.I. SnapShot: Abscisic Acid Signaling. Cell 2017, 171, 1708. [Google Scholar] [CrossRef] [PubMed]
- Jegla, T.; Busey, G.; Assmann, S.M. Evolution and Structural Characteristics of Plant Voltage-Gated K+ Channels. Plant Cell 2018, 30, 2898–2909. [Google Scholar] [CrossRef]
- Pan, W.; You, Y.; Shentu, J.-L.; Weng, Y.-N.; Wang, S.-T.; Xu, Q.-R.; Liu, H.-J.; Du, S.-T. Abscisic acid (ABA)-importing transporter 1 (AIT1) contributes to the inhibition of Cd accumulation via exogenous ABA application in Arabidopsis. J. Hazard. Mater. 2020, 391, 122189. [Google Scholar] [CrossRef]
- Shi, W.-G.; Liu, W.; Yu, W.; Zhang, Y.; Ding, S.; Li, H.; Mrak, T.; Kraigher, H.; Luo, Z.-B. Abscisic acid enhances lead translocation from the roots to the leaves and alleviates its toxicity in Populus × canescens. J. Hazard. Mater. 2019, 362, 275–285. [Google Scholar] [CrossRef]
- Hu, B.; Deng, F.; Chen, G.; Chen, X.; Gao, W.; Long, L.; Xia, J.; Chen, Z.-H. Evolution of Abscisic Acid Signaling for Stress Responses to Toxic Metals and Metalloids. Front. Plant Sci. 2020, 11, 909. [Google Scholar] [CrossRef]
- Tao, Q.; Jupa, R.; Liu, Y.; Luo, J.; Li, J.; Kováč, J.; Li, B.; Li, Q.; Wu, K.; Liang, Y.; et al. Abscisic acid-mediated modifications of radial apoplastic transport pathway play a key role in cadmium uptake in hyperaccumulator Sedum alfredii. Plant Cell Environ. 2018, 42, 1425–1440. [Google Scholar] [CrossRef]
- Cai, S.; Chen, G.; Wang, Y.; Huang, Y.; Marchant, D.B.; Wang, Y.; Yang, Q.; Dai, F.; Hills, A.; Franks, P.J.; et al. Evolutionary Conservation of ABA Signaling for Stomatal Closure. Plant Physiol. 2017, 174, 732–747. [Google Scholar] [CrossRef]
- Lu, Q.; Weng, Y.; You, Y.; Xu, Q.; Li, H.; Li, Y.; Liu, H.; Du, S. Inoculation with abscisic acid (ABA)-catabolizing bacteria can improve phytoextraction of heavy metal in contaminated soil. Environ. Pollut. 2020, 257, 113497. [Google Scholar] [CrossRef]
- Lu, Q.; Chen, S.; Li, Y.; Zheng, F.; He, B.; Gu, M. Exogenous abscisic acid (ABA) promotes cadmium (Cd) accumulation in Sedum alfredii Hance by regulating the expression of Cd stress response genes. Environ. Sci. Pollut. Res. 2020, 27, 8719–8731. [Google Scholar] [CrossRef]
- Wang, T.; Hua, Y.; Chen, M.; Zhang, J.; Guan, C.; Zhang, Z. Mechanism Enhancing Arabidopsis Resistance to Cadmium: The Role of NRT1.5 and Proton Pump. Front. Plant Sci. 2018, 9, 1892. [Google Scholar] [CrossRef]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-Type ATPase Heavy Metal Transporters with Roles in Essential Zinc Homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef]
- Mills, R.F.; Francini, A.; Da Rocha, P.S.F.; Baccarini, P.J.; Aylett, M.; Krijger, G.C.; Williams, L.E. The plant P1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett. 2004, 579, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Xia, J.; Mitani-Ueno, N.; Yokosho, K.; Ma, J.F. Preferential Delivery of Zinc to Developing Tissues in Rice Is Mediated by P-Type Heavy Metal ATPase OsHMA2. Plant Physiol. 2013, 162, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase Allowing Cd/Zn/Co/Pb Vacuolar Storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.F.; Ando, T.; Yano, M.; Ma, J.F. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef]
- Chao, D.-Y.; Silva, A.; Baxter, I.; Huang, Y.S.; Nordborg, M.; Danku, J.; Lahner, B.; Yakubova, E.; Salt, D.E. Genome-Wide Association Studies Identify Heavy Metal ATPase3 as the Primary Determinant of Natural Variation in Leaf Cadmium in Arabidopsis thaliana. PLoS Genet. 2012, 8, e1002923. [Google Scholar] [CrossRef]
- Liu, C.; Gao, Z.; Shang, L.; Yang, C.; Ruan, B.; Zeng, D.; Guo, L.; Zhao, F.; Huang, C.; Qian, Q. Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and Japonica rice. J. Integr. Plant Biol. 2020, 62, 314–329. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, K.-F.; Poysa, V.; Shi, C.; Zhou, Y.-H. A Single Point Mutation in GmHMA3 Affects Cadimum (Cd) Translocation and Accumulation in Soybean Seeds. Mol. Plant 2012, 5, 1154–1156. [Google Scholar] [CrossRef][Green Version]
- Lin, S.-H.; Kuo, H.-F.; Canivenc, G.; Lin, C.-S.; Lepetit, M.; Hsu, P.-K.; Tillard, P.; Lin, H.-L.; Wang, Y.-Y.; Tsai, C.-B.; et al. Mutation of the Arabidopsis NRT1.5 Nitrate Transporter Causes Defective Root-to-Shoot Nitrate Transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar] [CrossRef]
- Li, J.-Y.; Fu, Y.-L.; Pike, S.M.; Bao, J.; Tian, W.; Zhang, Y.; Chen, C.-Z.; Zhang, Y.; Li, H.-M.; Huang, J.; et al. TheArabidopsisNitrate Transporter NRT1.8 Functions in Nitrate Removal from the Xylem Sap and Mediates Cadmium Tolerance. Plant Cell 2010, 22, 1633–1646. [Google Scholar] [CrossRef]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB Transcription Factor Superfamily of Arabidopsis: Expression Analysis and Phylogenetic Comparison with the Rice MYB Family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.; Cheng, D.; Zhang, G.; Zhu, D.; Chen, Y.; Tan, M. The role of ZmWRKY4 in regulating maize antioxidant defense under cadmium stress. Biochem. Biophys. Res. Commun. 2017, 482, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, Q.; Wang, Y.; Wu, T.; Yang, Y.; Zhang, X.; Han, Z.; Xu, X. Ethylene response factor AtERF72 negatively regulates Arabidopsis thaliana response to iron deficiency. Biochem. Biophys. Res. Commun. 2017, 491, 862–868. [Google Scholar] [CrossRef]
- Tiwari, P.; Indoliya, Y.; Chauhan, A.S.; Singh, P.; Singh, P.K.; Singh, P.C.; Srivastava, S.; Pande, V.; Chakrabarty, D. Auxin-salicylic acid cross-talk ameliorates OsMYB–R1 mediated defense towards heavy metal, drought and fungal stress. J. Hazard. Mater. 2020, 399, 122811. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhou, J.; Jie, Y.; Xing, H.; Zhong, Y.; Yu, W.; She, W.; Ma, Y.; Liu, Z.; Zhang, Y. A Ramie bZIP Transcription Factor BnbZIP2 Is Involved in Drought, Salt, and Heavy Metal Stress Response. DNA Cell Biol. 2016, 35, 776–786. [Google Scholar] [CrossRef]
- An, X.; Chen, J.; Jin, G. Transcriptome profiling of kenaf (Hibiscus cannabinus L.) under plumbic stress conditions implies the involvement of NAC transcription factors regulating reactive oxygen species-dependent programmed cell death. PeerJ 2020, 8, e8733. [Google Scholar] [CrossRef]
- Cai, Y.; Li, Y.; Liang, G. FIT and bHLH Ib transcription factors modulate iron and copper crosstalk in Arabidopsis. Plant Cell Environ. 2021, 44, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Xian, P.; Wang, H.; Lin, R.; Lian, T.; Cheng, Y.; Ma, Q.; Nian, H. Transcription Factor GmWRKY142 Confers Cadmium Resistance by Up-Regulating the Cadmium Tolerance 1-Like Genes. Front. Plant Sci. 2020, 11, 724. [Google Scholar] [CrossRef]
- Sheng, Y.; Yan, X.; Huang, Y.; Han, Y.; Zhang, C.; Ren, Y.; Fan, T.; Xiao, F.; Liu, Y.; Cao, S. The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 2018, 42, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, C.; Du, J.; Liu, H.; Cui, Y.; Zhang, Y.; He, Y.; Wang, Y.; Chu, C.; Feng, Z.; et al. Co-Overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-Enhanced Cadmium Tolerance via Increased Cadmium Sequestration in Roots and Improved Iron Homeostasis of Shoots. Plant Physiol. 2012, 158, 790–800. [Google Scholar] [CrossRef]
- Wang, N.; Cui, Y.; Liu, Y.; Fan, H.; Du, J.; Huang, Z.; Yuan, Y.; Wu, H.; Ling, H.-Q. Requirement and Functional Redundancy of Ib Subgroup bHLH Proteins for Iron Deficiency Responses and Uptake in Arabidopsis thaliana. Mol. Plant 2013, 6, 503–513. [Google Scholar] [CrossRef]
- Sanz-Carbonell, A.; Marques, M.C.; Bustamante, A.; Fares, M.A.; Rodrigo, G.; Gomez, G. Inferring the regulatory network of the miRNA-mediated response to biotic and abiotic stress in melon. BMC Plant Biol. 2019, 19, 78. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and Their Regulatory Roles in Plant–Environment Interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.; Wani, S. MicroRNAs As Potential Targets for Abiotic Stress Tolerance in Plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef]
- Siddiqui, Z.; Abbas, Z.K.; Ansari, M.W.; Khan, M.N. The role of miRNA in somatic embryogenesis. Genomics 2019, 111, 1026–1033. [Google Scholar] [CrossRef]
- Liang, G.; Yu, D. Reciprocal regulation among miR395,APSandSULTR2;1inArabidopsis thaliana. Plant Signal. Behav. 2010, 5, 1257–1259. [Google Scholar] [CrossRef]
- Meng, J.G.; Zhang, X.D.; Tan, S.K.; Zhao, K.X.; Yang, Z.M. Genome-wide identification of Cd-responsive NRAMP transporter genes and analyzing expression of NRAMP 1 mediated by miR167 in Brassica napus. BioMetals 2017, 30, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, Y.; Jiang, Z.; Wang, F.; Jiang, Q.; Sun, J.; Chen, Z.; Zhu, C. MicroRNA268 Overexpression Affects Rice Seedling Growth under Cadmium Stress. J. Agric. Food Chem. 2017, 65, 5860–5867. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Villalobos, L.I.A.C.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nat. Cell Biol. 2007, 446, 640–645. [Google Scholar] [CrossRef] [PubMed]
- García, J.J.; Martínez-Ballarín, E.; Millán-Plano, S.; Allué, J.; Albendea, C.; Fuentes, L.; Escanero, J. Effects of trace elements on membrane fluidity. J. Trace Elements Med. Biol. 2005, 19, 19–22. [Google Scholar] [CrossRef]
- Singh, P.; Sharma, V.K.; Singha, S.; Sakai, V.G.; Mukhopadhyay, R.; Das, R.; Pal, S.K. Unraveling the Role of Monoolein in Fluidity and Dynamical Response of a Mixed Cationic Lipid Bilayer. Langmuir 2019, 35, 4682–4692. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Liu, L.; Barkla, B.J. Membrane Lipid Remodeling in Response to Salinity. Int. J. Mol. Sci. 2019, 20, 4264. [Google Scholar] [CrossRef]
- Cacas, J.-L.; Buré, C.; Grosjean, K.; Gerbeau-Pissot, P.; Lherminier, J.; Rombouts, Y.; Maes, E.; Bossard, C.; Gronnier, J.; Furt, F.; et al. Revisiting Plant Plasma Membrane Lipids in Tobacco: A Focus on Sphingolipids. Plant Physiol. 2016, 170, 367–384. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Huang, X.; Kropat, J.; Henras, A.; Merchant, S.S.; Dickson, R.C.; Chanfreau, G.F. Sphingolipid Signaling Mediates Iron Toxicity. Cell Metab. 2012, 16, 90–96. [Google Scholar] [CrossRef]
- Corbacho, J.; Inês, C.; Paredes, M.A.; Labrador, J.; Cordeiro, A.M.; Gallardo, M.; Gomez-Jimenez, M.C. Modulation of sphingolipid long-chain base composition and gene expression during early olive-fruit development, and putative role of brassinosteroid. J. Plant Physiol. 2018, 231, 383–392. [Google Scholar] [CrossRef]
- Liu, N.-J.; Hou, L.-P.; Bao, J.-J.; Wang, L.-J.; Chen, X.-Y. Sphingolipid metabolism, transport and functions in plants: Recent progress and future perspectives. Plant Commun. 2021, 2, 100214. [Google Scholar] [CrossRef]
- Magnin-Robert, M.; Le Bourse, D.; Markham, J.E.; Dorey, S.; Clément, C.; Baillieul, F.; Dhondt-Cordelier, S. Modifications of sphingolipid content affect tolerance to hemibiotrophic and necrotrophic pathogens by modulating plant defense responses in Arabidopsis. Plant Physiol. 2015, 169, 2255–2274. [Google Scholar] [CrossRef]
- Michell, R.H.; Wakelam, M. Second Messengers: Sphingolipid signalling. Curr. Biol. 1994, 4, 370–373. [Google Scholar] [CrossRef]
- Coursol, S.; Fan, L.-M.; Le Stunff, H.; Spiegel, S.; Gilroy, S.; Assmann, S.M. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nat. Cell Biol. 2003, 423, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, G.L. The Fluid—Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40years. Biochim. et Biophys. Acta (BBA)-Biomembr. 2014, 1838, 1451–1466. [Google Scholar] [CrossRef]
- Tycko, R. BIOMOLECULAR SOLID STATE NMR: Advances in Structural Methodology and Applications to Peptide and Protein Fibrils. Annu. Rev. Phys. Chem. 2001, 52, 575–606. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.D.; Lorigan, G.A. Electron Paramagnetic Resonance as a Tool for Studying Membrane Proteins. Biomolecules 2020, 10, 763. [Google Scholar] [CrossRef]
- Bolla, J.R.; Corey, R.; Sahin, C.; Gault, J.; Hummer, A.; Hopper, J.T.S.; Lane, D.P.; Drew, D.; Allison, T.M.; Stansfeld, P.J.; et al. A Mass-Spectrometry-Based Approach to Distinguish Annular and Specific Lipid Binding to Membrane Proteins. Angew. Chem. Int. Ed. 2020, 59, 3523–3528. [Google Scholar] [CrossRef]
- Putta, P.; Creque, E.; Piontkivska, H.; Kooijman, E.E. Lipid−protein interactions for ECA1 an N-ANTH domain protein involved in stress signaling in plants. Chem. Phys. Lipids 2020, 231, 104919. [Google Scholar] [CrossRef]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Lee, A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta Biomembr. 2004, 1666, 62–87. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.G. Lipid–protein interactions. Biochem. Soc. Trans. 2011, 39, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, T.A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nat. Cell Biol. 2007, 450, 663–669. [Google Scholar] [CrossRef]
- Von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006, 7, 909–918. [Google Scholar] [CrossRef]
- Lange, C.; Nett, J.H.; Trumpower, B.L.; Hunte, C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001, 20, 6591–6600. [Google Scholar] [CrossRef]
- Eghiaian, F. Lipid Chirality Revisited: A Change in Lipid Configuration Transforms Membrane-Bound Protein Domains. Biophys. J. 2015, 108, 2757–2758. [Google Scholar] [CrossRef]
- Ho, H.; Miu, A.; Alexander, M.K.; Garcia, N.K.; Oh, A.; Zilberleyb, I.; Reichelt, M.; Austin, C.D.; Tam, C.; Shriver, S.; et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nat. Cell Biol. 2018, 557, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Kuk, A.C.Y.; Hao, A.; Guan, Z.; Lee, S.-Y. Visualizing conformation transitions of the Lipid II flippase MurJ. Nat. Commun. 2019, 10, 1736. [Google Scholar] [CrossRef] [PubMed]
- Weinert, T.; Skopintsev, P.; James, D.; Dworkowski, F.; Panepucci, E.; Kekilli, D.; Furrer, A.; Brünle, S.; Mous, S.; Ozerov, D.; et al. Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography. Science 2019, 365, 61–65. [Google Scholar] [CrossRef]
- Tilegenova, C.; Cortes, D.M.; Jahovic, N.; Hardy, E.; Hariharan, P.; Guan, L.; Cuello, L.G. Structure, function, and ion-binding properties of a K+ channel stabilized in the 2,4-ion–bound configuration. Proc. Natl. Acad. Sci. USA 2019, 116, 16829–16834. [Google Scholar] [CrossRef]
- Neutze, R.; Brändén, G.; Schertler, G.F. Membrane protein structural biology using X-ray free electron lasers. Curr. Opin. Struct. Biol. 2015, 33, 115–125. [Google Scholar] [CrossRef]
- Fotiadis, D. Atomic force microscopy for the study of membrane proteins. Curr. Opin. Biotechnol. 2012, 23, 510–515. [Google Scholar] [CrossRef]
- Liao, X.; Zhang, B.; Blatt, M.R.; Jenkins, G.I. A FRET method for investigating dimer/monomer status and conformation of the UVR8 photoreceptor. Photochem. Photobiol. Sci. 2019, 18, 367–374. [Google Scholar] [CrossRef]
- Gupta, K.; Donlan, J.; Hopper, J.T.S.; Uzdavinys, P.; Landreh, M.; Struwe, W.; Drew, D.; Baldwin, A.J.; Stansfeld, P.; Robinson, C. The role of interfacial lipids in stabilizing membrane protein oligomers. Nat. Cell Biol. 2017, 541, 421–424. [Google Scholar] [CrossRef]
- Muallem, S.; Chung, W.Y.; Jha, A.; Ahuja, M. Lipids at membrane contact sites: Cell signaling and ion transport. EMBO Rep. 2017, 18, 1893–1904. [Google Scholar] [CrossRef]
- Tronin, A.Y.; Maciunas, L.J.; Grasty, K.C.; Loll, P.J.; Ambaye, H.A.; Parizzi, A.A.; Lauter, V.; Geragotelis, A.D.; Freites, J.A.; Tobias, D.J.; et al. Voltage-Dependent Profile Structures of a Kv-Channel via Time-Resolved Neutron Interferometry. Biophys. J. 2019, 117, 751–766. [Google Scholar] [CrossRef]
- Yang, F.; Zheng, J. Understand spiciness: Mechanism of TRPV1 channel activation by capsaicin. Protein Cell 2017, 8, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Liu, Z.; Li, J. From membrane tension to channel gating: A principal energy transfer mechanism for mechanosensitive channels. Protein Sci. 2016, 25, 1954–1964. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.S. Store-operated calcium channels: From function to structure and back again. Cold Spring Harb. Perspect. Biol. 2020, 12, a035055. [Google Scholar] [CrossRef] [PubMed]
- Uehara, C.; Takeda, K.; Ibuki, T.; Furuta, T.; Hoshi, N.; Tanudjaja, E.; Uozumi, N. Analysis of Arabidopsis TPK2 and KCO3 reveals structural properties required for K+ channel function. Channels 2020, 14, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Deshmukh, R.; Muthamilarasan, M.; Rani, R.; Prasad, M. Versatile roles of aquaporin in physiological processes and stress tolerance in plants. Plant Physiol. Biochem. 2020, 149, 178–189. [Google Scholar] [CrossRef]
- García-Rubio, D.L.; de la Mora, M.; Cerecedo, D.; Blesa, J.M.S.; Villagrán-Muniz, M. An optical-based biosensor of the epithelial sodium channel as a tool for diagnosing hypertension. Biosens. Bioelectron. 2020, 157, 112151. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.J.; Baenziger, J.E. Ion channels as lipid sensors: From structures to mechanisms. Nat. Chem. Biol. 2020, 16, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Choveau, F.S.; Abderemane-Ali, F.; Coyan, F.C.; Es-Salah-Lamoureux, Z.; Baró, I.; Loussouarn, G. Opposite Effects of the S4–S5 Linker and PIP2 on Voltage-Gated Channel Function: KCNQ1/KCNE1 and Other Channels. Front. Pharmacol. 2012, 3, 125. [Google Scholar] [CrossRef]
- Urquhart, W.; Chin, K.; Ung, H.; Moeder, W.; Yoshioka, K. The cyclic nucleotide-gated channels AtCNGC11 and 12 are involved in multiple Ca2+-dependent physiological responses and act in a synergistic manner. J. Exp. Bot. 2011, 62, 3671–3682. [Google Scholar] [CrossRef]
- Jarratt-Barnham, E.; Wang, L.; Ning, Y.; Davies, J. The Complex Story of Plant Cyclic Nucleotide-Gated Channels. Int. J. Mol. Sci. 2021, 22, 874. [Google Scholar] [CrossRef]
- Moon, J.Y.; Belloeil, C.; Ianna, M.L.; Shin, R. Arabidopsis CNGC Family Members Contribute to Heavy Metal Ion Uptake in Plants. Int. J. Mol. Sci. 2019, 20, 413. [Google Scholar] [CrossRef]
- Kefauver, J.M.; Ward, A.B.; Patapoutian, A. Discoveries in structure and physiology of mechanically activated ion channels. Nat. Cell Biol. 2020, 587, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Iida, H.; Furuichi, T.; Nakano, M.; Toyota, M.; Sokabe, M.; Tatsumi, H. New candidates for mechano-sensitive channels potentially involved in gravity sensing in Arabidopsis thaliana. Plant Biol. 2013, 16, 39–42. [Google Scholar] [CrossRef]
- Shepherd, V.; Beilby, M.; Shimmen, T. Mechanosensory ion channels in charophyte cells: The response to touch and salinity stress. Eur. Biophys. J. 2002, 31, 341–355. [CrossRef]
- Stieger, B.; Steiger, J.; Locher, K.P. Membrane lipids and transporter function. Biochim. et Biophys. Acta (BBA)-Mol. Basis Dis. 2021, 1867, 166079. [Google Scholar] [CrossRef] [PubMed]
- Neumann, J.; Rose-Sperling, D.; Hellmich, U.A. Diverse relations between ABC transporters and lipids: An overview. Biochim. et Biophys. Acta (BBA)-Biomembr. 2017, 1859, 605–618. [Google Scholar] [CrossRef]
- Ogasawara, F.; Kodan, A.; Ueda, K. ABC proteins in evolution. FEBS Lett. 2020, 594, 3876–3881. [Google Scholar] [CrossRef]
- Aye, I.L.; Singh, A.T.; Keelan, J.A. Transport of lipids by ABC proteins: Interactions and implications for cellular toxicity, viability and function. Chem. Interactions 2009, 180, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Spector, A.; Yorek, M. Membrane lipid composition and cellular function. J. Lipid Res. 1985, 26, 1015–1035. [Google Scholar] [CrossRef]
- Sytar, O.; Ghosh, S.; Malinska, H.; Zivcak, M.; Brestic, M. Physiological and molecular mechanisms of metal accumulation in hyperaccumulator plants. Physiol. Plant. 2020, 173, 148–166. [Google Scholar] [CrossRef]
- Casares, D.; Escribá, P.V.; Rosselló, C.A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef]
- Muller, M.P.; Jiang, T.; Sun, C.; Lihan, M.; Pant, S.; Mahinthichaichan, P.; Trifan, A.; Tajkhorshid, E. Characterization of Lipid–Protein Interactions and Lipid-Mediated Modulation of Membrane Protein Function through Molecular Simulation. Chem. Rev. 2019, 119, 6086–6161. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, C.; Guo, J.; Xu, C. Chapter Three-Ionic protein-lipid interactions at the plasma membrane regulate the structure and function of immunoreceptors. In Advances in Immunology; Dong, C., Jiang, Z., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 144, pp. 65–85. [Google Scholar] [CrossRef]
- Wang, P.; Chao, D. Phytoremediation of heavy metal contamination and related molecular mechanisms in plants. Sheng Wu Gong Cheng Xue Bao 2020, 36, 426–435. [Google Scholar]
- Zhang, Y.; Zheng, J. Bioinformatics of Metalloproteins and Metalloproteomes. Molecules 2020, 25, 3366. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-Y.; Yamaki, T.; Yamaji, N.; Ko, D.; Jung, K.H.; Fujii-Kashino, M.; An, G.; Martinoia, E.; Lee, Y.; Ma, J.F. A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. USA 2014, 111, 15699–15704. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Lu, Y.; Zhang, X.; Yang, G.; Chao, D.; Wang, Z.; Shi, M.; Chen, J.; Chao, D.-Y.; Li, R.; et al. The ABC transporter ABCG36 is required for cadmium tolerance in rice. J. Exp. Bot. 2019, 70, 5909–5918. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Peng, Z.; Li, J.; Huang, W.; Liu, Y.; Wang, X.; Xie, S.; Sun, L.; Han, E.; et al. Ectopic Expression of Poplar ABC Transporter PtoABCG36 Confers Cd Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 3293. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, P.; Zanella, L.; De Paolis, A.; Di Litta, D.; Cecchetti, V.; Falasca, G.; Barbieri, M.; Altamura, M.M.; Costantino, P.; Cardarelli, M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 3815–3829. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-Y.; Bovet, L.; Kushnir, S.; Noh, E.W.; Martinoia, E.; Lee, Y. AtATM3 Is Involved in Heavy Metal Resistance in Arabidopsis. Plant Physiol. 2006, 140, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.; Qin, H.; Lin, X.; Tang, D.; Wu, Z.; Luo, W.; Shen, Y.; Dong, F.; Wang, Y.; et al. A rice chloroplast-localized ABC transporter ARG1 modulates cobalt and nickel homeostasis and contributes to photosynthetic capacity. New Phytol. 2020, 228, 163–178. [Google Scholar] [CrossRef]
- Wang, J.; Liang, S.; Xiang, W.; Dai, H.; Duan, Y.; Kang, F.; Chai, T. A repeat region from the Brassica juncea HMA4 gene BjHMA4R is specifically involved in Cd2+ binding in the cytosol under low heavy metal concentrations. BMC Plant Biol. 2019, 19, 89. [Google Scholar] [CrossRef]
- Deng, F.; Yamaji, N.; Xia, J.; Ma, J.F. A Member of the Heavy Metal P-Type ATPase OsHMA5 Is Involved in Xylem Loading of Copper in Rice. Plant Physiol. 2013, 163, 1353–1362. [Google Scholar] [CrossRef]
- Cai, H.; Huang, S.; Che, J.; Yamaji, N.; Ma, J.F. The tonoplast-localized transporter OsHMA3 plays an important role in maintaining Zn homeostasis in rice. J. Exp. Bot. 2019, 70, 2717–2725. [Google Scholar] [CrossRef]
- Wang, X.; Zhi, J.; Liu, X.; Zhang, H.; Liu, H.; Xu, J. Transgenic tobacco plants expressing a P1B-ATPase gene from Populus tomentosa Carr. (PtoHMA5) demonstrate improved cadmium transport. Int. J. Biol. Macromol. 2018, 113, 655–661. [Google Scholar] [CrossRef]
- Migocka, M.; Papierniak-Wygladala, A.; Maciaszczyk-Dziubinska, E.; Posyniak, E.; Kosieradzka, A. Molecular and biochemical properties of two P 1B2 -ATPases, CsHMA3 and CsHMA4, from cucumber. Plant Cell Environ. 2014, 38, 1127–1141. [Google Scholar] [CrossRef]
- Bernal, M.; Testillano, P.S.; Alfonso, M.; Risueño, M.D.C.; Picorel, R.; Yruela, I. Identification and subcellular localization of the soybean copper P1B-ATPase GmHMA8 transporter. J. Struct. Biol. 2007, 158, 46–58. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Liu, Y.; Yu, K.; Zhou, Y. GmHMA3 sequesters Cd to the root endoplasmic reticulum to limit translocation to the stems in soybean. Plant Sci. 2018, 270, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Pedas, P.; Schiller, M.; Vincze, E.; Mills, R.F.; Borg, S.; Møller, A.; Schjoerring, J.K.; Williams, L.E.; Baekgaard, L.; et al. Barley HvHMA1 Is a Heavy Metal Pump Involved in Mobilizing Organellar Zn and Cu and Plays a Role in Metal Loading into Grains. PLoS ONE 2012, 7, e49027. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Zheng, L.; Li, Y.; Zhou, X.; Li, J.; Gu, D.; Xu, E.; Lu, Y.; Chen, X.; et al. The Intracellular Transporter AtNRAMP6 Is Involved in Fe Homeostasis in Arabidopsis. Front. Plant Sci. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Thomine, S.; Lelièvre, F.; Debarbieux, E.; Schroeder, J.I.; Barbier-Brygoo, H. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J. 2003, 34, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, X.; Yao, Q.; Long, D.; Fan, X.; Kang, H.; Zeng, J.; Sha, L.; Zhang, H.; Zhou, Y.; et al. Overexpression of TtNRAMP6 enhances the accumulation of Cd in Arabidopsis. Gene 2019, 696, 225–232. [Google Scholar] [CrossRef]
- Lu, M.; Wang, Z.; Fu, S.; Yang, G.; Shi, M.; Lu, Y.; Wang, X.; Xia, J. Functional characterization of the SbNrat1 gene in sorghum. Plant Sci. 2017, 262, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi-Masuno, T.; Shitan, N.; Sugiyama, A.; Takanashi, K.; Inaba, S.; Kaneko, S.; Yazaki, K. The Crotalaria juncea metal transporter CjNRAMP1 has a high Fe uptake activity, even in an environment with high Cd contamination. Int. J. Phytoremediation 2018, 20, 1427–1437. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Dwivedi, S.; Singh, M.; Tripathi, R.D.; Trivedi, P.K. Expression inArabidopsisand cellular localization reveal involvement of rice NRAMP, OsNRAMP1, in arsenic transport and tolerance. Plant Cell Environ. 2014, 37, 140–152. [Google Scholar] [CrossRef]
- Wu, D.; Yamaji, N.; Yamane, M.; Kashino-Fujii, M.; Sato, K.; Ma, J.F. The HvNramp5 Transporter Mediates Uptake of Cadmium and Manganese, But Not Iron. Plant Physiol. 2016, 172, 1899–1910. [Google Scholar] [CrossRef]
- Liu, X.S.; Feng, S.J.; Zhang, B.Q.; Wang, M.Q.; Cao, H.W.; Rono, J.K.; Chen, X.; Yang, Z.M. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 2019, 19, 283. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells 2010, 29, 551–558. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Mitani-Ueno, N.; Kashino, M.; Ma, J.F. A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 2015, 84, 374–384. [Google Scholar] [CrossRef]
- Huang, S.; Sasaki, A.; Yamaji, N.; Okada, H.; Mitani-Ueno, N.; Ma, J.F. The ZIP Transporter Family Member OsZIP9 Contributes To Root Zinc Uptake in Rice under Zinc-Limited Conditions. Plant Physiol. 2020, 183, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Maślińska-Gromadka, K.; Barabasz, A.; Palusińska, M.; Kozak, K.; Antosiewicz, D. Suppression of NtZIP4A/B Changes Zn and Cd Root-to-Shoot Translocation in a Zn/Cd Status-Dependent Manner. Int. J. Mol. Sci. 2021, 22, 5355. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, X.; Zhou, X.; Li, Y.; Yang, W.; Chen, R. Improving Zinc and Iron Accumulation in Maize Grains Using the Zinc and Iron Transporter ZmZIP5. Plant Cell Physiol. 2019, 60, 2077–2085. [Google Scholar] [CrossRef]
- Yang, F.; Gao, Y.; Liu, J.; Chen, Z.; de Dios, V.R.; Gao, Q.; Zhang, M.; Peng, Z.; Yao, Y. Metal tolerance protein MTP6 is involved in Mn and Co distribution in poplar. Ecotoxicol. Environ. Saf. 2021, 226, 112868. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Gruber, B.D.; Pittman, J.K.; White, R.G.; Leung, H.; Miao, Y.; Jiang, L.; Ryan, P.R.; Richardson, A.E. A role for theAtMTP11gene of Arabidopsis in manganese transport and tolerance. Plant J. 2007, 51, 198–210. [Google Scholar] [CrossRef]
- Gu, D.; Zhou, X.; Ma, Y.; Xu, E.; Yu, Y.; Liu, Y.; Chen, X.; Zhang, W. Expression of a Brassica napus metal transport protein (BnMTP3) in Arabidopsis thaliana confers tolerance to Zn and Mn. Plant Sci. 2021, 304, 110754. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Q.; Xu, W.; Zhao, H.; Guo, F.; Wang, P.; Wang, Y.; Ni, D.; Wang, M.; Wei, C. Identification of MTP gene family in tea plant (Camellia sinensis L.) and characterization of CsMTP8.2 in manganese toxicity. Ecotoxicol. Environ. Saf. 2020, 202, 110904. [Google Scholar] [CrossRef]
- Migocka, M.; Małas, K.; Maciaszczyk-Dziubinska, E.; Papierniak-Wygladala, A.; Posyniak, E.; Garbiec, A. Retracted: Cucumber metal tolerance protein 7 (CsMTP7) is involved in the accumulation of Fe in mitochondria under Fe excess. Plant J. 2018, 95, 988–1003. [Google Scholar] [CrossRef] [PubMed]
- Migocka, M.; Papierniak-Wygladala, A.; Kosieradzka, A.; Posyniak, E.; Maciaszczyk-Dziubinska, E.; Biskup, R.; Garbiec, A.; Marchewka, T. Retracted: Cucumber metal tolerance protein Cs MTP 9 is a plasma membrane H + -coupled antiporter involved in the Mn 2+ and Cd 2+ efflux from root cells. Plant J. 2015, 84, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Li, J.; Li, J.; Li, Y.; Gu, D.; Chen, C.; Cui, J.; Chen, X.; Zhang, W. OsMTP11, a trans-Golgi network localized transporter, is involved in manganese tolerance in rice. Plant Sci. 2018, 274, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Wongkaew, A.; Asayama, K.; Kitaiwa, T.; Nakamura, S.-I.; Kojima, K.; Stacey, G.; Sekimoto, H.; Yokoyama, T.; Ohkama-Ohtsu, N. AtOPT6 Protein Functions in Long-Distance Transport of Glutathione in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1443–1451. [Google Scholar] [CrossRef]
- Hu, Y.T.; Ming, F.; Chen, W.W.; Yan, J.Y.; Xu, Z.Y.; Li, G.X.; Xu, C.Y.; Yang, J.L.; Zheng, S.J. TcOPT3, a Member of Oligopeptide Transporters from the Hyperaccumulator Thlaspi caerulescens, Is a Novel Fe/Zn/Cd/Cu Transporter. PLoS ONE 2012, 7, e38535. [Google Scholar] [CrossRef]
- Gendre, D.; Czernic, P.; Conéjéro, G.; Pianelli, K.; Briat, J.-F.; Lebrun, M.; Mari, S. TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J. 2006, 49, 1–15. [Google Scholar] [CrossRef]
- Feng, S.; Tan, J.; Zhang, Y.; Liang, S.; Xiang, S.; Wang, H.; Chai, T. Isolation and characterization of a novel cadmium-regulated Yellow Stripe-Like transporter (SnYSL3) in Solanum nigrum. Plant Cell Rep. 2016, 36, 281–296. [Google Scholar] [CrossRef]
- Dai, J.; Wang, N.; Xiong, H.; Qiu, W.; Nakanishi, H.; Kobayashi, T.; Nishizawa, N.K.; Zuo, Y. The Yellow Stripe-Like (YSL) Gene Functions in Internal Copper Transport in Peanut. Genes 2018, 9, 635. [Google Scholar] [CrossRef]
- Zheng, L.; Fujii, M.; Yamaji, N.; Sasaki, A.; Yamane, M.; Sakurai, I.; Sato, K.; Ma, J.F. Isolation and Characterization of a Barley Yellow Stripe-Like Gene, HvYSL5. Plant Cell Physiol. 2011, 52, 765–774. [Google Scholar] [CrossRef]
- Koike, S.; Inoue, H.; Mizuno, D.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004, 39, 415–424. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Xia, J.; Ma, J.F. OsYSL6 Is Involved in the Detoxification of Excess Manganese in Rice. Plant Physiol. 2011, 157, 1832–1840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lu, W.; Yang, Y.; Shen, Z.; Ma, J.F.; Zheng, L. OsYSL16 is Required for Preferential Cu Distribution to Floral Organs in Rice. Plant Cell Physiol. 2018, 59, 2039–2051. [Google Scholar] [CrossRef]
- Röth, S.; Fulcher, L.J.; Sapkota, G.P. Advances in targeted degradation of endogenous proteins. Cell. Mol. Life Sci. 2019, 76, 2761–2777. [Google Scholar] [CrossRef]
- Moreno, L.A.; Omidi, M.; Wurlitzer, M.; Luthringer, B.; Helmholz, H.; Schluter, H.; Willumeit, R.; Fügenschuh, A. Understanding Protein Networks using Vester’s Sensitivity Model. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 17, 1440–1450. [Google Scholar] [CrossRef] [PubMed]
- Herzberg, M.; Schüttau, M.; Reimers, M.; Große, C.; Hans-Günther-Schlegel, H.-G.-S.; Nies, D.H. Synthesis of nickel–iron hydrogenase in Cupriavidus metallidurans is controlled by metal-dependent silencing and un-silencing of genomic islands. Metallomics 2015, 7, 632–649. [Google Scholar] [CrossRef]
- Barwinska-Sendra, A.; Waldron, K.J. The Role of Intermetal Competition and Mis-Metalation in Metal Toxicity. Adv. Microb. Physiol. 2017, 70, 315–379. [Google Scholar] [CrossRef]
- Tran, J.B.; Krężel, A. InterMetalDB: A Database and Browser of Intermolecular Metal Binding Sites in Macromolecules with Structural Information. J. Proteome Res. 2021, 20, 1889–1901. [Google Scholar] [CrossRef]
- Putignano, V.; Rosato, A.; Banci, L.; Andreini, C. MetalPDB in 2018: A database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2018, 46, D459–D464. [Google Scholar] [CrossRef] [PubMed]
- Dennis, K.K.; Uppal, K.; Liu, K.H.; Ma, C.; Liang, B.; Go, Y.-M.; Jones, D.P. Phytochelatin database: A resource for phytochelatin complexes of nutritional and environmental metals. Database 2019, 2019, baz083. [Google Scholar] [CrossRef]
- Jayakanthan, M.; Muthukumaran, J.; Chandrasekar, S.; Chawla, K.; Punetha, A.; Sundar, D. ZifBASE: A database of zinc finger proteins and associated resources. BMC Genom. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Su, Y.; Yan, Y.-H.; Peng, J.-Y.; Dai, Q.-Q.; Ning, X.-L.; Zhu, C.-L.; Fu, C.; A McDonough, M.; Schofield, C.J.; et al. MeLAD: An integrated resource for metalloenzyme-ligand associations. Bioinformatics 2019, 36, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Ireland, S.M.; Martin, A.C.R. ZincBind—the database of zinc binding sites. Database 2019, 2019, baz006. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Cooper, D.R.; Porebski, P.J.; Shabalin, I.G.; Handing, K.B.; Minor, W. CheckMyMetal: A macromolecular metal-binding validation tool. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Holliday, G.L.; Andreini, C.; Fischer, J.D.; Rahman, S.A.; Almonacid, D.E.; Williams, S.T.; Pearson, W.R. MACiE: Exploring the diversity of biochemical reactions. Nucleic Acids Res. 2011, 40, D783–D789. [Google Scholar] [CrossRef]
- Zhang, Y.; Gladyshev, V.N. dbTEU: A protein database of trace element utilization. Bioinformatics 2010, 26, 700–702. [Google Scholar] [CrossRef]
- Valasatava, Y.; Rosato, A.; Banci, L.; Andreini, C. MetalPredator: A web server to predict iron–sulfur cluster binding proteomes. Bioinformatics 2016, 32, 2850–2852. [Google Scholar] [CrossRef]
- Andreini, C.; Bertini, I.; Cavallaro, G.; Decaria, L.; Rosato, A. A Simple Protocol for the Comparative Analysis of the Structure and Occurrence of Biochemical Pathways across Superkingdoms. J. Chem. Inf. Model. 2011, 51, 730–738. [Google Scholar] [CrossRef]
- Ajitha, M.; Sundar, K.; Arul, M.S.; Arumugam, S. Development of METAL-ACTIVE SITE and ZINCCLUSTER tool to predict active site pockets. Proteins 2018, 86, 322–331. [Google Scholar] [CrossRef]
- Lu, C.; Liu, Z.; Zhang, E.; He, F.; Ma, Z.; Wang, H. MPLs-Pred: Predicting Membrane Protein-Ligand Binding Sites Using Hybrid Sequence-Based Features and Ligand-Specific Models. Int. J. Mol. Sci. 2019, 20, 3120. [Google Scholar] [CrossRef]
- Sánchez-Aparicio, J.E.; Tiessler-Sala, L.; Velasco-Carneros, L.; Roldán-Martín, L.; Sciortino, G.; Maréchal, J.D. BioMetAll: Identifying Metal-Binding Sites in Proteins from Backbone Preorganization. J. Chem. Inf. Model. 2021, 61, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, M. SECISearch3 and Seblastian: In-Silico Tools to Predict SECIS Elements and Selenoproteins. Springer Protoc. Handb. 2017, 1661, 3–16. [Google Scholar] [CrossRef]
- Levy, R.; Edelman, M.; Sobolev, V. Prediction of 3D metal binding sites from translated gene sequences based on remote-homology templates. Proteins: Struct. Funct. Bioinform. 2009, 76, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Xie, D. MIonSite: Ligand-specific prediction of metal ion-binding sites via enhanced AdaBoost algorithm with protein sequence information. Anal. Biochem. 2019, 566, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-F.; Cheng, C.-W.; Shih, C.-S.; Hwang, J.-K.; Yu, C.-S.; Lu, C.-H. MIB: Metal Ion-Binding Site Prediction and Docking Server. J. Chem. Inf. Model. 2016, 56, 2287–2291. [Google Scholar] [CrossRef]



| Membrane Protein Families | Proteins | Species | Metal Ion (Metal Complex) | References |
|---|---|---|---|---|
| ABCs | OsABCC1 | Oryza sativa | As | [182] |
| OsABCG36 | Oryza sativa | Cd | [183] | |
| PtoABCG36 | Oryza sativa | Cd | [184] | |
| AtABCC3 | Arabidopsis thaliana | Cd | [185] | |
| AtATM3 | Arabidopsis thaliana | Cd, Pb | [186] | |
| AtPDR8 | Arabidopsis thaliana | Cd, Pb | [187] | |
| ARG1 | Oryza sativa | Co, Ni | [188] | |
| HMAs | BjHMA4 | Brassica juncea | Cd | [189] |
| OsHMA5 | Oryza sativa | Cu | [190] | |
| OsHMA3 | Oryza sativa | Zn | [191] | |
| PtoHMA5 | Populus tomentosa | Cd | [192] | |
| CsHMA3/4 | Cucumis sativus | Cd, Pb, Zn | [193] | |
| GmHMA8 | Glycine max | Cu | [194] | |
| GmHMA3 | Glycine max | Cd | [195] | |
| HvHMA1 | Hordeum vulgare | Zn, Cu | [196] | |
| NRAMPs | AtNRAMP6/3 | Arabidopsis thaliana | Fe | [197,198] |
| TtNRAMP6 | Triticum turgidum | Cd | [199] | |
| SbNrat1 | Sorghum bicolor | Al | [200] | |
| CjNRAMP1 | Crotalaria juncea | Fe, Cd | [201] | |
| OsNRAMP1 | Oryza sativa | As | [202] | |
| HvNramp5 | Hordeum vulgare | Mn, Cd | [203] | |
| AhNRAMP1 | Arachis hypogaea | Mn, Zn | [50] | |
| ZIPs | OsZIP1 | Oryza sativa | Zn, Cu, Cd | [204] |
| OsZIP9/3/8 | Oryza sativa | Zn | [205,206,207] | |
| NtZIP4A/B | Nicotiana tabacum | Zn, Cd | [208] | |
| ZmZIP5 | Zea mays | Zn, Fe | [209] | |
| VsRIT1 | Vicia sativa | Cd | [44] | |
| MTPs | PtrMTP6 | Populus trichocarpa | Mn, Co | [210] |
| AtMTP11 | Arabidopsis thaliana | Mn | [211] | |
| BnMTP3 | Brassica napus | Mn, Zn | [212] | |
| CsMTP8.2 | Camellia sinensis | Mn | [213] | |
| CsMTP7 | Camellia sinensi | Fe | [214] | |
| CsMTP9 | Camellia sinensi | Mn, Cd | [215] | |
| OsMTP11 | Oryza sativa | Mn | [216] | |
| OPTs | AtOPT6 | Arabidopsis thaliana | Cd-GSH | [217] |
| TcOPT3 | Thlaspi caerulescens | Fe, Zn, Cd, Cu | [218] | |
| TcYSL3 | Thlaspi caerulescens | Fe, Ni-NA | [219] | |
| SnYSL3 | Solanum nigrum | Fe, Cu, Zn, Cd-NA | [220] | |
| AhYSL3.1 | Arachis hypogaea | Cu | [221] | |
| HvYSL5 | Hordeum vulgare | Fe-MA | [222] | |
| OsYSL2 | Oryza sativa | Fe-NA, Mn-NA | [223] | |
| OsYSL6 | Oryza sativa | Mn-NA | [224] | |
| OsYSL16 | Oryza sativa | Cu-NA | [225] |
| Databases and Tools | Description | Website | References |
|---|---|---|---|
| Metalloprotein databases | |||
| InterMetalDB | Database and browser of intermolecular metal binding sites in macromolecules with structural information | intermetaldb.biotech.uni.wroc.pl/ (accessed on 12 November 2021) | [230] |
| MetalPDB | Database of metal sites in biological macromolecular structures | metalpdb.cerm.unifi.it/ (accessed on 12 November 2021) | [231] |
| PyCDB | Resource for phytochelatin complexes of nutritional and environmental metals | kuppal.shinyapps.io/pycdb (accessed on 12 November 2021) | [232] |
| ZifBASE | Database of zinc finger proteins and associated resources | web.iitd.ac.in/~sundar/zifbase (accessed on 12 November 2021) | [233] |
| MeLAD | Integrated resource for metalloenzyme-ligand associations | melad.ddtmlab.org/ (accessed on 12 November 2021) | [234] |
| ZincBind | Database of zinc binding sites, automatically generated from the Protein Data Bank. | zincbind.net/ (accessed on 12 November 2021) | [235] |
| CheckMyMetal | Database of metal binding site validation server | https://cmm.minorlab.org/ (accessed on 12 November 2021) | [236] |
| MACiE | Exploring the diversity of biochemical reactions | www.ebi.ac.uk/thornton-srv/databases/MACiE/ (accessed on 12 November 2021) | [237] |
| dbTEU | DataBase of Trace Element Utilization | gladyshevlab.bwh.harvard.edu/trace_element (accessed on 12 November 2021) | [238] |
| Computational tools | |||
| MetalPredator | Fe/(Fe-S) | metalweb.cerm.unifi.it/tools/metalpredator/ (accessed on 12 November 2021) | [239] |
| RDGB | Fe, Cu, Zn and other metals | www.cerm.unifi.it/ (accessed on 12 November 2021) | [240] |
| ZINCCLUSTER | Zn | www.metalactive.in/ (accessed on 12 November 2021) | [241] |
| MPLs-Pred | Metal ions | icdtools.nenu.edu.cn/mpls_pred (accessed on 12 November 2021) | [242] |
| BioMetAll | identification of conformational changes that alter the formation of metal-binding sites; Metalloenzyme design | github.com/insilichem/biometall (accessed on 12 November 2021) | [243] |
| SECISearch3 and Seblastian | Se | seblastian.crg.eu/ (accessed on 12 November 2021) | [244] |
| SeqCHED | Mg, Co, Ni, Ca | oca.weizmann.ac.il/oca-bin/ocamain (accessed on 12 November 2021) | [245] |
| MIonSite | Mg, Mn, Fe, Cu, Co, Cd, Ni | github.com/LiangQiaoGu/MIonSite.git (accessed on 12 November 2021) | [246] |
| MIB | Mn, Co, Zn, Ni, Hg, Cd | bioinfo.cmu.edu.tw/MIB/ (accessed on 12 November 2021) | [247] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, D.; Saleem, M.; He, T.; He, G. The Mechanism of Metal Homeostasis in Plants: A New View on the Synergistic Regulation Pathway of Membrane Proteins, Lipids and Metal Ions. Membranes 2021, 11, 984. https://doi.org/10.3390/membranes11120984
Wu D, Saleem M, He T, He G. The Mechanism of Metal Homeostasis in Plants: A New View on the Synergistic Regulation Pathway of Membrane Proteins, Lipids and Metal Ions. Membranes. 2021; 11(12):984. https://doi.org/10.3390/membranes11120984
Chicago/Turabian StyleWu, Danxia, Muhammad Saleem, Tengbing He, and Guandi He. 2021. "The Mechanism of Metal Homeostasis in Plants: A New View on the Synergistic Regulation Pathway of Membrane Proteins, Lipids and Metal Ions" Membranes 11, no. 12: 984. https://doi.org/10.3390/membranes11120984
APA StyleWu, D., Saleem, M., He, T., & He, G. (2021). The Mechanism of Metal Homeostasis in Plants: A New View on the Synergistic Regulation Pathway of Membrane Proteins, Lipids and Metal Ions. Membranes, 11(12), 984. https://doi.org/10.3390/membranes11120984







