Abstract
Methionine sulfoxide reductases are found in all domains of life and are important in reversing the oxidative damage of the free and protein forms of methionine, a sulfur containing amino acid particularly sensitive to reactive oxygen species (ROS). Archaea are microbes of a domain of life distinct from bacteria and eukaryotes. Archaea are well known for their ability to withstand harsh environmental conditions that range from habitats of high ROS, such as hypersaline lakes of intense ultraviolet (UV) radiation and desiccation, to hydrothermal vents of low concentrations of dissolved oxygen at high temperature. Recent evidence reveals the methionine sulfoxide reductases of archaea function not only in the reduction of methionine sulfoxide but also in the ubiquitin-like modification of protein targets during oxidative stress, an association that appears evolutionarily conserved in eukaryotes. Here is reviewed methionine sulfoxide reductases and their distribution and function in archaea.
1. Introduction
Reactive oxygen species (ROS), such as singlet oxygen (1O2), hydrogen peroxide (H2O2), superoxide anion (O−2) and hydroxyl radical (HO•), can cause widespread damage to cells. Proteins, amino acids, lipids, nucleic acids, and carbohydrates are generally susceptible to ROS damage [1,2]. The sulfur containing amino acids, methionine (Met) and cysteine (Cys), whether in protein or free form, are particularly sensitive to oxidation by ROS [3,4]. Met oxidation leads to the formation of diastereoisomers of methionine sulfoxide (Met-S-O and Met-R-O) which can be further oxidized to methionine sulfone (Figure 1). The accumulation of these oxidized methionine derivatives leads to protein carbonylation, aggregation and/or degradation. While methionine sulfone is irreversible, MetO can be repaired to Met by the action of methionine sulfoxide reductase (MSR) enzymes [5].
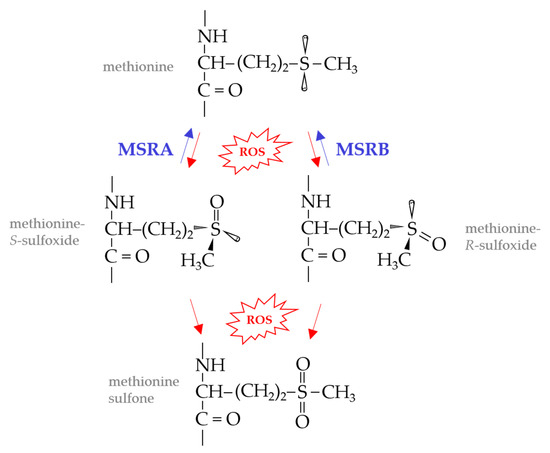
Figure 1.
Methionine oxidation and repair. Methionine, whether in proteins or as a free amino acid, is readily oxidized to a mixture of methionine sulfoxide diasteromers by reactive oxygen species (ROS) and other oxidants. Of methionine sulfoxide reductase (MSR) enzymes, MSRA stereospecifically reduces methionine-S-sulfoxide, whereas MSRB is specific for the R-form of methionine sulfoxide. Methionine sulfoxides, if not repaired, may be further oxidized by ROS to produce methionine sulfone or radicals.
MSR enzymes are of structurally distinct families and substrate specificity. MSRA of the IPR036509 superfamily catalyzes the stereospecific reduction of Met-S-O in free and protein forms (Figure 1) and can reduce N-Ac-L-MetO, dimethylsulfoxide (DMSO), L-ethionine sulfoxide and sulindac [6,7,8]. MSRB of the IPR028427 family reduces free and protein forms of Met-R-O [9]. fRMSR of the GAF (cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA)-like domain superfamily (IPR029016) reduces only the free form of Met-R-O [10,11,12]. MSRP of the molybdopterin-dependent sulfite oxidase family reduces free and protein forms of Met-R-O and Met-S-O [13,14] and can reduce dimethylsulfoxide (DMSO), trimethylamine-N-oxide (TMAO) and phenylmethyl sulfoxide in vitro [15]. Members of the molybdopterin-dependent DMSO reductase family, such as BisC [16,17], DmsA [18], TorZ/MSRZ [19] and BisZ [20], reduce free and/or protein forms of MetO in addition to other substrates such as biotin sulfoxide, nicotinamide-N-oxide, adenosine-N-oxide, DMSO and/or TMAO.
Common to MSRA, MSRB and fRMSR is the use of an active site cysteine (CysA) to catalyze the nucleophilic attack of the S atom of the MetO substrate (Figure 2) [21]. This attack results in the formation of a tetrahedral transition state which is rearranged to release Met (the product) and form a CysA sulfenic acid. To resolve this inactive state, a resolving cysteine (CysR) or reductant such as glutathione (YS−) serves as the nucleophile to attack the S atom of the CysA sulfenic acid (Figure 2, upper vs. lower). This attack releases water and forms a disulfide bond. In the CysR mechanism, an intradisulfide bond is formed between the CysA and CysR residues that can be rearranged by other Cys residues on the enzyme. If a separate thiol molecule is used to resolve the CysA sulfenic acid, an interdisulfide bond is formed. Ultimately, the inter- and intra-disulfide bonds are reduced by thiol relay systems such as nicotinamide adenine dinucleotide (phosphate) hydrogen [NAD(P)H]-dependent thioredoxin reductase (TrxR)/thioredoxin (Trx) or glutathione reductase (GR)/glutathione (GSH)/glutaredoxin (Grx) systems [21]. This reduction recycles the MSR enzyme back to an active state.
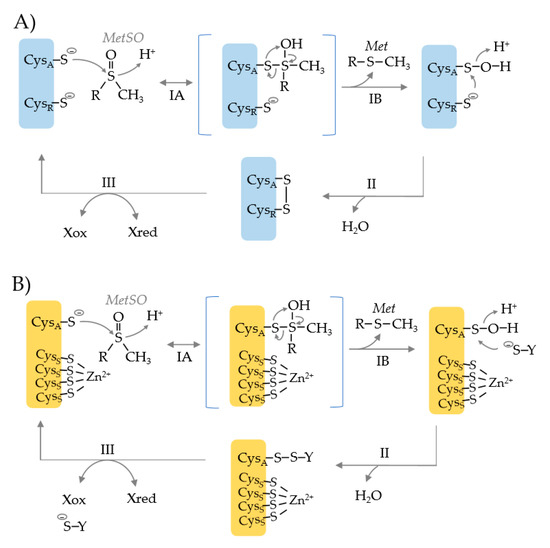
Figure 2.
Catalytic mechanism of methionine sulfoxide reductases that use an active site cysteine nucleophile (CysA) and a resolving cysteine (CysR) (blue) or a single CysA active site (orange). (A) CysA attacks the S atom of the methionine sulfoxide (MetSO) substrate resulting in the formation of a tetrahedral transition state (step IA). The intermediate is rearranged to release the product methionine (Met) and form a CysA sulfenic acid (step IB). CysR attacks the S atom of the sulfenic acid, resulting in the release of water and the formation of an intradisulfide bond (CysA-CysR) (step II). The disulfide bond can be rearranged by other Cys residues (not shown), but ultimately must be reduced to regenerate the MSR enzyme (step III), where X represents a thiol relay system such as NAD(P)H > thioredoxin reductase (TrxR) > thioredoxin (Trx). (B) MSR enzymes that use a CysA nucleophile but rely upon a resolving agent (−S-Y), as exemplified by the Methanothermobacter thermoautotrophicus Zn2+-type MSRB (MTH711). Only the active site nucleophile (CysA) is directly involved in catalysis. The four cysteine residues (CysS) that coordinate the Zn2+ ion are used for structural integrity. The CysA sulfenic acid intermediate is resolved by formation of an interdisulfide bond with −S-Y (step II) that is reduced by Xred (step III).
The reduction of MetO by molybdopterin-dependent enzymes relies upon two distinct prosthetic groups (Figure 3). Enzymes of the DMSO reductase family use bis-MGD (molybdopterin guanine dinucleotide), a complex of Mo and two molybdopterin guanine dinucleotide (MGD) cofactors, for the redox-active prosthetic group, while members of the sulfide oxidase family coordinate a di-oxo form of the molybdenum cofactor (di-oxo Moco) as the redox-active center [22]. The source of electrons used for molybdopterin-dependent reduction of MetO varies. For example, the DMSO reductase family member BisC of Rhodobacter sphaeroides can use electrons directly from NAD(P)H [23], whereas E. coli BisC relies upon a protein-(SH)2 and flavoprotein relay system [24]. The sulfide oxidase family protein MSRP, which coordinates di-oxo Moco through a conserved Cys residue [15], resides in the periplasmic space of gram-negative bacteria [13]. MSRP reduces MetO using electrons from MSRQ, an integral b-type heme transmembrane protein of the NADPH oxidase family [13,14], which shuttles electrons from the quinone pool [13,14].
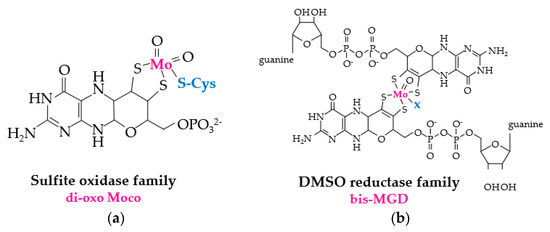
Figure 3.
Molybdopterin prosthetic groups of MSR enzymes. (a) di-oxo Moco of sulfite oxidase family members such as MSRP (where Cys that coordinates the prosthetic group is represented in blue). (b) bis-MGD (molybdopterin guanine dinucleotide) prosthetic group of DMSO (dimethylsulfoxide) reductase family members such as BisC, BisZ, DmsA and TorZ/MSRZ which display MetO reductase activity. X (in blue) represents the additional ligand of bis-MGD which can be a serine, a cysteine, a selenocysteine, an aspartate or a hydroxide and/or water molecule.
2. An Archaeal Perspective on Methionine Sulfoxide Repair
Archaea are microbes of a domain of life distinct from bacteria and eukaryotes [25]. Archaea are well known for their ability to withstand harsh environmental conditions that range from habitats of high ROS, such as hypersaline lakes with intense UV radiation and desiccation, to hydrothermal vents with low concentrations of dissolved oxygen at the high temperature [26]. Life evolved in an anaerobic world; thus, early archaea were relatively free from the damaging effects of ROS [27] and likely used a Wood–Ljungdahl pathway to generate energy and assimilate carbon (acetyl-CoA) from H2 and CO2 [28,29]. As oxygen levels increased over time, archaea and other microbes were challenged to develop mechanisms to repair and protect against ROS damage. Most archaea today, including strict anaerobes, have at least one antioxidant enzyme to protect and/or repair against ROS damage such as superoxide dismutase, catalase, peroxidiredoxin, superoxide reductase and/or MSR enzymes [30,31] (Table S1). The archaeal MSR enzymes are of particular interest in terms of evolutionary history [25,28,29] and how protein homeostasis is maintained in extreme habitats [26], as these enzymes are recently linked to ubiquitin-like protein modification pathways that are associated with oxidative stress and sulfur mobilization [32].
3. Archaeal Methionine Sulfoxide Reductase Homologs
The MSR homologs of archaea include: (i) MSRA and MSRB, which are prevalent in archaea but notably absent from most (hyper)thermophiles, (ii) fRMSR and MSRP, which are noted in archaea but are not common, and (iii) molybdopterin (MPT)/tungstopterin (WPT) oxidoreductase (OR) enzymes of the sulfite oxidase and DMSO reductase families (e.g., DMSO reductase, formate dehydrogenase, assimilatory nitrate reductase and formylmethanofuran dehydrogenase), which are widespread in archaea but are not known to function in MetO reduction [33,34,35,36] (Table S1).
The MSRA and MSRB homologs of archaea are stand-alone and fusion proteins (Figure 4A, Table S2). While most archaeal MSRA/B homologs are cytosolic, a subset have transmembrane spanning domains and signals for protein translocation via the general secretory (Sec) and/or twin arginine translocation (TAT) pathways. Of the archaeal fusion proteins, the MSRAB, MSRBA, MSRA-Trx-MSRB and MSRA-Grx share a general domain architecture with proteins of bacteria (e.g., MSRAB, MSRBA, and Trx-MSRAB) [37] and certain eukaryotes (e.g., Entamoeba invadens MSRAB, UniProt S0B0R4). The Trx/Grx domains likely facilitate a thiol relay to reduce MSRA/B, while the fusion of MSRA to MSRB may enhance the catalytic efficiency of MetO reduction [38]. One MSR homolog that appears unique to archaea is the fusion of MSRB to an N-terminal adenylation (AANH) domain (IPR020536) (Figure 4B). The AANH domain includes a conserved cysteine that is used by tRNA sulfurtransferases to catalyze the nucleophilic attack of ATP and form an adenylated tRNA intermediate during the transfer of sulfur to tRNA [39]. The archaeal AANH-MSRB homolog is missing the THUMP (thiouridine synthase, RNA methylase and pseudouridine synthase domain) domain used by the tRNA sulfurtransferases to bind tRNA [40]. Thus, while the archaeal MSRB-AANH is predicted to catalyze the reduction of MetO and the adenylation of a substrate (X), the identity of X has yet to be determined. In certain Thaumarchaeota, MSRB is fused to a ThyX-like domain (Figure 4C). ThyX is a flavin-dependent thymidylate synthase that uses reduced flavin to relay a methylene from a folate carrier to the deoxyuridine monophosphate (dUMP) acceptor to form deoxythymidine monophosphate (dTMP) [41]. While the archaeal ThyX-MSRB likely binds flavin and reduces MetO, the enzyme is not predicted synthesize dTMP as the Ser required for ThyX activity [42,43] is not conserved (Figure 4C). Thus, an unusual mechanism of electron transfer may occur in some archaeal MSR enzymes in which a flavin group, bound by a ThyX-like domain, may relay electrons for MSRB-mediated reduction of MetO.
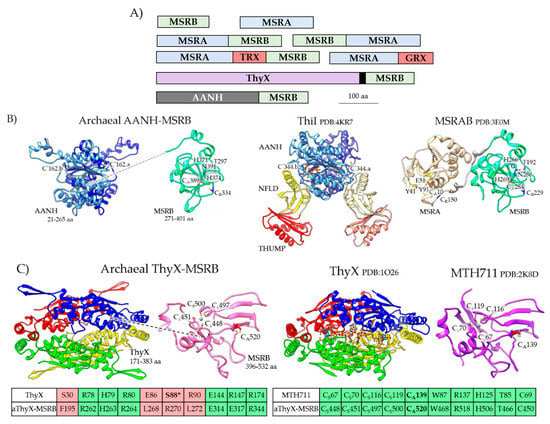
Figure 4.
Archaeal MSRA/B stand-alone and fusion protein homologs. (A) Protein domain architecture of archaeal MSRA/B homologs. The black box indicates the coiled coil domain that links the ThyX and MSRB domains. (B) Archaeal AANH-MSRB (e.g., Uniprot: A0A2D6JNB0) has a predicted structural fold related to the adenylation (AANH) domain of tRNA sulfurtransferases such as Thermotoga maritima ThiI (Protein Data Bank or PDB: 4KR7) and the MSRB domain of Streptococcus pneumoniae MSRAB (PDB:3E0M). The active site cysteine nucleophiles of the AANH (Cys*162) and MsrB (CysA389) domains as well as the resolving Cys residue of MsrB (CysR334) are conserved. The N-terminal ferredoxin-like domain (NFLD) and RNA binding THUMP (thiouridine synthases, RNA methylases and pseudouridine synthases) domain of ThiI are not conserved. (C) Archaeal aThyX-MSRB (Uniprot A0A075GM99) has a predicted structural fold related to Thermotoga maritima flavin-dependent thymidylate synthase (ThyX, PDB: 1O26) and the Zn2+-binding MSRB of Methanothermobacter thermautotrophicus (MTH711, PDB: 2K8D). The ThyX flavin-binding site and MSRB CysA nucleophile and CysS Zn2+ coordinating residues are conserved; however, the serine residue (S88*) required for ThyX catalytic activity is not conserved in the aThyX-MSRB. TRX: thioredoxin domain; GRX: glutaredoxin domain.
4. Archaeal MSR Enzymes Characterized
The archaeal MSR enzymes that are purified and biochemically characterized are summarized below and in Table S3.
An fRMSR enzyme is characterized from the thermophilic archaeon Thermoplasma acidophilum (Ta) [44]. TafRMSR is the only archaeal MSR enzyme to date that can use an NADPH > Trx > TrxR system to reduce MetO in vitro [44]. Similarly to other fRMSR enzymes, TafRMSR reduces the free form of Met-R-O, not the protein MetO or Met-S-O forms [44]. TafRMSR has three cysteine residues (Cys15, Cys60 and Cys84). Of these, Cys60 and Cys84 are critical for MetO reductase activity, whereas, Cys15 is unimportant [44]. Cys84 forms the active site nucleophile (CysA) [44]. Cys60, while important for activity, does not form a disulfide bond with Cys84 and, thus, is not considered a resolving cysteine (CysR) [44]. TafRMSR is instead proposed to use a separate thiol to resolve the CysA active site. By contrast, bacterial and yeast fRMSR enzymes form intrachain disulfide bonds between catalytic and resolving cysteine residues (CysA-CysR) in the catalytic mechanism of MetO reduction [10,45,46,47].
MSRA/B homologs are not common among the hyperthermophiles; however, an MSRAB fusion is described from the hyperthermophilic archaeon Thermococcus kodakarensis (Tk). The Tk MSRAB enzyme reduces free and protein forms of MetO, with steroselectivity of the MSRA and MSRB domains as observed in bacteria and eukaryotes [48]. Tk MSRAB does not bind Zn2+ and is instead proposed to be acquired by horizontal gene transfer from bacteria [48]. Consistent with this evolutionary history, the Tk MSRAB is less active and less abundant at 85 °C (the optimal growth temperature) compared to temperatures below 80 °C where dissolved oxygen is higher [48]. Hydrothermal vent communities encounter drastic thermal gradients from the hot venting fluid and cold seawater; thus, MSRAB is thought to provide a selective advantage to Thermococcus species that prevail in vent ecosystems [49] where ROS damage may increase as temperatures drop.
MSRB enzymes that coordinate Zn2+ and require formation of an intramolecular thiol to regenerate the CysA active site are common in archaea and exemplified by MTH711 from the thermophilic methanogen Methanobacterium thermoautotrophicus [50]. Zn2+-containing MSRBs, such as MTH711, are apparent prototypes of the MSRB enzymes that lost the metal later in evolution [51]. Like other MSRBs, MTH711 can reduce free and protein forms of Met-R-O [50]. The CysA of MTH711 is directly involved in catalysis and can be reduced in vitro by dithiothreitol (DTT) or cysteine [50]. By contrast, the four cysteine residues (CysS) that coordinate the Zn2+ ion are only required for structural integrity, and the two non-Zn2+-binding Cys residues that reside outside the catalytic center are not needed for catalysis [50]. Thus, the CysA sulfenic acid intermediate is proposed to be resolved by a thiol that is distinct from the MSRB enzyme [50,52].
Stand-alone MSRA and Zn2+-type MSRB enzymes are recently characterized from the halophilic archaeon Haloferax volcanii. These MSR enzymes reduce the peptide mimic, dabsyl-Met-(R/S)-O, with DTT serving as the reductant [32]. Futher analysis of MSRA reveals it requires a conserved active site nucleophile (CysA, Cys13) and an invariant glutamate (Glu56, presumed to bind the MetO oxygen atom) for its activity [32]. The MetO reductase activity of MSRA is inhibited by DMSO [32], a competitive active site inhibitor based on analogy to yeast MSRA which reduces DMSO to DMS (dimethylsulfide) [53,54]. Surprisingly, DMSO stimulates an MSRA-dependent ubiquitin-like protein (Ubl) modification system in this archaeon as described below [32].
5. MSRA and Its Function in Ubiquitin-Like Protein Modification
In addition to its role as a MetO reductase, the H. volcanii MSRA has an apparent ‘moonlighting’ function in ubiquitin-like (Ubl) modification [32]. Archaea mediate Ubl modification by a mechanism that is related to eukaryotic ubiquitination [55,56]. An E1-like enzyme adenylates the Ubl and forms an E1~Ubl thioester intermediate prior to Ubl modification [57,58]. Most archaea are missing homologs of the classical E2 ubiquitin-conjugating and E3 ubiquitin-ligase enzymes of eukaryotic ubiquitination [56]. Thus, the latter stages of archaeal Ubl modification are unclear. In the presence of DMSO, an inhibitor of MetO reductase activity, MSRA stimulates the E1-dependent Ubl modification of target proteins [32] and is itself a target of Ubl modification [59]. This reaction is unaffected by (excess or limiting) DTT suggesting the MSRA mechanism occurs independent of its oxidase or reductase activities [32]. Further studies are needed to clarify the precise role of MSRA in this process.
6. MSRA/B Regulation in Archaea
Archaeal MSRA/B enzymes are regulated at the transcript, protein, and post-translational levels (Table S4). In Halobacterium salinarum (known for its purple membrane), the transcript levels of MSRA and MSRB are up during conditions of severe oxidative stress [60,61]. The H. salinarum msrB is one of the core genes of the RosR (reactive oxygen species transcriptional regulator) regulon that includes superoxide dismutase, Trx-like, and other related genes [60]. MSRA transcript levels are also found to be up in Sulfolobus solfataricus and Sulfolobus acidocaldarius (crenarchaeota from acidic hot springs) after UV irradiation [62], a condition known to generate ROS [63]. Likewise, MSRAB abundance is up at the protein level after exposure of T. kodakarensis to saturating oxygen, low temperature or high salt [48,64]. As discussed earlier, oxidative stress is associated with temperature downshifts from 80 °C due to the increase in dissolved oxygen concentration. Hyposaline and hypersaline conditions stimulate antioxidant responses in algae and plants [65] and appear to do so in archaea based on the Tk MSRAB response [64]. The MSRA and MSRB of H. volcanii are Ubl modified (sampylated) in the presence of the mild oxidant DMSO [59]. DMSO exposure reduces the level of unmodified MSRA and increases the level of Ubl-modified MSRA [32]. Thus, Ubl modification appears to target MSRA for proteolysis and may serve to autoregulate the oxidative stress response of this haloarchaeon.
7. Protein Disulfide Relay Systems of Archaea
Archaeal protein disulfide relay systems may provide the reductant for MetO reduction by MSR enzymes that use a CysA nucleophile. Archaeal NAD(P)H-dependent protein disulfide oxidoreductase relay systems which could serve this role include: (i) Trx or Grx/TrxR [66,67], (ii) protein disulfide oxidoreductase (PDO)/TrxR [68], (iii) methanoredoxin (Mrx)/coenzyme M disulfide reductase (CoMR) [69,70] and (iv) bis-γ-glutamylcystine reductase (BggR) [71,72] (Figure 5). An archaeal F420-dependent TrxR is also described [73]. Of these disulfide relay systems, Grx/TrxR can resolve the disulfide bond of peroxiredoxins (Prx), such as the alkyl hydroperoxide reductase AhpC [74]. Furthermore, PDO/TrxR and Mrx/CoMR can reduce the disulfide bonds of oxidized proteins [68,70]. While these systems are not yet linked to archaeal MSR enzymes, TafRMSR can use an NADPH-dependent E. coli Trx/human TrxR system to reduce MetO [44], suggesting that a native Trx/TrxR may resolve the CysA active site of TafRMSR. Trx and Grx domains are fused to MSRA/B in some archaea (see above) providing further evidence for disulfide relay systems.
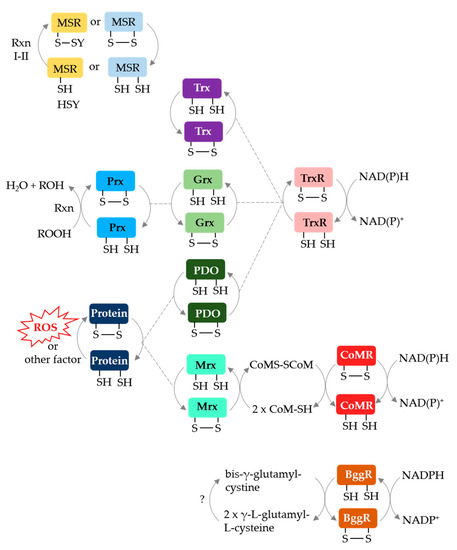
Figure 5.
Archaeal protein disulfide relay systems which may provide reductant for MetO reduction by MSR enzymes that use a CysA mechanism. See text for details. Prx: peroxiredoxin; PDO: protein disulfide oxidoreductase; NAD(P)H: nicotinamide adenine dinucleotide (phosphate) hydrogen; CoMR: coenzyme M disulfide reductase; BggR: bis-γ-glutamylcystine reductase; CoMS-SCoM: coenzyme M disulfide.
8. Physiological Roles of Archaeal MSR Enzymes
In general, loss of MSR activity can reduce cell viability, increase ROS production, and promote protein carbonylation [75]. Archaea are no exception. H. volcanii ΔmsrA (when compared to wild type, ΔmsrB and ΔmsrA msrA+) has a pronounced decrease in the abundance of Ubl modified proteins that form after cells are exposed to the mild oxidant DMSO; subsequent transfer of these cells to severe oxidative stress (hypochlorite or H2O2) results in reduced cell viability [32]. Thus, the archaeal MSRA appears important in responses to oxidative stress that are physiologically distinct from MSRB [32].
9. Conclusions and Future Perspectives
Archaeal MSR enzymes are full of surprises. The dearth of MSRA/B homologs in hyperthermophiles and low temperature preference of the Thermococcus MSRAB suggest that Met oxidation may not be a limiting factor for extremophiles that live at high temperature and pressure. Alternatively, these archaea use an MSR enzyme that has yet to be discovered. The finding that TafRMSR and MSRB-type MTH711 use an active site CysA that is reduced by an external thiol and not a CysR suggests that archaea use a streamlined mechanism for resolving the active site. The most recent finding—that H. volcanii MSRA has a moonlighting function in Ubl modification—is also surprising and helped guide the insight that MSRA competes for capturing ubiquitin and promotes ubiquitination in mammals [76]. Whether archaeal MSR enzymes can extend the lifespan of an organism remains to be determined. Expression of fRMSR enzymes lost during evolution can lead to an increased lifespan in animals [77]. Likewise, expression of MSRA, together with DMSO, can also extend an organism’s life [78,79]. Thus, archaeal MSR enzymes, including those yet to be discovered, may hold a key to the fountain of youth.
Supplementary Materials
The following are available online at http://www.mdpi.com/2076-3921/7/10/124/s1, Table S1: Phylogenetic distribution of methionine sulfoxide reductase homologs compared to 20S proteasome, superoxide dismutase and other oxidative stress response homologs. Table S2: Domain architecture of MSRA- and MSRB-type methionine sulfoxide reductase homologs among Archaea. Table S3: Archaeal methionine sulfoxide reductases biochemically and/or structurally characterized. Table S4: Archaeal methionine sulfoxide reductases regulated at the protein and/or transcript level.
Funding
Funds for this project were awarded to J.M.-F. through the Bilateral Research Council Biotechnology and Biological Research Council (BBSRC) and the U.S. National Science Foundation Directorate for Biological Sciences program (NSF/BIO 1642283), the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences and Biosciences, Physical Biosciences Program (DOE DE-FG02-05ER15650) and the National Institutes of Health (NIH R01 GM57498).
Conflicts of Interest
The author declares no conflict of interest.
References
- Moller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Alcock, L.J.; Perkins, M.V.; Chalker, J.M. Chemical methods for mapping cysteine oxidation. Chem. Soc. Rev. 2018, 47, 231–268. [Google Scholar] [CrossRef] [PubMed]
- Vogt, W. Oxidation of methionyl residues in proteins: Tools, targets, and reversal. Free Radic. Biol. Med. 1995, 18, 93–105. [Google Scholar] [CrossRef]
- Weissbach, H.; Etienne, F.; Hoshi, T.; Heinemann, S.H.; Lowther, W.T.; Matthews, B.; St John, G.; Nathan, C.; Brot, N. Peptide methionine sulfoxide reductase: Structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 2002, 397, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Weissbach, H.; Brot, N. Cloning the expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 2095–2099. [Google Scholar] [CrossRef] [PubMed]
- Etienne, F.; Resnick, L.; Sagher, D.; Brot, N.; Weissbach, H. Reduction of sulindac to its active metabolite, sulindac sulfide: Assay and role of the methionine sulfoxide reductase system. Biochem. Biophys. Res. Commun. 2003, 312, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Weissbach, H.; Resnick, L.; Brot, N. Methionine sulfoxide reductases: History and cellular role in protecting against oxidative damage. Biochim. Biophys. Acta 2005, 1703, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Tarrago, L.; Watanabe, Y.; Kaya, A.; Lee, B.C.; Tran, U.; Nishiyama, R.; Fomenko, D.E.; Gladyshev, V.N.; Tran, L.S. Diversity of plant methionine sulfoxide reductases B and evolution of a form specific for free methionine sulfoxide. PLoS ONE 2013, 8, e65637. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Johnson, L.C.; Weissbach, H.; Brot, N.; Lively, M.O.; Lowther, W.T. Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. Proc. Natl. Acad. Sci. USA 2007, 104, 9597–9602. [Google Scholar] [CrossRef] [PubMed]
- Grimaud, R.; Ezraty, B.; Mitchell, J.K.; Lafitte, D.; Briand, C.; Derrick, P.J.; Barras, F. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 2001, 276, 48915–48920. [Google Scholar] [CrossRef] [PubMed]
- Moskovitz, J.; Singh, V.K.; Requena, J.; Wilkinson, B.J.; Jayaswal, R.K.; Stadtman, E.R. Purification and characterization of methionine sulfoxide reductases from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochem. Biophys. Res. Commun. 2002, 290, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Gennaris, A.; Ezraty, B.; Henry, C.; Agrebi, R.; Vergnes, A.; Oheix, E.; Bos, J.; Leverrier, P.; Espinosa, L.; Szewczyk, J.; et al. Repairing oxidized proteins in the bacterial envelope using respiratory chain electrons. Nature 2015, 528, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Juillan-Binard, C.; Picciocchi, A.; Andrieu, J.P.; Dupuy, J.; Petit-Hartlein, I.; Caux-Thang, C.; Vives, C.; Niviere, V.; Fieschi, F. A Two-component NADPH oxidase (NOX)-like system in bacteria is involved in the electron transfer chain to the methionine sulfoxide reductase MsrP. J. Biol. Chem. 2017, 292, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- Loschi, L.; Brokx, S.J.; Hills, T.L.; Zhang, G.; Bertero, M.G.; Lovering, A.L.; Weiner, J.H.; Strynadka, N.C. Structural and biochemical identification of a novel bacterial oxidoreductase. J. Biol. Chem. 2004, 279, 50391–50400. [Google Scholar] [CrossRef] [PubMed]
- Ezraty, B.; Bos, J.; Barras, F.; Aussel, L. Methionine sulfoxide reduction and assimilation in Escherichia coli: New role for the biotin sulfoxide reductase BisC. J. Bacteriol. 2005, 187, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Pollock, V.V.; Barber, M.J. Biotin sulfoxide reductase. Heterologous expression and characterization of a functional molybdopterin guanine dinucleotide-containing enzyme. J. Biol. Chem. 1997, 272, 3355–3362. [Google Scholar] [CrossRef] [PubMed]
- Hilton, J.C.; Temple, C.A.; Rajagopalan, K.V. Re-design of Rhodobacter sphaeroides dimethyl sulfoxide reductase. Enhancement of Adenosine N1-Oxide Reductase Activity. J. Biol. Chem. 1999, 274, 8428–8436. [Google Scholar] [CrossRef] [PubMed]
- Dhouib, R.; Othman, D.S.; Lin, V.; Lai, X.J.; Wijesinghe, H.G.; Essilfie, A.T.; Davis, A.; Nasreen, M.; Bernhardt, P.V.; Hansbro, P.M.; et al. A novel, molybdenum-containing methionine sulfoxide reductase supports survival of Haemophilus influenzae in an in vivo model of infection. Front. Microbiol. 2016, 7, 1743. [Google Scholar] [CrossRef] [PubMed]
- Gon, S.; Patte, J.C.; Mejean, V.; Iobbi-Nivol, C. The torYZ (yecK bisZ) operon encodes a third respiratory trimethylamine N-oxide reductase inEscherichia coli. J. Bacteriol. 2000, 182, 5779–5786. [Google Scholar] [CrossRef] [PubMed]
- Boschi-Muller, S.; Branlant, G. Methionine sulfoxide reductase: Chemistry, substrate binding, recycling process and oxidase activity. Bioorg. Chem. 2014, 57, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Hille, R. Molybdenum and tungsten in biology. Trends Biochem. Sci. 2002, 27, 360–367. [Google Scholar] [CrossRef]
- Pollock, V.V.; Barber, M.J. Kinetic and mechanistic properties of biotin sulfoxide reductase. Biochemistry 2001, 40, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Del Campillo-Campbell, A.; Dykhuizen, D.; Cleary, P.P. Enzymic reduction of d-biotin d-sulfoxide to d-biotin. Methods Enzymol. 1979, 62, 379–385. [Google Scholar] [PubMed]
- Eme, L.; Spang, A.; Lombard, J.; Stairs, C.W.; Ettema, T.J.G. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 2017, 15, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Chaban, B.; Ng, S.Y.; Jarrell, K.F. Archaeal habitats—From the extreme to the ordinary. Can. J. Microbiol. 2006, 52, 73–116. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Diagnosing oxidative stress in bacteria: Not as easy as you might think. Curr. Opin. Microbiol. 2015, 24, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Sousa, F.L.; Thiergart, T.; Landan, G.; Nelson-Sathi, S.; Pereira, I.A.; Allen, J.F.; Lane, N.; Martin, W.F. Early bioenergetic evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130088. [Google Scholar] [CrossRef] [PubMed]
- Adam, P.S.; Borrel, G.; Brochier-Armanet, C.; Gribaldo, S. The growing tree of Archaea: New perspectives on their diversity, evolution and ecology. ISME J. 2017, 11, 2407–2425. [Google Scholar] [CrossRef] [PubMed]
- Slesak, I.; Slesak, H.; Zimak-Piekarczyk, P.; Rozpadek, P. Enzymatic antioxidant systems in early anaerobes: Theoretical considerations. Astrobiology 2016, 16, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Weissbach, H. Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases. Biol. Rev. Camb. Philos. Soc. 2008, 83, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Adams, Z.; Liu, R.; Hepowit, N.L.; Wu, Y.; Bowmann, C.F.; Moskovitz, J.; Maupin-Furlow, J.A. Methionine sulfoxide reductase A (MsrA) and its function in ubiquitin-like protein modification in Archaea. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Espinosa, R.M.; Marhuenda-Egea, F.C.; Bonete, M.J. Assimilatory nitrate reductase from the haloarchaeon Haloferax mediterranei: Purification and characterisation. FEMS Microbiol. Lett. 2001, 204, 381–385. [Google Scholar] [CrossRef]
- White, W.B.; Ferry, J.G. Identification of formate dehydrogenase-specific mRNA species and nucleotide sequence of the fdhC gene of Methanobacterium formicicum. J. Bacteriol. 1992, 174, 4997–5004. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.A.; DasSarma, S. Genomic analysis of anaerobic respiration in the archaeon Halobacterium sp. strain NRC-1: Dimethyl sulfoxide and trimethylamine N-oxide as terminal electron acceptors. J. Bacteriol. 2005, 187, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Hochheimer, A.; Schmitz, R.A.; Thauer, R.K.; Hedderich, R. The tungsten formylmethanofuran dehydrogenase from Methanobacterium thermoautotrophicum contains sequence motifs characteristic for enzymes containing molybdopterin dinucleotide. Eur. J. Biochem. 1995, 234, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Galperin, M.Y. Genome annotation and analysis. In Sequence-Evolution-Function: Computational Approaches in Comparative Genomics; Kluwer Academic: Boston, MA, USA, 2003. [Google Scholar]
- Chen, B.; Markillie, L.M.; Xiong, Y.; Mayer, M.U.; Squier, T.C. Increased catalytic efficiency following gene fusion of bifunctional methionine sulfoxide reductase enzymes from Shewanella oneidensis. Biochemistry 2007, 46, 14153–14161. [Google Scholar] [CrossRef] [PubMed]
- You, D.; Xu, T.; Yao, F.; Zhou, X.; Deng, Z. Direct evidence that ThiI is an ATP pyrophosphatase for the adenylation of uridine in 4-thiouridine biosynthesis. ChemBioChem 2008, 9, 1879–1882. [Google Scholar] [CrossRef] [PubMed]
- Waterman, D.G.; Ortiz-Lombardía, M.; Fogg, M.J.; Koonin, E.V.; Antson, A.A. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 2006, 356, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Mishanina, T.V.; Yu, L.; Karunaratne, K.; Mondal, D.; Corcoran, J.M.; Choi, M.A.; Kohen, A. An unprecedented mechanism of nucleotide methylation in organisms containing thyX. Science 2016, 351, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Myllykallio, H.; Lipowski, G.; Leduc, D.; Filee, J.; Forterre, P.; Liebl, U. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 2002, 297, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Mathews, I.I.; Deacon, A.M.; Canaves, J.M.; McMullan, D.; Lesley, S.A.; Agarwalla, S.; Kuhn, P. Functional analysis of substrate and cofactor complex structures of a thymidylate synthase-complementing protein. Structure 2003, 11, 677–690. [Google Scholar] [CrossRef]
- Kim, H.S.; Kwak, G.H.; Lee, K.; Jo, C.H.; Hwang, K.Y.; Kim, H.Y. Structural and biochemical analysis of a type II free methionine-R-sulfoxide reductase from Thermoplasma acidophilum. Arch. Biochem. Biophys. 2014, 560, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Gruez, A.; Libiad, M.; Boschi-Muller, S.; Branlant, G. Structural and biochemical characterization of free methionine-R-sulfoxide reductase from Neisseria meningitidis. J. Biol. Chem. 2010, 285, 25033–25043. [Google Scholar] [CrossRef] [PubMed]
- Bong, S.M.; Kwak, G.H.; Moon, J.H.; Lee, K.S.; Kim, H.S.; Kim, H.Y.; Chi, Y.M. Structural and kinetic analysis of free methionine-R-sulfoxide reductase from Staphylococcus aureus: Conformational changes during catalysis and implications for the catalytic and inhibitory mechanisms. J. Biol. Chem. 2010, 285, 25044–25052. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.S.; Burden, L.M.; Hurley, J.H. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000, 19, 5288–5299. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, E.; Shinka, Y.; Fukui, T.; Atomi, H.; Imanaka, T. Methionine sulfoxide reductase from the hyperthermophilic archaeon Thermococcus kodakaraensis, an enzyme designed to function at suboptimal growth temperatures. J. Bacteriol. 2007, 189, 7134–7144. [Google Scholar] [CrossRef] [PubMed]
- Holden, J.F.; Takai, K.; Summit, M.; Bolton, S.; Zyskowski, J.; Baross, J.A. Diversity among three novel groups of hyperthermophilic deep-sea Thermococcus species from three sites in the northeastern Pacific Ocean. FEMS Microbiol. Ecol. 2001, 36, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Carella, M.; Becher, J.; Ohlenschläger, O.; Ramachandran, R.; Gührs, K.H.; Wellenreuther, G.; Meyer-Klaucke, W.; Heinemann, S.H.; Görlach, M. Structure-function relationship in an archaebacterial methionine sulphoxide reductase B. Mol. Microbiol. 2011, 79, 342–358. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.A.; Koc, A.; Cerny, R.L.; Gladyshev, V.N. Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 2002, 277, 37527–37535. [Google Scholar] [CrossRef] [PubMed]
- Carella, M.; Ohlenschläger, O.; Ramachandran, R.; Görlach, M. 1H, 13C and 15N resonance assignment of a zinc-binding methionine sulfoxide reductase type-B from the thermophilic archeabacterium Methanothermobacter thermoautotrophicus. Biomol. NMR Assign. 2010, 4, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Kwak, G.H.; Choi, S.H.; Kim, H.Y. Dimethyl sulfoxide elevates hydrogen peroxide-mediated cell death in Saccharomyces cerevisiae by inhibiting the antioxidant function of methionine sulfoxide reductase A. BMB Rep. 2010, 43, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Kwak, G.H.; Choi, S.H.; Kim, J.R.; Kim, H.Y. Inhibition of methionine sulfoxide reduction by dimethyl sulfoxide. BMB Rep. 2009, 42, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Humbard, M.; Miranda, H.; Lim, J.; Krause, D.; Pritz, J.; Zhou, G.; Chen, S.; Wells, L.; Maupin-Furlow, J. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 2010, 463, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Maupin-Furlow, J.A. Prokaryotic ubiquitin-like protein modification. Annu. Rev. Microbiol. 2014, 68, 155–175. [Google Scholar] [CrossRef] [PubMed]
- Hepowit, N.L.; de Vera, I.M.; Cao, S.; Fu, X.; Wu, Y.; Uthandi, S.; Chavarria, N.E.; Englert, M.; Su, D.; Sӧll, D.; et al. Mechanistic insight into protein modification and sulfur mobilization activities of noncanonical E1 and associated ubiquitin-like proteins of Archaea. FEBS J. 2016, 283, 3567–3586. [Google Scholar] [CrossRef] [PubMed]
- Miranda, H.; Nembhard, N.; Su, D.; Hepowit, N.; Krause, D.; Pritz, J.; Phillips, C.; Söll, D.; Maupin-Furlow, J. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc. Natl. Acad. Sci. USA 2011, 108, 4417–4422. [Google Scholar] [CrossRef] [PubMed]
- Dantuluri, S.; Wu, Y.; Hepowit, N.L.; Chen, H.; Chen, S.; Maupin-Furlow, J.A. Proteome targets of ubiquitin-like samp1ylation are associated with sulfur metabolism and oxidative stress in Haloferax volcanii. Proteomics 2016, 16, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Gillum, N.; Boyd, J.L.; Schmid, A. The RosR transcription factor is required for gene expression dynamics in response to extreme oxidative stress in a hypersaline-adapted archaeon. BMC Genom. 2012, 13, 351. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Van, P.T.; Busch, C.R.; Robinson, C.K.; Pan, M.; Pang, W.L.; Reiss, D.J.; DiRuggiero, J.; Baliga, N.S. Coordination of frontline defense mechanisms under severe oxidative stress. Mol. Syst. Biol. 2010, 6, 393. [Google Scholar] [CrossRef] [PubMed]
- Gotz, D.; Paytubi, S.; Munro, S.; Lundgren, M.; Bernander, R.; White, M.F. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 2007, 8, R220. [Google Scholar] [CrossRef] [PubMed]
- Ravanat, J.L.; Douki, T. UV and ionizing radiations induced DNA damage, differences and similarities. Radiat. Phys. Chem. 2016, 128, 92–102. [Google Scholar] [CrossRef]
- Jia, B.; Liu, J.; Van Duyet, L.; Sun, Y.; Xuan, Y.H.; Cheong, G.W. Proteome profiling of heat, oxidative, and salt stress responses in Thermococcus kodakarensis KOD1. Front. Microbiol. 2015, 6, 605. [Google Scholar] [CrossRef] [PubMed]
- Jahnke, L.S.; White, A.L. Long-term hyposaline and hypersaline stresses produce distinct antioxidant responses in the marine alga Dunaliella tertiolecta. J. Plant Physiol. 2003, 160, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.K.; Jung, H.C.; Kang, S.G.; Lee, H.S. Redox regulation of SurR by protein disulfide oxidoreductase in Thermococcus onnurineus NA1. Extremophiles 2017, 21, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Susanti, D.; Loganathan, U.; Compton, A.; Mukhopadhyay, B. A reexamination of thioredoxin reductase from Thermoplasma acidophilum, a thermoacidophilic euryarchaeon, identifies it as an NADH-dependent enzyme. ACS Omega 2017, 2, 4180–4187. [Google Scholar] [CrossRef] [PubMed]
- Kashima, Y.; Ishikawa, K. A hyperthermostable novel protein-disulfide oxidoreductase is reduced by thioredoxin reductase from hyperthermophilic archaeon Pyrococcus horikoshii. Arch. Biochem. Biophys. 2003, 418, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.G.; Rouviere, P.E. Purification and characterization of the reduced-nicotinamide-dependent 2,2′-dithiodiethanesulfonate reductase from Methanobacterium thermoautotrophicum delta H. J. Bacteriol. 1990, 172, 6435–6441. [Google Scholar] [CrossRef] [PubMed]
- Yenugudhati, D.; Prakash, D.; Kumar, A.K.; Kumar, R.S.; Yennawar, N.H.; Yennawar, H.P.; Ferry, J.G. Structural and biochemical characterizations of methanoredoxin from Methanosarcina acetivorans, a glutaredoxin-like enzyme with coenzyme M-dependent protein disulfide reductase activity. Biochemistry 2016, 55, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, A.R.; Fahey, R.C. The function of γ-glutamylcysteine and bis-γ-glutamylcystine reductase in Halobacterium halobium. J. Biol. Chem. 1989, 264, 719–725. [Google Scholar] [PubMed]
- Sundquist, A.R.; Fahey, R.C. The novel disulfide reductase bis-γ-glutamylcystine reductase and dihydrolipoamide dehydrogenase from Halobacterium halobium: Purification by immobilized-metal-ion affinity chromatography and properties of the enzymes. J. Bacteriol. 1988, 170, 3459–3467. [Google Scholar] [CrossRef] [PubMed]
- Susanti, D.; Loganathan, U.; Mukhopadhyay, B. A novel F420-dependent thioredoxin reductase gated by low potential FAD: A tool for redox regulation in an anaerobe. J. Biol. Chem. 2016, 291, 23084–23100. [Google Scholar] [CrossRef] [PubMed]
- Kashima, Y.; Ishikawa, K. Alkyl hydroperoxide reductase dependent on thioredoxin-like protein from Pyrococcus horikoshii. J. Biochem. 2003, 134, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Si, M.; Zhang, L.; Chaudhry, M.T.; Ding, W.; Xu, Y.; Chen, C.; Akbar, A.; Shen, X.; Liu, S.J. Corynebacterium glutamicum methionine sulfoxide reductase A uses both mycoredoxin and thioredoxin for regeneration and oxidative stress resistance. Appl. Environ. Microbiol. 2015, 81, 2781–2796. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, B.; Rankin, C.L.; Toyo-Oka, K.; Richter, M.L.; Maupin-Furlow, J.A.; Moskovitz, J. Methionine sulfoxide reductase A (MsrA) mediates the ubiquitination of 14-3-3 protein isotypes in brain. Free Radic. Biol. Med. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.C.; Lee, H.M.; Kim, S.; Avanesov, A.S.; Lee, A.; Chun, B.H.; Vorbruggen, G.; Gladyshev, V.N. Expression of the methionine sulfoxide reductase lost during evolution extends Drosophila lifespan in a methionine-dependent manner. Sci. Rep. 2018, 8, 1010. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.L.; Wu, P.F.; Wang, S.; Zhang, J.J.; Shen, Z.C.; Luo, H.; Chen, H.; Long, L.H.; Chen, J.G.; Wang, F. Dimethyl sulfide protects against oxidative stress and extends lifespan via a methionine sulfoxide reductase A-dependent catalytic mechanism. Aging Cell 2017, 16, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Salmon, A.B.; Kim, G.; Liu, C.; Wren, J.D.; Georgescu, C.; Richardson, A.; Levine, R.L. Effects of transgenic methionine sulfoxide reductase A (MsrA) expression on lifespan and age-dependent changes in metabolic function in mice. Redox Biol. 2016, 10, 251–256. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).