Machine Learning Strategies for Forecasting Mannosylerythritol Lipid Production Through Fermentation: A Proof-of-Concept
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
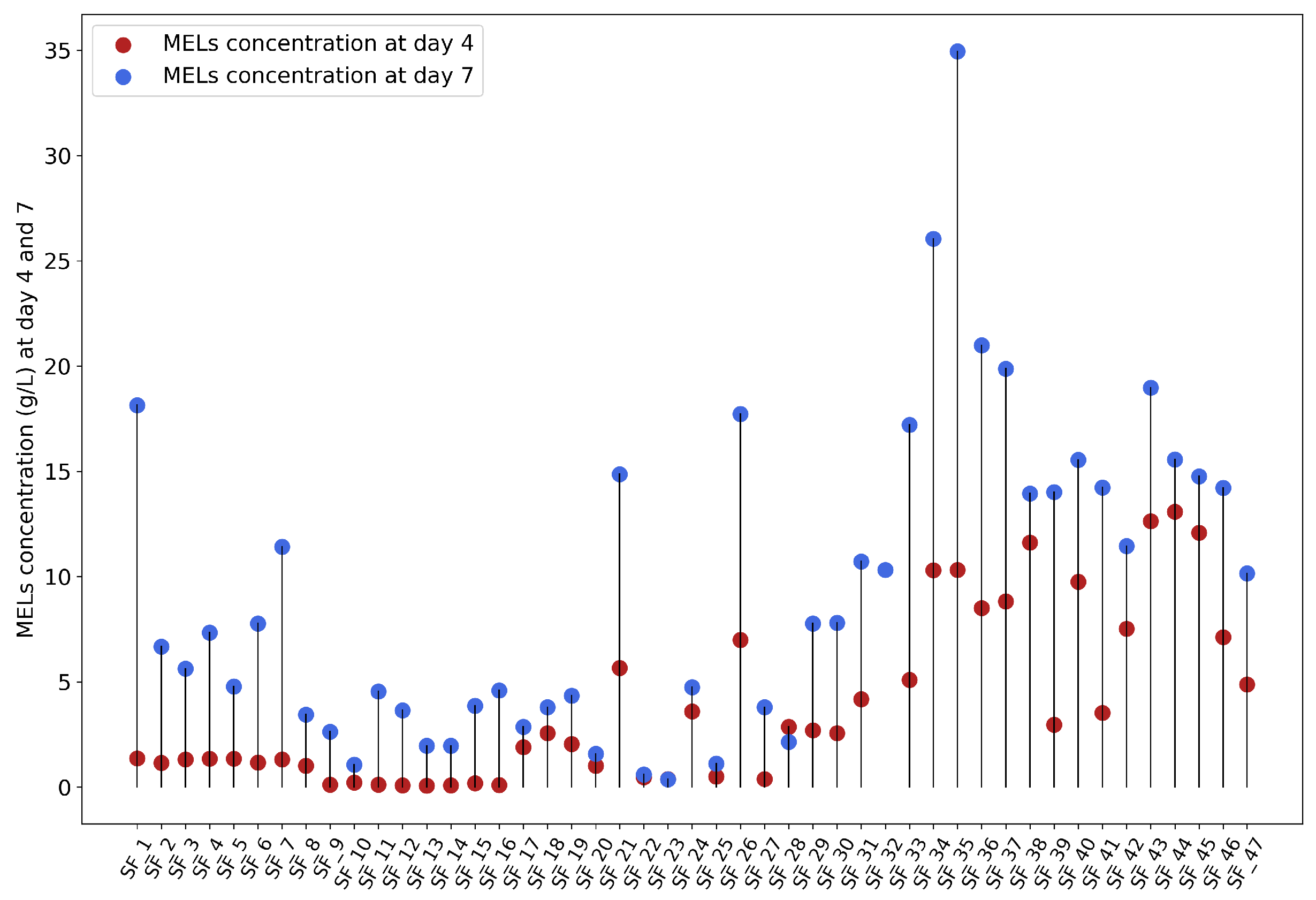
2.1. Dataset
- -
- Experiment 1 included 18 samples, a duration of 18 days, and a volume of 50 mL. D-glucose or glycerol (or simply no hydrophilic carbon source) was used in combination with a hydrophobic carbon source, such as soybean oil, rapeseed oil, or waste frying oil.
- -
- Experiment 2 included 10 samples, a duration of 10 days, and a volume of 50 mL. Waste frying oil was used as the hydrophobic carbon source, while D-glucose and cheese whey (CW) were used as the hydrophilic carbon sources.
- -
- Experiment 3 comprised 8 samples, a duration of 7 days, and the consistent use of D-glucose and waste frying oil as the hydrophilic and hydrophobic carbon sources, respectively. Four different volumes were tested (200 mL, 100 mL, 50 mL, and 25 mL), with each set of conditions run in duplicate.
- -
- Experiment 4 included 11 samples, with each running for 10 days with a volume of 50 mL. D-glucose was used as the hydrophilic carbon source, while the hydrophobic carbon sources were varied between residual fish oil and sunflower oil.
2.2. Data Preprocessing
2.3. Feature Engineering
2.4. Machine Learning Techniques
3. Results and Discussion
3.1. MEL Production Forecasting: Day 4
3.2. MEL Production Forecasting: Day 7
3.3. Benchmarking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Adaptive moment estimation | ADAM |
| Analysis of variance | ANOVA |
| Cheese whey | CW |
| Coefficient of determination | R2 |
| Decision tree | DT |
| Emulsification index | E24 |
| Least Absolute Shrinkage and Selection Operator | LASSO |
| Machine learning | ML |
| Mannosylerythritol lipids | MELs |
| Mean absolute error | MAE |
| Mean squared error | MSE |
| Neural network | NN |
| Principal component analysis | PCA |
| Radial basis function | RBF |
| Random forest | RF |
| Recursive feature elimination | RFE |
| Root mean squared error | RMSE |
| Support vector machine | SVM |
References
- Sharma, R.; Garg, P.; Kumar, P.; Bhatia, S.K.; Kulshrestha, S. Microbial fermentation and its role in quality improvement of fermented foods. Fermentation 2020, 6, 106. [Google Scholar] [CrossRef]
- Eastham, J.L.; Leman, A.R. Precision fermentation for food proteins: Ingredient innovations, bioprocess considerations, and outlook—A mini-review. Curr. Opin. Food Sci. 2024, 58, 101194. [Google Scholar] [CrossRef]
- Formenti, L.R.; Nørregaard, A.; Bolic, A.; Hernandez, D.Q.; Hagemann, T.; Heins, A.L.; Larsson, H.; Mears, L.; Mauricio-Iglesias, M.; Krühne, U.; et al. Challenges in industrial fermentation technology research. Biotechnol. J. 2014, 9, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Nagtode, V.S.; Cardoza, C.; Yasin, H.K.A.; Mali, S.N.; Tambe, S.M.; Roy, P.; Singh, K.; Goel, A.; Amin, P.D.; Thorat, B.R.; et al. Green surfactants (biosurfactants): A petroleum-free substitute for Sustainability—Comparison, applications, market, and future prospects. ACS Omega 2023, 8, 11674–11699. [Google Scholar] [CrossRef]
- Rebello, S.; Asok, A.K.; Mundayoor, S.; Jisha, M. Surfactants: Toxicity, remediation and green surfactants. Environ. Chem. Lett. 2014, 12, 275–287. [Google Scholar] [CrossRef]
- Farias, C.B.B.; Almeida, F.C.; Silva, I.A.; Souza, T.C.; Meira, H.M.; Rita de Cássia, F.; Luna, J.M.; Santos, V.A.; Converti, A.; Banat, I.M.; et al. Production of green surfactants: Market prospects. Electron. J. Biotechnol. 2021, 51, 28–39. [Google Scholar] [CrossRef]
- Zhou, Y.; Harne, S.; Amin, S. Optimization of the Surface Activity of Biosurfactant–Surfactant Mixtures. J. Cosmet. Sci. 2019, 70, 127. [Google Scholar]
- Coelho, A.L.S.; Feuser, P.E.; Carciofi, B.A.M.; de Andrade, C.J.; de Oliveira, D. Mannosylerythritol lipids: Antimicrobial and biomedical properties. Appl. Microbiol. Biotechnol. 2020, 104, 2297–2318. [Google Scholar] [CrossRef]
- de Andrade, C.J.; Coelho, A.L.; Feuser, P.E.; de Andrade, L.M.; Carciofi, B.A.; de Oliveira, D. Mannosylerythritol lipids: Production, downstream processing, and potential applications. Curr. Opin. Biotechnol. 2022, 77, 102769. [Google Scholar] [CrossRef]
- Kitamoto, D.; Haneishi, K.; Nakahara, T.; Tabuchi, T. Production of mannosylerythritol lipids by Candida antarctica from vegetable oils. Agric. Biol. Chem. 1990, 54, 37–40. [Google Scholar] [CrossRef][Green Version]
- Rau, U.; Nguyen, L.; Schulz, S.; Wray, V.; Nimtz, M.; Roeper, H.; Koch, H.; Lang, S. Formation and analysis of mannosylerythritol lipids secreted by Pseudozyma aphidis. Appl. Microbiol. Biotechnol. 2005, 66, 551–559. [Google Scholar] [CrossRef]
- Saikia, R.R.; Deka, H.; Goswami, D.; Lahkar, J.; Borah, S.N.; Patowary, K.; Baruah, P.; Deka, S. Achieving the best yield in glycolipid biosurfactant preparation by selecting the proper carbon/nitrogen ratio. J. Surfactants Deterg. 2014, 17, 563–571. [Google Scholar] [CrossRef]
- Joice, P.A.; Parthasarathi, R. Optimization of biosurfactant production from Pseudomonas aeruginosa PBSC1. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 140–151. [Google Scholar] [CrossRef]
- Xia, W.J.; Luo, Z.b.; Dong, H.P.; Yu, L.; Cui, Q.F.; Bi, Y.Q. Synthesis, characterization, and oil recovery application of biosurfactant produced by indigenous Pseudomonas aeruginosa WJ-1 using waste vegetable oils. Appl. Biochem. Biotechnol. 2012, 166, 1148–1166. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.F.; Barreiros, R.; Oliveira, A.C.; Ferreira, F.C.; Faria, N.T. Moesziomyces spp. cultivation using cheese whey: New yeast extract-free media, β-galactosidase biosynthesis and mannosylerythritol lipids production. Biomass Convers. Biorefinery 2024, 14, 6783–6796. [Google Scholar] [CrossRef]
- Agostinho, S.P.; A. Branco, M.; ES Nogueira, D.; Diogo, M.M.; S. Cabral, J.M.; N. Fred, A.L.; V. Rodrigues, C.A. Unsupervised analysis of whole transcriptome data from human pluripotent stem cells cardiac differentiation. Sci. Rep. 2024, 14, 3110. [Google Scholar]
- Helleckes, L.M.; Hemmerich, J.; Wiechert, W.; von Lieres, E.; Grünberger, A. Machine learning in bioprocess development: From promise to practice. Trends Biotechnol. 2023, 41, 817–835. [Google Scholar] [CrossRef]
- Cybenko, G. Approximation by superpositions of a sigmoidal function. Math. Control Signals Syst. 1989, 2, 303–314. [Google Scholar] [CrossRef]
- Thibault, J.; Van Breusegem, V.; Chéruy, A. On-line prediction of fermentation variables using neural networks. Biotechnol. Bioeng. 1990, 36, 1041–1048. [Google Scholar] [CrossRef]
- Zhang, A.H.; Zhu, K.Y.; Zhuang, X.Y.; Liao, L.X.; Huang, S.Y.; Yao, C.Y.; Fang, B.S. A robust soft sensor to monitor 1, 3-propanediol fermentation process by Clostridium butyricum based on artificial neural network. Biotechnol. Bioeng. 2020, 117, 3345–3355. [Google Scholar] [CrossRef]
- Cutler, A.; Cutler, D.R.; Stevens, J.R. Random forests. Ensemble Mach. Learn. Methods Appl. 2012, 157–175. [Google Scholar] [CrossRef]
- Zhang, W.; Li, J.; Liu, T.; Leng, S.; Yang, L.; Peng, H.; Jiang, S.; Zhou, W.; Leng, L.; Li, H. Machine learning prediction and optimization of bio-oil production from hydrothermal liquefaction of algae. Bioresour. Technol. 2021, 342, 126011. [Google Scholar] [CrossRef] [PubMed]
- Jovic, S.; Guresic, D.; Babincev, L.; Draskovic, N.; Dekic, V. Comparative efficacy of machine-learning models in prediction of reducing uncertainties in biosurfactant production. Bioprocess Biosyst. Eng. 2019, 42, 1695–1699. [Google Scholar] [CrossRef]
- de Andrade Bustamante, R.; de Oliveira, J.S.; Dos Santos, B.F. Modeling biosurfactant production from agroindustrial residues by neural networks and polynomial models adjusted by particle swarm optimization. Environ. Sci. Pollut. Res. 2023, 30, 6466–6491. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, M.F.; Coelho, T.; Reis, A.; Gouveia, L.; Faria, N.T.; Ferreira, F.C. Production of Mannosylerythritol Lipids Using Oils from Oleaginous Microalgae: Two Sequential Microorganism Culture Approach. Microorganisms 2022, 10, 2390. [Google Scholar] [CrossRef]
- Keković, P.; Borges, M.; Faria, N.T.; Ferreira, F.C. Towards Mannosylerythritol Lipids (MELs) for Bioremediation: Effects of NaCl on M. antarcticus Physiology and Biosurfactant and Lipid Production; Ecotoxicity of MELs. J. Mar. Sci. Eng. 2022, 10, 1773. [Google Scholar] [CrossRef]
- Kachrimanidou, V.; Alexandri, M.; Nascimento, M.F.; Alimpoumpa, D.; Torres Faria, N.; Papadaki, A.; Castelo Ferreira, F.; Kopsahelis, N. Lactobacilli and Moesziomyces Biosurfactants: Toward a Closed-Loop Approach for the Dairy Industry. Fermentation 2022, 8, 517. [Google Scholar] [CrossRef]
- Faria, N.T.; Nascimento, M.F.; Ferreira, F.A.; Esteves, T.; Santos, M.V.; Ferreira, F.C. Substrates of Opposite Polarities and Downstream Processing for Efficient Production of the Biosurfactant Mannosylerythritol Lipids from Moesziomyces spp. Appl. Biochem. Biotechnol. 2023, 195, 6132–6149. [Google Scholar] [CrossRef]
- Potdar, K.; Pardawala, T.S.; Pai, C.D. A comparative study of categorical variable encoding techniques for neural network classifiers. Int. J. Comput. Appl. 2017, 175, 7–9. [Google Scholar] [CrossRef]
- Lin, W.C.; Tsai, C.F. Missing value imputation: A review and analysis of the literature (2006–2017). Artif. Intell. Rev. 2020, 53, 1487–1509. [Google Scholar] [CrossRef]
- Sharma, V. A study on data scaling methods for machine learning. Int. J. Glob. Acad. Sci. Res. 2022, 1, 31–42. [Google Scholar] [CrossRef]
- Rahmat, F.; Zulkafli, Z.; Ishak, A.J.; Abdul Rahman, R.Z.; Stercke, S.D.; Buytaert, W.; Tahir, W.; Ab Rahman, J.; Ibrahim, S.; Ismail, M. Supervised feature selection using principal component analysis. Knowl. Inf. Syst. 2024, 66, 1955–1995. [Google Scholar] [CrossRef]
- Bejani, M.; Gharavian, D.; Charkari, N.M. Audiovisual emotion recognition using ANOVA feature selection method and multi-classifier neural networks. Neural Comput. Appl. 2014, 24, 399–412. [Google Scholar] [CrossRef]
- Nasiri, H.; Alavi, S.A. A Novel Framework Based on Deep Learning and ANOVA Feature Selection Method for Diagnosis of COVID-19 Cases from Chest X-Ray Images. Comput. Intell. Neurosci. 2022, 2022, 4694567. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, R.; Rohini, R. LASSO: A feature selection technique in predictive modeling for machine learning. In Proceedings of the 2016 IEEE International Conference on Advances in Computer Applications (ICACA), Coimbatore, India, 24 October 2016; pp. 18–20. [Google Scholar] [CrossRef]
- Ghosh, P.; Azam, S.; Jonkman, M.; Karim, A.; Shamrat, F.J.M.; Ignatious, E.; Shultana, S.; Beeravolu, A.R.; De Boer, F. Efficient prediction of cardiovascular disease using machine learning algorithms with relief and LASSO feature selection techniques. IEEE Access 2021, 9, 19304–19326. [Google Scholar] [CrossRef]
- Spearman, C. The Proof and Measurement of Association Between Two Things, oAmerican J. Psychol 1904, 15, 72–101. [Google Scholar] [CrossRef]
- Gopika, N.; ME, A.M.K. Correlation based feature selection algorithm for machine learning. In Proceedings of the 2018 3rd International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 15–16 October 2018; pp. 692–695. [Google Scholar] [CrossRef]
- Yan, K.; Zhang, D. Feature selection and analysis on correlated gas sensor data with recursive feature elimination. Sens. Actuators B Chem. 2015, 212, 353–363. [Google Scholar] [CrossRef]
- Zhang, F.; O’Donnell, L.J. Chapter 7—Support vector regression. In Machine Learning; Mechelli, A., Vieira, S., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 123–140. [Google Scholar] [CrossRef]
- Werbos, P.J. Backpropagation through time: What it does and how to do it. Proc. IEEE 1990, 78, 1550–1560. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems, 2015. Software. Available online: https://www.tensorflow.org/ (accessed on 19 March 2025).
- Chollet, F. Keras. 2015. Software. Available online: https://keras.io (accessed on 19 March 2025).
- Ahmad, Z.; Crowley, D.; Marina, N.; Jha, S.K. Estimation of biosurfactant yield produced by Klebseilla sp. FKOD36 bacteria using artificial neural network approach. Measurement 2016, 81, 163–173. [Google Scholar]



| Model | Hyperparameter | Day 4 | Day 7 |
|---|---|---|---|
| Random Forest | Number of estimators | 10 | 1 |
| Criterion | Absolute error | Squared error | |
| Maximum depth | None | None | |
| Minimum sample split | 3 | 3 | |
| Maximum features | Sqrt | Sqrt | |
| Bootstrap | True | True | |
| Support Vector Machine | Kernel | RBF | Polynomial |
| Epsilon | 0.001 | 0.001 | |
| Gamma | 1 | 0.001 | |
| C | 0.1 | 75 | |
| Neural Network | Number of layers and neurons | [64, 32, 16] | [128, 64] |
| Loss function | MSE | MSE | |
| Output layer | 1 | 4 | |
| Batch size | 8 | 4 |
| All Features | RFE | Correlation | LASSO | ANOVA | PCA | |
|---|---|---|---|---|---|---|
| Neural Network | 1.77 | 12.88 | 0.69 | 2.83 | 2.63 | 1.63 |
| Random Forest | 4.45 | 9.22 | 11.01 | 7.35 | 3.45 | 4.15 |
| Support Vector | 29.12 | 29.52 | 28.24 | 30.76 | 30.04 | 28.64 |
| Experimental Values (g/L) | RF Predictions (g/L) ± Error | NN Predictions (g/L) ± Error | SVM Predictions (g/L) ± Error |
|---|---|---|---|
| 0.08 | 0.14 | 0.00 | 1.41 |
| ± 0.06 | ± 0.08 | ± 1.33 | |
| 4.17 | 7.19 | 4.59 | 1.54 |
| ± 3.02 | ± 0.42 | ± 2.63 | |
| 3.60 | 5.24 | 3.11 | 1.67 |
| ± 1.64 | ± 0.49 | ± 1.93 | |
| 13.08 | 10.16 | 10.81 | 1.91 |
| ± 2.92 | ± 2.27 | ± 11.17 | |
| 9.76 | 9.92 | 10.00 | 1.58 |
| ± 0.16 | ± 0.24 | ± 8.18 | |
| 1.17 | 3.32 | 1.42 | 1.24 |
| ± 2.15 | ± 0.25 | ± 0.07 | |
| 0.12 | 2.00 | 0.59 | 1.50 |
| ± 1.88 | ± 0.47 | ± 1.38 | |
| 0.38 | 0.50 | 1.20 | 1.61 |
| ± 0.12 | ± 0.82 | ± 1.23 | |
| 6.99 | 4.62 | 6.56 | 1.76 |
| ± 2.37 | ± 0.43 | ± 5.23 | |
| 8.82 | 8.32 | 8.52 | 1.91 |
| ± 0.50 | ± 0.30 | ± 6.91 |
| All Features | RFE | Correlation | LASSO | ANOVA | PCA | |
|---|---|---|---|---|---|---|
| Neural Network | 7.01 | 3.76 | 6.47 | 10.31 | 1.63 | 4.2 |
| Random Forest | 12.99 | 13.82 | 27.07 | 9.48 | 59.39 | 27.96 |
| Support Vector | 44.21 | 37.36 | 41.34 | 45.01 | 36.63 | 36.25 |
| Experimental Values (g/L) | RF Predictions (g/L) ± Error | NN Predictions (g/L) ± Error | SVM Predictions (g/L) ± Error |
|---|---|---|---|
| 15.45 | 10.16 | 15.64 | 6.73 |
| ± 5.29 | ± 0.19 | ± 8.72 | |
| 1.97 | 4.23 | 3.62 | 5.84 |
| ± 2.26 | ± 1.65 | ± 3.87 | |
| 3.65 | 4.23 | 4.77 | 5.73 |
| ± 0.58 | ± 1.12 | ± 2.08 | |
| 4.34 | 4.74 | 6.71 | 6.58 |
| ± 0.40 | ± 2.37 | ± 2.24 | |
| 10.32 | 13.83 | 11.79 | 7.06 |
| ± 3.51 | ± 1.47 | ± 3.26 | |
| 7.77 | 5.62 | 9.24 | 7.74 |
| ± 2.15 | ± 1.47 | ± 0.03 | |
| 11.42 | 5.62 | 10.74 | 6.74 |
| ± 5.80 | ± 0.68 | ± 4.68 | |
| 20.99 | 18397 | 20.75 | 7.74 |
| ± 2.02 | ± 0.24 | ± 13.25 | |
| 7.77 | 5.62 | 8.67 | 6.89 |
| ± 2.15 | ± 0.90 | ± 0.88 | |
| 13.94 | 13.01 | 14.93 | 6.72 |
| ± 0.93 | ± 0.99 | ± 7.22 |
| Random Forest | Neural Network | Support Vector Machine | |
|---|---|---|---|
| R2—day 4 | 0.82 | 0.96 | −0.47 |
| R2—day 7 | 0.70 | 0.95 | −0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vares, C.A.; Agostinho, S.P.; Fred, A.L.N.; Faria, N.T.; Rodrigues, C.A.V. Machine Learning Strategies for Forecasting Mannosylerythritol Lipid Production Through Fermentation: A Proof-of-Concept. Appl. Sci. 2025, 15, 3709. https://doi.org/10.3390/app15073709
Vares CA, Agostinho SP, Fred ALN, Faria NT, Rodrigues CAV. Machine Learning Strategies for Forecasting Mannosylerythritol Lipid Production Through Fermentation: A Proof-of-Concept. Applied Sciences. 2025; 15(7):3709. https://doi.org/10.3390/app15073709
Chicago/Turabian StyleVares, Carolina A., Sofia P. Agostinho, Ana L. N. Fred, Nuno T. Faria, and Carlos A. V. Rodrigues. 2025. "Machine Learning Strategies for Forecasting Mannosylerythritol Lipid Production Through Fermentation: A Proof-of-Concept" Applied Sciences 15, no. 7: 3709. https://doi.org/10.3390/app15073709
APA StyleVares, C. A., Agostinho, S. P., Fred, A. L. N., Faria, N. T., & Rodrigues, C. A. V. (2025). Machine Learning Strategies for Forecasting Mannosylerythritol Lipid Production Through Fermentation: A Proof-of-Concept. Applied Sciences, 15(7), 3709. https://doi.org/10.3390/app15073709






