Microwave-Assisted Synthesis, Lipophilicity and In Vitro Antimicrobial Activity of Hydrazide-Hydrazones of Phenylacetic Acid
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemistry
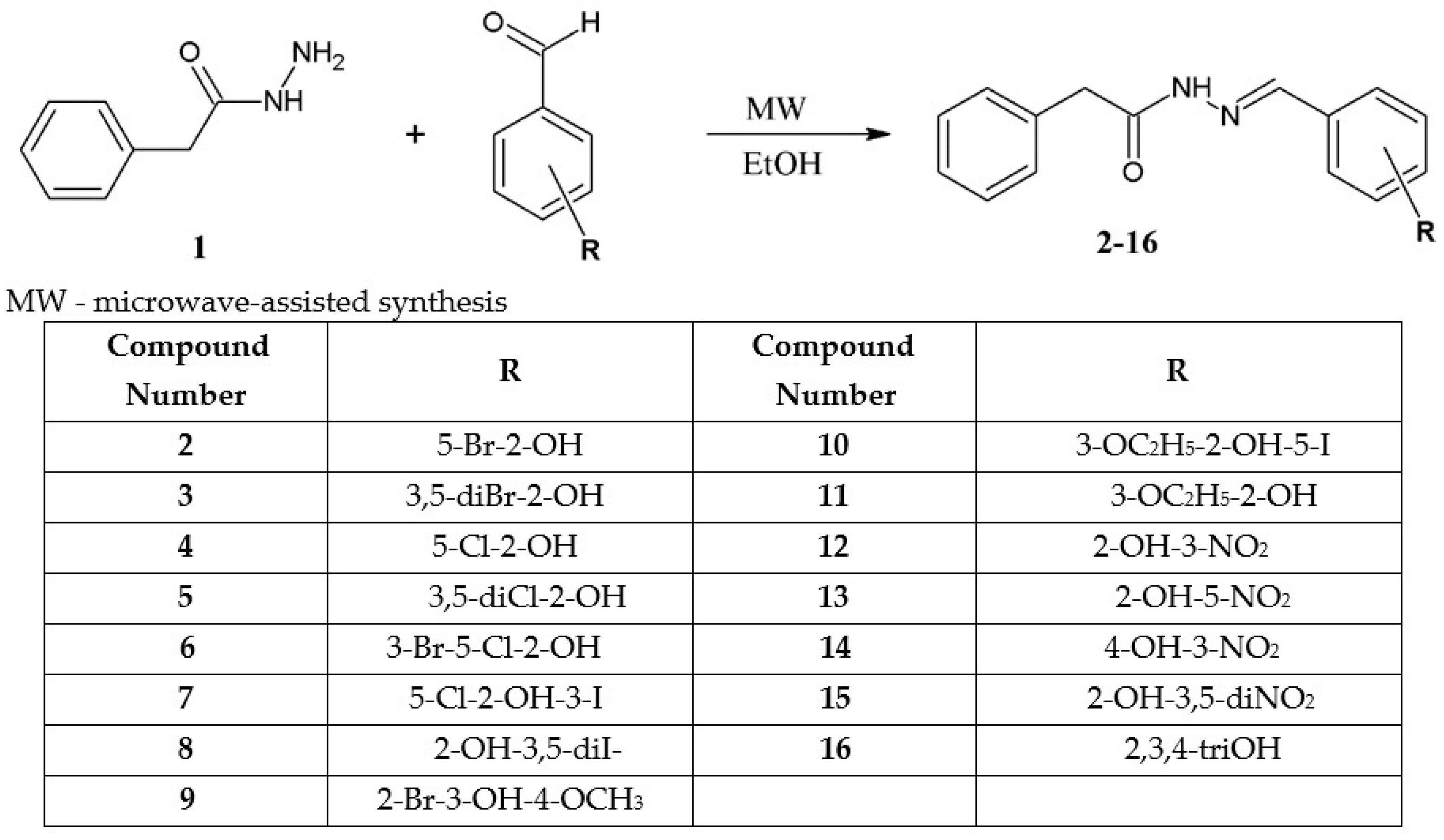
Detailed Procedure for the Synthesis of Hydrazide-Hydrazones of Phenylacetic Acid (2–16)
2.2. Lipophilicity
2.3. Microbiology
In Vitro Antimicrobial Assay
3. Results
3.1. Chemistry
3.2. Lipophilicity
- (1)
- acetonitrile: logPEXP = (RM0 − 0.6348)/0.5749; r2 = 0.9568
- (2)
- 1,4-dioxane: logPEXP = (RM0 − 0.2591)/0.7899; r2 = 0.9098
- (3)
- methanol: logPEXP = (RM0 − 0.1883)/1.0124; r2 = 0.9568
3.3. Microbiology
4. Discussion
4.1. Chemistry
4.2. Lipophilicity
4.3. Microbiology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thota, S.; Rodrigues, D.A.; Pinheiro, P.d.S.M.; Lima, L.M.; Fraga, C.A.M.; Barreiro, E.J. N-Acylhydrazones as drugs. Bioorg. Med. Chem. Lett. 2018, 28, 2797–2806. [Google Scholar] [CrossRef]
- Wahid, S.; Jahangir, S.; Versiani, M.A.; Khan, K.M.; Sung, Y.Y.; Iqbal, J.; Wadood, A.; Rehman, A.U.; Uzair, M.; Khan, I.A. Biology-oriented drug synthesis of nitrofurazone derivatives: Their α-glucosidase inhibitory activity and molecular docking studies. Arab. J. Chem. 2022, 15, 103806. [Google Scholar] [CrossRef]
- Zuma, N.H.; Smit, F.J.; Seldon, R.; Aucamp, J.; Jordaan, A.; Warner, D.F.; N’Da, D.D. Single-step synthesis and in vitro anti-mycobacterial activity of novel nitrofurantoin analogues. Bioorg. Chem. 2020, 96, 103587. [Google Scholar] [CrossRef]
- Mohammadi, M.; Attaran, B.; Malekzadeh, R.; Graham, D.Y. Furazolidone, an Underutilized Drug for H. pylori Eradication: Lessons from Iran. Diges. Dis. Sci. 2017, 62, 1890–1896. [Google Scholar] [CrossRef]
- Zhang, J.; Rong, C.; Yan, C.; Chen, J.; Yang, W.; Yu, L.; Dai, H. Risk factors of furazolidone-associated fever. PLoS ONE 2022, 17, e0266763. [Google Scholar] [CrossRef]
- Luo, Y.; Zhao, X.; Releken, Y.; Yang, Z.; Pei, F.; Kang, P. Hemostatic and Anti-Inflammatory Effects of Carbazochrome Sodium Sulfonate in Patients Undergoing Total Knee Arthroplasty: A Randomized Controlled Trial. J. Arthroplast. 2020, 35, 61–68. [Google Scholar] [CrossRef]
- Bailly, C. Toward a repositioning of the antibacterial drug nifuroxazide for cancer treatment. Drug Disc. Today 2019, 24, 1359–6446. [Google Scholar] [CrossRef]
- Whyte, C.J.; Rosini, J.M. Dantrolene for treatment of suspected neuroleptic malignant syndrome. J. Emerg. Nurs. 2018, 44, 207–209. [Google Scholar] [CrossRef]
- Chakraborty, P.; Massé, S.; Azam, M.A.; Thollon, C.; Niri, A.; Lai, P.F.H.; Bouly, M.; Riazi, S.; Nanthakumar, K. Effects of azumolene on arrhythmia substrate in a model of recurrent long-duration ventricular fibrillation. Biochem. Biophys. Res. Commun. 2022, 600, 123–129. [Google Scholar] [CrossRef]
- Popiołek, Ł.; Biernasiuk, A. Synthesis and investigation of antimicrobial activities of nitrofurazone analogues containing hydrazide-hydrazone moiety. Saudi Pharm. J. 2017, 25, 1097–1102. [Google Scholar] [CrossRef]
- Popiołek, Ł. Hydrazide-hydrazones as potential antimicrobial agents: Overview of the literature since 2010. Med. Chem. Res. 2017, 26, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Popiołek, Ł. Updated Information on Antimicrobial Activity of Hydrazide–Hydrazones. Int. J. Mol. Sci. 2021, 22, 9389. [Google Scholar] [CrossRef] [PubMed]
- Ajani, O.O.; Iyaye, K.T.; Audu, O.Y.; Olorunshola, S.J.; Kuye, A.O.; Olanrewaju, I.O. Microwave Assisted Synthesis and Antimicrobial Potential of Quinoline-Based 4-Hydrazide-Hydrazone Derivatives. J. Heterocycl. Chem. 2018, 55, 302–312. [Google Scholar] [CrossRef]
- Guilherme, F.D.; Simonetti, J.E.; Folquitto, L.R.S.; Reis, A.C.C.; Oliver, J.C.; Dias, A.L.T.; Dias, D.F.; Carvalho, D.T.; Brandão, G.C.; Belarmino de Souza, T. Synthesis, chemical characterization and antimicrobial activity of new acylhydrazones derived from carbohydrates. J. Mol. Struct. 2019, 1184, 349–356. [Google Scholar] [CrossRef]
- Ajani, O.O.; Iyaye, K.T.; Aderohunmu, D.V.; Olanrewaju, I.O.; Germann, M.W.; Olorunshola, S.J.; Bello, B.L. Microwave-assisted synthesis and antibacterial propensity of N′-s-benzylidene-2-propylquinoline-4-carbohydrazide and N′-((s-1H-pyrrol-2-yl)methylene)-2-propylquinoline-4-carbohydrazide motifs. Arab. J. Chem. 2020, 13, 1809–1820. [Google Scholar] [CrossRef]
- Aarjane, M.; Aouidate, A.; Slassi, S.; Amine, A. Synthesis, antibacterial evaluation, in silico ADMET and molecular docking studies of new N-acylhydrazone derivatives from acridone. Arab. J. Chem. 2020, 13, 6236–6245. [Google Scholar] [CrossRef]
- Dascalu, A.-E.; Ghinet, A.; Lipka, E.; Furman, C.; Rigo, B.; Feyulle, A.; Billamboz, M. Design, synthesis and evaluation of hydrazine and acyl hydrazone derivatives of 5-pyrrolidin-2-one as antifungal agents. Bioorg. Med. Chem. Lett. 2020, 30, 127220. [Google Scholar] [CrossRef]
- Dwivedi, D.K.; Sahu, A.; Dighade, S.J.; Agrawal, R.K. Design, synthesis, and antimicrobial evaluation of some nifuroxazide analogs against nosocomial infection. J. Heterocycl. Chem. 2020, 57, 1666–1671. [Google Scholar] [CrossRef]
- Rohane, S.H.; Chauhan, A.J.; Fuloria, N.K.; Fuloria, S. Synthesis and in vitro antimycobacterial potential of novel hydrazones of eugenol. Arab. J. Chem. 2020, 13, 4495–4504. [Google Scholar] [CrossRef]
- Lu, Y.-J.; Zhao, Z.-D.; Chen, Y.-X.; Wang, J.; Xu, S.-C.; Gu, Y. Synthesis and biological activity of pyridine acylhydrazone derivatives of isopimaric acid. J. Asian Nat. Prod. Res. 2021, 23, 545–555. [Google Scholar] [CrossRef]
- Gao, H.; Li, J.-Q.; Kang, P.-W.; Chigan, J.-Z.; Wang, H.; Liu, L.; Xu, Y.-S.; Zhai, L.; Yang, K.-W. N-acylhydrazones confer inhibitory efficacy against New Delhi metallo-β-lactamase-1. Bioorg. Chem. 2021, 114, 105138. [Google Scholar] [CrossRef]
- Popiołek, Ł.; Tuszyńska, K.; Biernasiuk, A. Searching for novel antimicrobial agents among hydrazide-hydrazones of 4-iodosalicylic acid. Biomed. Pharmacother. 2022, 153, 113302. [Google Scholar] [CrossRef]
- Filho, J.M.d.S.; Castro, M.V.B.d.S. Synthesis, structural characterization, and antimicrobial activity of novel ferrocene-N-acyl hydrazones designed by means of molecular simplification strategy. Celebrating the 100th anniversary of the birth of Professor Paulo Freire. J. Organomet. Chem. 2022, 979, 122488. [Google Scholar] [CrossRef]
- Barbier, T.; Barbry, A.; Magand, J.; Badiou, C.; Davy, F.; Baudouin, A.; Queneau, Y.; Dumitrescu, O.; Lina, G.; Soulère, L. Synthesis and Biological Evaluation of Benzo[b]thiophene Acylhydrazones as Antimicrobial Agents against Multidrug-Resistant Staphylococcus aureus. Biomolecules 2022, 12, 131. [Google Scholar] [CrossRef]
- Angelova, V.T.; Pencheva, T.; Vassilev, N.; Yovkova, E.K.; Mihaylova, R.; Petrov, B.; Valcheva, V. Development of New Antimycobacterial Sulfonyl Hydrazones and 4-Methyl-1,2,3-thiadiazole-Based Hydrazone Derivatives. Antibiotics 2022, 11, 562. [Google Scholar] [CrossRef]
- Cui, P.; Cai, M.; Meng, Y.; Yang, Y.; Song, H.; Liu, Y.; Wang, Q. Design, synthesis and biological activities of echinopsine derivatives containing acylhydrazone moiety. Sci. Rep. 2022, 12, 2935. [Google Scholar] [CrossRef]
- Yeye, E.O.; Adeniyi-Akee, M.A.l.; Ahmed, S.A.; Aboaba, S.A. In silico studies and antimicrobial investigation of synthesized novel N-acylhydrazone derivatives of indole. Sci. Afr. 2023, 19, e01463. [Google Scholar] [CrossRef]
- Ince, U.; Han, M.İ. Investigation of Antimicrobial Activity of Some Ethylparaben Hydrazide-Hydrazone Derivatives. Turk. J. Pharm. Sci. 2023, 20, 35–38. [Google Scholar] [CrossRef]
- Fernandes, G.d.S.; Moreno-Viguri, E.; Santivañez-Veliz, M.; Paucar, R.; Chin, C.M.; Pérez-Silanes, S.; dos Santosa, J.L. A Comparative Study of Conventional and Microwave-Assisted Synthesis of Quinoxaline 1,4-di-N-oxide N-acylhydrazones Derivatives Designed as Antitubercular Drug Candidates. J. Heterocycl. Chem. 2017, 54, 2380–2388. [Google Scholar] [CrossRef]
- Demirci, S.; Mermer, A.; Ak, G.; Aksakal, F.; Colak, N.; Demirbas, A.; Ayaz, F.A.; Demirbas, N. Conventional and Microwave-assisted Total Synthesis, Antioxidant Capacity, Biological Activity, and Molecular Docking Studies of New Hybrid Compounds. J. Heterocycl. Chem. 2017, 54, 1785–1805. [Google Scholar] [CrossRef]
- Sravanthi, K.; Snehalatha, P.; Subhashini, N.J.P. Microwave Assisted Green Synthesis of Pyrazole, 1,2,3-Triazole Based Novel Benzohydrazones and Their Antibacterial Activities. J. Heterocycl. Chem. 2018, 55, 508–516. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, B.C.V.; Chandra; Revanasiddappa, H.D. Crystal structure, Hirshfeld analysis and HSA interaction studies of N’-[(E)-(5-bromothiophen-2-yl)methylidene]-3-hydroxynaphthalene-2-carbohydrazide. J. Mol. Struct. 2019, 1189, 343–351. [Google Scholar] [CrossRef]
- Al-Sodies, S.A.; Aouad, M.R.; Ihmaid, S.; Aljuhani, A.; Messali, M.; Ali, I.; Rezki, N. Microwave and conventional synthesis of ester based dicationic pyridinium ionic liquids carrying hydrazone linkage: DNA binding, anticancer and docking studies. J. Mol. Struct. 2020, 1189, 127756. [Google Scholar] [CrossRef]
- Komsta, Ł.; Skibiński, R.; Berecka, A.; Gumieniczek, A.; Radkiewicz, B.; Radoń, M. Revisiting thin-layer chromatography as a lipophilicity determination tool—A comparative study on several techniques with a model solute set. J. Pharm. Biomed. Anal. 2010, 53, 911–918. [Google Scholar] [PubMed]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST). Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. EUCAST discussion document E. Dis 5.1. Clin. Microbiol. Infect. 2003, 9, 1–7. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. M27-S4; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Biernasiuk, A.; Kawczyńska, M.; Berecka-Rycerz, A.; Rosada, B.; Gumieniczek, A.; Malm, A.; Dzitko, K.; Łączkowski, K.Z. Synthesis, antimicrobial activity, and determination of the lipophilicity of ((cyclohex-3-enylmethylene)hydrazinyl)thiazole derivatives. Med. Chem. Res. 2019, 28, 2023–2036. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar]
- O’Donnell, F.; Smyth, T.J.; Ramachandran, V.N.; Smyth, W.F. A study of the antimicrobial activity of selected synthetic and naturally occurring quinolines. Int. J. Antimicrob. Agents. 2010, 35, 30–38. [Google Scholar]
- Liu, Y.; Li, M.; Zhang, Y.; Wu, C.; Yang, K.; Gao, S.; Zheng, M.; Li, X.; Li, H.; Chen, L. Structure based discovery of novel hexokinase 2 inhibitors. Bioorg. Chem. 2020, 96, 103609. [Google Scholar] [CrossRef]
- Dahlgren, M.K.; Zetterström, C.E.; Gylfe, Å.; Linusson, A.; Elofsson, M. Statistical molecular design of a focused salicylidene acylhydrazide library and multivariate QSAR of inhibition of type III secretion in the Gram-negative bacterium Yersinia. Bioorg. Med. Chem. 2010, 18, 2686–2703. [Google Scholar] [CrossRef]
- Szklarzewicz, J.; Jurowska, A.; Hodorowicz, M.; Kazek, G.; Głuch-Lutwin, M.; Sapa, J.; Papież, M. Tridentate ONO ligands in vanadium(III-V) complexes—Synthesis, characterization and biological activity. J. Mol. Struct. 2021, 1224, 129205. [Google Scholar] [CrossRef]
- Ramzan, S.; Rahim, S.; Hussain, S.T.; Holt, K.B.; Cockcroft, J.K.; Muhammad, N.; Ur-Rehman, Z.; Nawaz, A.; Shujah, S. Synthesis, characterization, X-ray structure, DNA binding, antioxidant and docking study of new organotin(IV) complexes. Appl. Organomet. Chem. 2023, 37, e7161. [Google Scholar] [CrossRef]
- Jasińska, A.; Szklarzewicz, J.; Jurowska, A.; Hodorowicz, M.; Kazek, G.; Mordyl, B.; Głuch-Lutwin, M. V(III) and V(IV) Schiff base complexes as potential insulin-mimetic compounds—Comparison, characterization and biological activity. Polyhedron 2022, 215, 115682. [Google Scholar] [CrossRef]
- Melnyk, P.; Leroux, V.; Sergheraert, C.; Grellier, P. Design, synthesis and in vitro antimalarial activity of an acylhydrazone library. Bioorg. Med. Chem. Lett. 2006, 16, 31–35. [Google Scholar] [CrossRef]
- Kim, B.-K.; Ko, H.; Jeon, E.-S.; Ju, E.-S.; Jeong, L.S.; Kim, Y.-C. 2,3,4-Trihydroxybenzyl-hydrazide analogues as novel potent coxsackievirus B3 3C protease inhibitors. Eur. J. Med. Chem. 2016, 120, 202–216. [Google Scholar] [CrossRef]
- Eros, D.; Kovesdi, I.; Orfi, L.; Takacs-Novak, K.; Ascady, G.; Keri, G. Reliability of logP Predictions Based on Calculated Molecular Descriptors: A Critical Review. Curr. Med. Chem. 2002, 9, 1819–1828. [Google Scholar]

| RM0 | S | r2 | φ | |
|---|---|---|---|---|
| Acetonitrile-Water | ||||
| 2-aminophenol | 1.1768 | −0.02 | 0.9964 | 54.37 |
| salicylamide | 1.2269 | −0.03 | 0.9970 | 44.77 |
| 4-dimethylaminebenzaldehyde | 1.5319 | −0.02 | 0.9843 | 62.20 |
| eugenol | 2.0104 | −0.03 | 0.9879 | 62.44 |
| thymol | 2.4306 | −0.04 | 0.9890 | 68.33 |
| phenyl salicylate | 2.9514 | −0.04 | 0.9873 | 79.68 |
| 1,4-Dioxane-Water | ||||
| 2-aminophenol | 1.0844 | −0.02 | 0.9916 | 47.21 |
| salicylamide | 0.9964 | −0.02 | 0.9957 | 45.60 |
| 4-dimethylaminebenzaldehyde | 1.6406 | −0.03 | 0.9928 | 56.20 |
| eugenol | 1.9741 | −0.03 | 0.9940 | 66.90 |
| thymol | 2.5440 | −0.04 | 0.9828 | 69.90 |
| phenyl salicylate | 3.6480 | −0.05 | 0.9940 | 73.84 |
| Methanol-Water | ||||
| 2-aminophenol | 0.9785 | −0.02 | 0.9975 | 58.90 |
| salicylamide | 1.1377 | −0.02 | 0.9817 | 56.60 |
| 4-dimethylaminebenzaldehyde | 2.1143 | −0.03 | 0.9891 | 75.02 |
| eugenol | 2.7002 | −0.04 | 0.9937 | 75.91 |
| thymol | 3.2151 | −0.04 | 0.9957 | 80.56 |
| phenyl salicylate | 4.2259 | −0.05 | 0.9724 | 88.04 |
| Compound No | Acetonitrile-Water | 1,4-Dioxane-Water | Methanol-Water | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RM0 | S | r2 | φ | RM0 | S | r2 | φ | RM0 | S | r2 | φ | |
| 2 | 2.7793 | −0.04 | 0.9722 | 70.86 | 3.1301 | −0.05 | 0.9803 | 64.17 | 4.0434 | −0.05 | 0.9931 | 83.00 |
| 3 | 3.1071 | −0.04 | 0.9792 | 76.11 | 3.7815 | −0.06 | 0.9717 | 67.54 | 4.8978 | −0.06 | 0.9913 | 87.08 |
| 4 | 2.4385 | −0.04 | 0.9849 | 68.03 | 3.2336 | −0.05 | 0.9927 | 61.91 | 3.9436 | −0.05 | 0.9862 | 80.80 |
| 5 | 2.8437 | −0.04 | 0.9872 | 73.71 | 3.8941 | −0.06 | 0.9997 | 65.10 | 4.7399 | −0.06 | 0.9886 | 84.77 |
| 6 | 2.9758 | −0.04 | 0.9918 | 74.58 | 3.9957 | −0.06 | 0.9955 | 65.42 | 4.7418 | −0.04 | 0.9983 | 86.00 |
| 7 | 3.1485 | −0.04 | 0.9863 | 76.34 | 4.0481 | −0.06 | 0.9978 | 66.01 | 4.8895 | −0.05 | 0.9913 | 87.24 |
| 8 | 3.3686 | −0.04 | 0.9747 | 79.02 | 4.0034 | −0.06 | 0.9846 | 68.82 | 5.0271 | −0.06 | 0.9979 | 88.90 |
| 9 | 2.0621 | −0.04 | 0.9916 | 58.52 | 2.4781 | −0.04 | 0.9939 | 58.01 | 3.1511 | −0.04 | 0.9866 | 76.25 |
| 10 | 2.1722 | −0.04 | 0.9873 | 61.40 | 2.6718 | −0.05 | 0.9926 | 59.00 | 3.5089 | −0.04 | 0.9943 | 78.19 |
| 11 | 2.1966 | −0.04 | 0.9834 | 62.39 | 2.4623 | −0.04 | 0.9957 | 58.57 | 3.3256 | −0.06 | 0.9932 | 77.28 |
| 12 | 2.3122 | −0.04 | 0.9858 | 62.26 | 2.7184 | −0.05 | 0.9985 | 60.10 | 3.3190 | −0.06 | 0.9913 | 78.44 |
| 13 | 2.5229 | −0.04 | 0.9916 | 63.57 | 2.9981 | −0.05 | 0.9932 | 59.95 | 3.5991 | −0.05 | 0.9902 | 79.83 |
| 14 | 2.1061 | −0.04 | 0.9838 | 59.31 | 2.4686 | −0.04 | 0.9988 | 58.77 | 3.1517 | −0.04 | 0.9891 | 76.62 |
| 15 | 0.8263 | −0.02 | 0.9903 | 40.06 | 1.0641 | −0.02 | 0.9882 | 45.35 | 2.4159 | −0.04 | 0.9783 | 65.56 |
| 16 | 1.2273 | −0.03 | 0.9732 | 43.00 | 1.0413 | −0.03 | 0.9948 | 45.60 | 1.7280 | −0.03 | 0.9809 | 63.13 |
| Compound Number | LogPacetonitrile | LogP1.4-dioxane | LogPmethanol |
|---|---|---|---|
| 2 | 3.73 | 3.63 | 3.81 |
| 3 | 4.30 | 4.46 | 4.65 |
| 4 | 3.14 | 3.77 | 3.71 |
| 5 | 3.84 | 4.60 | 4.50 |
| 6 | 4.07 | 4.73 | 4.50 |
| 7 | 4.37 | 4.80 | 4.64 |
| 8 | 4.76 | 4.74 | 4.78 |
| 9 | 2.48 | 2.81 | 2.93 |
| 10 | 2.67 | 3.05 | 3.28 |
| 11 | 2.72 | 2.79 | 3.10 |
| 12 | 2.92 | 3.11 | 3.09 |
| 13 | 3.28 | 3.47 | 3.37 |
| 14 | 2.56 | 2.80 | 2.93 |
| 15 | 0.33 | 1.02 | 2.20 |
| 16 | 1.03 | 0.99 | 1.52 |
| Species | MIC (MBC or MFC) [µg/mL] and {MBC/MIC or MFC/MIC} Values of the Studied Compounds and Positive Controls | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 9 | CIP/VA */ NY ** | NIT | CFX | APC | ||
| Gram-positive bacteria | Staphylococcus aureus ATCC 43300 | 1000 (>1000) {>1} | - | 1000 (>1000) {>1} | - | - | - | - | 0.24 (0.24) {1} | 7.81 (15.62) | 0.49 | nd |
| Staphylococcus aureus ATCC 6538 | 1000 (>1000) {>1} | - | 1000 (>1000) {>1} | - | 1000 (>1000) {>1} | - | - | 0.24 (0.24) {1} | 15.62 (15.62) | nd | nd | |
| Staphylococcus aureus ATCC 29213 | 1000 (>1000) {>1} | 1000 (>1000) {>1} | - | - | 250 (>1000) {>4} | - | - | 0.48 (0.48) {1} | nd | nd | nd | |
| Staphylococcus epidermidis ATCC 12228 | - | 500 (>1000) {>2} | - | - | 62.5 (>1000) {>16} | - | - | 0.12 (0.12) {1} | 3.91 (7.81) | 0.24 | nd | |
| Enterococcus faecalis ATCC 29212 | 250 (>1000) {>4} | - | 250 (>1000) {>4} | - | 31.25 (>1000) {>32} | - | 1000 (>1000) {>1} | 0.98 * (1.95) {2} | nd | nd | nd | |
| Micrococcus luteus ATCC 10240 | 1000 (>1000) {>1} | 250 (>1000) {>4} | 250 (>1000) {>4} | 250 (>1000) {>4} | 31.25 (>1000) {>32} | - | 250 (>1000) {>4} | 0.98 (1.95) {2} | 62.5 (62.5) | 0.98 | nd | |
| Bacillus subtilis ATCC 6633 | - | - | - | - | 500 (>1000) {>2} | - | - | 0.03 (0.03) {1} | 3.91 (3.91) | 15.62 | 62.5 | |
| Bacillus cereus ATCC 10876 | 1000 (>1000) {>1} | 500 (>1000) {>2} | - | - | 250 (250) {1} | - | - | 0.06 (0.12) {2} | 7.81 (15.62) | 31.25 | nd | |
| Gram-negative bacteria | Bordetella bronchiseptica ATCC 4617 | - | - | - | - | - | - | - | 0.98 (0.98) {1} | 125 (>1000) | nd | nd |
| Klebsiella pneumoniae ATCC 13883 | - | - | - | - | - | - | - | 0.12 (0.24) {2} | 15.62 (31.25) | nd | nd | |
| Proteus mirabilis ATCC 12453 | - | - | - | - | - | - | - | 0.03 (0.03) {1) | 62.5 (125) | nd | nd | |
| Salmonella typhimurium ATCC 14028 | - | - | - | - | - | - | - | 0.06 (0.06) {1} | 31.25 (62.5) | nd | nd | |
| Escherichia coli ATCC 25922 | - | - | - | - | - | - | - | 0.004 (0.008) {2} | 7.81 (15.62) | nd | nd | |
| Pseudomonas aeruginosa ATCC 9027 | - | - | - | - | - | - | - | 0.48 (0.98) {2} | nd | nd | nd | |
| Fungi | Candida auris CDC 311903 | 1000 (>1000) {>1} | - | - | - | - | - | - | 0.48 ** (0.48) {1} | na | na | na |
| Candida albicans ATCC 2091 | 1000 (>1000) {>1} | - | - | - | 1000 (>1000) {>1} | - | - | 0.24 ** (0.24) {1} | na | na | na | |
| Candida albicans ATCC 10231 | 1000 (>1000) {>1} | 1000 (>1000) {>1} | 1000 (>1000) {>1} | - | 500 (>1000) {>2} | 1000 (>1000) {>1} | - | 0.48 ** (0.48) {1} | na | na | na | |
| Candida parapsilosis ATCC 22019 | 500 (>1000) {>2} | 500 (>1000) {>2} | 500 (>1000) {>2} | - | 1000 (>1000) {>1} | 1000 (>1000) {>1} | 1000 (>1000) {>1} | 0.24 ** (0.48) {2} | na | na | na | |
| Candida glabrata ATCC 90030 | 1000 (>1000) {>1} | - | - | - | 1000 (>1000) {>1} | - | - | 0.24 ** (0.48) {2} | na | na | na | |
| Candida krusei ATCC 14243 | - | - | - | - | - | - | - | 0.24 ** (0.24) {1} | na | na | na | |
| Species | MIC (MBC or MFC) [µg/mL] and {MBC/MIC or MFC/MIC} Values of the Studied Compounds and Positive Controls | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 11 | 12 | 13 | 14 | 15 | 16 | CIP/VA * /NY ** | NIT | CFX | APC | ||
| Gram-positive bacteria | Staphylococcus aureus ATCC 43300 | - | 15.62 (>1000) {>64} | 250 (>1000) {>4} | 15.62 (>1000) {>64} | - | - | 1.95 (1.95) {1} | 0.24 (0.24) {1} | 7.81 (15.62) | nd | nd |
| Staphylococcus aureus ATCC 6538 | - | 15.62 (>1000) {>64} | 250 (>1000) {>4} | 1.95 (>1000) {>512} | 500 (>1000) {>2} | 1000 (>1000) {>1} | 7.81 (31.25) {4} | 0.24 (0.24) {1} | 15.62 (15.62) | 0.98 | nd | |
| Staphylococcus aureus ATCC 29213 | - | 15.62 (>1000) {>64} | 250 (>1000) {>4} | 15.62 (>1000) {>64} | 125 (>1000) {>8} | - | 7.81 (7.81) {1} | 0.48 (0.48) {1} | nd | nd | nd | |
| Staphylococcus epidermidis ATCC 12228 | - | 62.5 (>1000) {>16} | 62.5 (>1000) {>16} | 7.81 (>1000) {>128} | 31.25 (>1000) {>32} | 500 (>1000) {>2} | 3.91 (3.91) {1} | 0.12 (0.12) {1} | 3.91 (7.81) | 0.24 | nd | |
| Enterococcus faecalis ATCC 29212 | - | 125 (>1000) {>8} | 500 (>1000) {>2} | 500 (>1000) {>2} | 1000 (>1000) {>1} | 125 (>1000) {>8} | 1000 (>1000) {>1} | 0.98 * (1.95) {2} | nd | nd | nd | |
| Micrococcus luteus ATCC 10240 | 500 (>1000) {>2} | 31.25 (>1000) {>32} | 125 (>1000) {>8} | 31.25 (>1000) {>32} | 500 (>1000) {>2} | 500 (>1000) {>2} | 125 (250) {2} | 0.98 (1.95) {2} | 62.5 (62.5) | 0.98 | nd | |
| Bacillus subtilis ATCC 6633 | - | 250 (>1000) {>4} | 125 (250) {2} | 250 (>1000) {>4} | 250 (>1000) {>4} | 1000 (>1000) {>1} | 500 (1000) {2} | 0.03 (0.03) {1} | 3.91 (3.91) | 15.62 | 62.5 | |
| Bacillus cereus ATCC 10876 | - | 62.5 (>1000) {>16} | 125 (>1000) {>8} | 250 (>1000) {>4} | 1000 (>1000) {>1} | - | 500 (500) {1} | 0.06 (0.12) {2} | 7.81 (15.62) | 31.25 | nd | |
| Gram-negative bacteria | Bordetella bronchiseptica ATCC 4617 | - | - | - | - | - | - | 1000 (>1000) {>1} | 0.98 (0.98) {1} | 125 (>1000) | nd | nd |
| Klebsiella pneumoniae ATCC 13883 | - | - | - | - | - | - | 31.25 (31.25) {1} | 0.12 (0.24) {2} | 15.62 (31.25) | nd | nd | |
| Proteus mirabilis ATCC 12453 | - | - | - | - | - | - | 500 (500) {1} | 0.03 (0.03) {1) | 62.5 (125) | nd | nd | |
| Salmonella typhimurium ATCC 14028 | - | - | - | - | - | - | 500 (500) {1} | 0.06 (0.06) {1} | 31.25 (62.5) | nd | nd | |
| Escherichia coli ATCC 25922 | - | - | - | - | - | - | 500 (1000) {2} | 0.004 (0.008) {2} | 7.81 (15.62) | nd | nd | |
| Pseudomonas aeruginosa ATCC 9027 | - | - | - | - | - | - | 500 (500) {1} | 0.48 (0.98) {2} | nd | nd | nd | |
| Fungi | Candida auris CDC 311903 | - | - | - | - | - | - | - | 0.48 ** (0.48) {1} | na | na | na |
| Candida albicans ATCC 2091 | - | - | - | - | - | - | 1000 (>1000) {>1} | 0.24 ** (0.24) {1} | na | na | na | |
| Candida albicans ATCC 10231 | - | - | - | - | - | - | 1000 (>1000) {>1} | 0.48 ** (0.48) {1} | na | na | na | |
| Candida parapsilosis ATCC 22019 | - | - | - | - | - | - | - | 0.24 ** (0.48) {2} | na | na | na | |
| Candida glabrata ATCC 90030 | - | - | - | - | - | - | - | 0.24 ** (0.48) {2} | na | na | na | |
| Candida krusei ATCC 14243 | - | - | - | - | - | - | - | 0.24 ** (0.24) {1} | na | na | na | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuć, M.; Berecka-Rycerz, A.; Biernasiuk, A.; Popiołek, Ł. Microwave-Assisted Synthesis, Lipophilicity and In Vitro Antimicrobial Activity of Hydrazide-Hydrazones of Phenylacetic Acid. Appl. Sci. 2025, 15, 3436. https://doi.org/10.3390/app15073436
Kuć M, Berecka-Rycerz A, Biernasiuk A, Popiołek Ł. Microwave-Assisted Synthesis, Lipophilicity and In Vitro Antimicrobial Activity of Hydrazide-Hydrazones of Phenylacetic Acid. Applied Sciences. 2025; 15(7):3436. https://doi.org/10.3390/app15073436
Chicago/Turabian StyleKuć, Magda, Anna Berecka-Rycerz, Anna Biernasiuk, and Łukasz Popiołek. 2025. "Microwave-Assisted Synthesis, Lipophilicity and In Vitro Antimicrobial Activity of Hydrazide-Hydrazones of Phenylacetic Acid" Applied Sciences 15, no. 7: 3436. https://doi.org/10.3390/app15073436
APA StyleKuć, M., Berecka-Rycerz, A., Biernasiuk, A., & Popiołek, Ł. (2025). Microwave-Assisted Synthesis, Lipophilicity and In Vitro Antimicrobial Activity of Hydrazide-Hydrazones of Phenylacetic Acid. Applied Sciences, 15(7), 3436. https://doi.org/10.3390/app15073436






