Abstract
Magnetic Resonance Angiography (MRA) is widely used for cerebrovascular assessment, with Time-of-Flight (TOF) MRA being a common non-contrast imaging technique. However, maximum intensity projection (MIP) images generated from TOF-MRA often include non-essential vascular structures such as external carotid branches, requiring manual editing for accurate visualization of intracranial arteries. This study proposes a deep learning-based semantic segmentation approach to automate the removal of these structures, enhancing MIP image clarity while reducing manual workload. Using DeepLab v3+, a convolutional neural network model optimized for segmentation accuracy, the method achieved an average Dice Similarity Coefficient (DSC) of 0.9615 and an Intersection over Union (IoU) of 0.9261 across five-fold cross-validation. The developed system processed MRA datasets at an average speed of 16.61 frames per second, demonstrating real-time feasibility. A dedicated software tool was implemented to apply the segmentation model directly to DICOM images, enabling fully automated MIP image generation. While the model effectively removed most external carotid structures, further refinement is needed to improve venous structure suppression. These results indicate that deep learning can provide an efficient and reliable approach for automated cerebrovascular image processing, with potential applications in clinical workflows and neurovascular disease diagnosis.
1. Introduction
1.1. Clinical Background and Challenges in Intracranial Aneurysm Detection
Intracranial aneurysms, commonly referred to as cerebral aneurysms, are abnormal dilations of the cerebral arteries often found at arterial bifurcations, including the anterior communicating artery and posterior communicating artery. Their formation can be saccular or fusiform in nature, and large aneurysms exceeding approximately 10–25 mm are at a notably higher risk of rupture leading to subarachnoid hemorrhage [1]. Subarachnoid hemorrhage is well known for its high mortality rate, and even in cases where patients survive the initial bleed, complications such as cerebral vasospasm, brain atrophy, and hydrocephalus can lead to significant morbidity. As a result, the likelihood of a favorable outcome following rupture remains only around 30–40%, posing a considerable threat to long-term quality of life.
Early detection [2] is therefore paramount for preventing rupture, but the challenge lies in the fact that most unruptured cerebral aneurysms remain asymptomatic, making them difficult to detect until they are incidentally found or until a catastrophic event occurs [3]. For many years, digital subtraction angiography (DSA) has served as a gold-standard imaging modality due to its high diagnostic accuracy and spatial resolution. However, it carries risks such as rebleeding during the procedure or the generation of micro-emboli, prompting a shift toward employing DSA primarily in therapeutic interventions and confirming final treatment decisions.
Meanwhile, less invasive techniques have been embraced for screening purposes, particularly CT angiography (CTA) and MR angiography (MRA). Of these, MRA is especially attractive in routine brain examinations because it does not require contrast agents, providing a noninvasive approach ideal for widespread screening. MRA is thus frequently used in brain check-up programs to identify potential intracranial aneurysms in asymptomatic individuals. Nevertheless, a known limitation is its lower spatial resolution relative to CTA, which complicates the detection of smaller aneurysms. Indeed, a past study has reported [4] that for aneurysms measuring 5 mm or less, the diagnostic yield is only about 80%, and for lesions of 2 mm or below, accurate identification becomes remarkably difficult. This underscores the critical need for improved detection and the reduction of missed diagnoses, particularly in smaller or more complex aneurysm morphologies.
1.2. Deep Learning for Medical Image Processing and the Need for Automated Preprocessing
In generating MRA images suitable for diagnosis, it is common to employ the maximum intensity projection (MIP) method, which reconstructs a three-dimensional view of the cerebrovascular structures from axial MRI scans by projecting the maximum signal values along various viewing angles. Although MRA-MIP images are valuable for clinical interpretation, they often include unwanted tissues or vascular structures such as external carotid arteries or surrounding fat tissue. As a result, manual editing is frequently performed to remove irrelevant anatomical features before the final images are ready for diagnostic review. This manual workflow can be time-consuming, leading to a delay in making images available for definitive interpretation.
Against this backdrop, recent advances in deep learning have opened up new possibilities for automated image analysis in various fields [5], including medical imaging [6,7,8,9,10,11,12,13], and have especially shown promise in the domain of neurosurgery [14]. Deep learning models, particularly convolutional neural networks (CNNs), have demonstrated remarkable performance in image classification tasks, object detection, and semantic segmentation [15], enabling highly accurate identification and localization of critical anatomical structures or pathological regions in brain scans.
Among these applications [16,17], segmentation techniques have been frequently employed in the detection and delineation of intracranial tumors, such as gliomas or meningiomas, because precise boundary estimation is crucial for treatment planning and surgical decision-making. Research in this area has leveraged large datasets and sophisticated network architectures to automatically distinguish tumorous tissue from healthy brain parenchyma, effectively reducing the workload of radiologists and improving the consistency of diagnostic outcomes.
Despite these successes, deep learning-based methodologies have not been broadly applied to what could be termed the “preprocessing stage” of medical image analysis. In typical clinical workflows, especially in MRI-based examinations, clinicians or radiographers often perform manual preprocessing tasks—such as excluding irrelevant tissues, removing background structures like the scalp or external carotid arteries, and adjusting contrast or brightness—to create images more suitable for further diagnostic interpretation. This manual approach is deeply ingrained in standard practice, largely because it has been the conventional method for ensuring that only pertinent anatomical regions are emphasized, and any extraneous data are eliminated. However, the very nature of such manual preprocessing implies variability in quality and outcome, as the level of precision may differ significantly among operators with different expertise and experience. Moreover, manual editing is time-consuming, adding to the overall waiting period before final diagnostic images can be reviewed by attending physicians or surgeons. As the demand for faster, more accurate imaging in neurosurgical procedures continues to rise, the potential benefits of automating these preliminary steps become increasingly evident. Yet, compared with tumor segmentation or lesion classification—fields where deep learning has made substantial inroads—the challenge of automating preprocessing has not garnered the same level of attention. Several factors might contribute to this gap. First, the datasets required to develop reliable preprocessing algorithms must include a variety of normal and pathological cases to ensure robust performance across different clinical scenarios, but such datasets are often harder to compile and annotate exhaustively for preprocessing purposes than for more targeted tasks like tumor detection. Second, the immediate clinical impact of image preprocessing may seem less apparent compared to tasks like diagnosing a malignant tumor or detecting a life-threatening intracranial hemorrhage, which can overshadow the perceived importance of refining workflows at earlier stages. Nonetheless, the automated removal of extraneous tissues and enhancement of regions of interest can play a critical role in improving the subsequent steps of image analysis, increasing both efficiency and diagnostic accuracy.
1.3. Research Objectives and Expected Contributions
In particular, for MRA images intended for the identification of intracranial aneurysms, automating preprocessing could reduce the time spent on laborious manual editing of MIP images. By training a suitable deep learning model—potentially one that combines multi-scale feature extraction with attention mechanisms to highlight vascular structures—critical vessels could be emphasized, while irrelevant tissues and overprojected structures are suppressed. Furthermore, the standardization that such an automated method would provide could mitigate inter-operator variability, leading to a more uniform level of image quality for diagnosis.
Given that cerebral vasculature is inherently complex, and that smaller aneurysms can be difficult to discern in lower-resolution or obscured images, an effective preprocessing step can have direct implications for the sensitivity and specificity of aneurysm detection algorithms downstream. By integrating these preprocessing capabilities with advanced detection or segmentation approaches specifically targeting aneurysms, there is an opportunity to create a more comprehensive pipeline that seamlessly delivers diagnostic-ready images.
This pipeline could then be integrated into existing picture archiving and communication systems (PACSs), reducing the turnaround time for radiologists and allowing for quicker therapeutic decision-making. Ultimately, automating image preprocessing for MRA, while less high-profile than tasks like tumor segmentation, has the potential to bridge a major gap in current deep learning applications within neurosurgery. If reliably implemented, such automation would not only expedite the creation of accurate and easily interpretable MIP images but also lessen the chance of operator-induced errors during the editing process. It could, in turn, amplify the benefits already seen in other deep learning applications, such as better detection of small aneurysms and improved quantification of vascular pathologies.
Previous studies have demonstrated the effectiveness of deep learning in cerebrovascular imaging, particularly in aneurysm detection [17,18,19,20], vessel segmentation [21,22,23,24], and MRA super-resolution [25]. However, most prior research has focused on downstream applications such as lesion detection rather than preprocessing tasks. By automating vascular preprocessing, our study aims to bridge this gap and improve the interpretability of MRA images.
Accordingly, the goal of this study is to develop a software application that integrates deep learning-based algorithms into the MRA processing pipeline to generate diagnostic-quality images of cerebral vessels, thereby mitigating the necessity for extensive manual preprocessing. By incorporating cutting-edge deep learning architectures with careful consideration of clinically relevant training data, we aim to produce a robust tool capable of removing superfluous structures, highlighting subtle vascular anomalies and ensuring that crucial diagnostic cues—like the presence of a small aneurysm—remain readily discernible. Such a tool could not only streamline the diagnostic workflow but also elevate the overall standard of cerebrovascular assessments, offering a new perspective on the role deep learning can play in neurosurgical imaging beyond the more common tasks of segmentation and lesion detection. Through the development and validation of this software, we hope to demonstrate that automation in the preprocessing stage has significant potential to enhance the quality, speed, and reliability of MRA-based diagnostic procedures, contributing to better patient outcomes and more efficient healthcare delivery.
In particular, while DeepLab v3+ is a proven semantic segmentation framework [26,27,28,29,30], our primary contribution lies in automating MRA preprocessing steps that have traditionally been carried out manually. Specifically, by establishing a dedicated ROI annotation approach for excluding external carotid artery branches and orbital tissue, we address challenges unique to TOF-MRA. Moreover, we have integrated these methods into a standalone software system that operates on DICOM data to generate segmentation masks and MIP images in an automated fashion. This system bridges the gap between deep learning-based segmentation and the clinical requirement of efficiently producing diagnostic-quality MIP images.
2. Materials and Methods
This section outlines the study design, data preparation procedures, and the core segmentation approach necessary to develop our automated MIP image generation system. Section 2.1 details the dataset composition and acquisition parameters that serve as the foundation for our research, essential for reproducibility and establishing the baseline imaging conditions. Section 2.2 then explains the image processing challenges specific to TOF-MRA that our approach aims to solve, particularly the need to remove external carotid artifacts that obscure clinical evaluation. Building on this problem statement, Section 2.3 describes our semantic segmentation framework and annotation methodology across different anatomical levels, which forms the technical core of our solution. Section 2.4 presents the DeepLab v3+ training protocols and evaluation metrics that quantify the effectiveness of our approach, while Section 2.5 details the assessment methodology for the resulting MIP images, which demonstrates the clinical value of our automated segmentation.
2.1. Subjects and Dataset Composition
This study utilized pre-acquired axial MRA images of the brain. The dataset consisted of 20 TOF-MRA cases, each containing approximately 160–172 axial slices, resulting in a total of approximately 3284 images. While the images were pre-acquired, we provide details on the acquisition parameters for reproducibility and to highlight the imaging conditions under which our segmentation model was trained. The MRI scanner’s acquisition parameters are summarized in Table 1.

Table 1.
MRI scanner’s acquisition parameters.
All TOF-MRA datasets were captured using standard clinical protocols that rely on the inflow effect of fresh spins rather than exogenous contrast agents. Following acquisition, the raw DICOM files were converted to 512 × 512 pixel JPEG format using “XTREK View” by J-MAC Corporation (J-Mac System, Inc., Sapporo, Japan). This conversion was performed to facilitate subsequent steps, including data annotation and network training. While JPEG compression can introduce minor artifacts, the goal of automating tissue removal in MIP images meant that slight losses in dynamic range were unlikely to significantly affect semantic segmentation performance. A workstation equipped with the Windows 11 operating system, an Intel Xeon Gold 5115 CPU (2.4 GHz, 10 cores, 20 threads) (Intel, Santa Clara, CA, USA), four NVIDIA Quadro P5000 GPUs (16 GB VRAM each) (NVIDIA, Santa Clara, CA, USA), and 128 GB of DDR4 2666 MHz RDIMM memory was employed for image processing and deep learning tasks.
These TOF-MRA datasets serve as the target for the image processing described in the next section, with the aim of clearly visualizing intracranial vessels crucial for evaluating the internal carotid system. The dataset characteristics directly influence the training and performance of the semantic segmentation model discussed later.
2.2. Image Processing
The primary focus of this study was to eliminate time-consuming manual steps involved in preparing MIP images for clinical evaluation. Using the dataset described in the previous section, MIP works by projecting the highest intensity signal along each line of sight across a stack of axial images, effectively highlighting high-intensity blood flow. However, TOF-MRA often includes bright signal contributions from branches of the external carotid artery, such as the superficial temporal artery, middle meningeal artery, and occipital artery, as shown in Figure 1, which can obscure or overlap intracranial arteries that are critical to the evaluation of the internal carotid system. Manual editing to remove these extraneous features is a standard practice but introduces operator-dependent variability and delays the final availability of diagnostic images. By employing deep learning-based semantic segmentation, this study aimed to automate and standardize this process, thereby providing clearer vascular images more quickly.
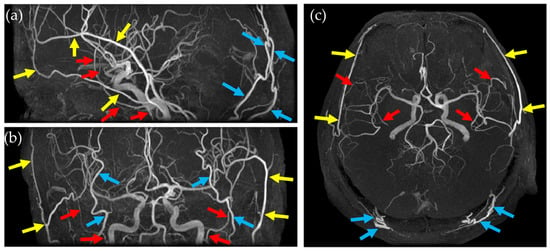
Figure 1.
MIP images from different views and arteries that hinder the evaluation of the internal carotid system. (a) Sagittal MIP image, (b) coronal MIP image, and (c) axial MIP image. Yellow arrow: superficial temporal artery, red arrow: middle meningeal artery, and light blue arrow: occipital artery.
2.3. Semantic Segmentation for Automatic MIP Image Generation
In this study, we employed DeepLab v3+ [31] as our semantic segmentation framework for automatic MIP image generation. DeepLab v3+ was selected based on its ability to capture multi-scale contextual information through atrous spatial pyramid pooling, as well as its strong performance in medical image segmentation tasks [32]. Compared with other architectures, such as U-Net [33] or a fully 3D convolutional network [34], DeepLab v3+ offers a favorable balance between segmentation accuracy and computational efficiency [35,36,37]. For instance, U-Net-based methods are widely used but can sometimes lose fine vessel boundaries, particularly in small datasets, while 3D CNNs can better capture volumetric features but often require significantly more computational resources and larger training sets. An in-house software was developed in MATLAB (v2024b; The MathWorks, Inc., Natick, MA, USA) to allow manual creation of Regions of Interest (ROIs) on the axial images. The manual segmentation process was performed by a radiological technologist (To.Y.). The annotation criteria were as follows: at the basal ganglia level, external carotid artery branches and non-intracranial structures were excluded; at the orbital level, fat tissue around the optic nerves was removed to reduce bright signal contamination; at the temporal–occipital junction level, only the internal carotid artery and vertebrobasilar system were retained. Each slice was independently reviewed by three experienced radiological technologists (Ta.Y., S.I. and H.S.) to ensure accuracy and consistency. Using this tool, the operator could visually identify and outline regions containing only the relevant intracranial vessels, excluding tissue and vascular structures deemed unnecessary for the final MIP representation. The 20 sets of axial images were thus annotated by drawing ROIs on each slice, guided by anatomical knowledge and specific exclusion criteria for unwanted structures.
To ensure clarity, ROI creation was tailored to different slice levels, which broadly corresponded to the cranial anatomy from the basal ganglia level of the head down to the skull base. These anatomical levels were selected to systematically exclude non-intracranial structures while preserving the integrity of the cerebrovascular network, ensuring that all critical intracranial vessels were retained while minimizing interference from external carotid branches. Representative examples (Figure 2, Figure 3, Figure 4 and Figure 5) illustrate how the ROIs were manually delineated.
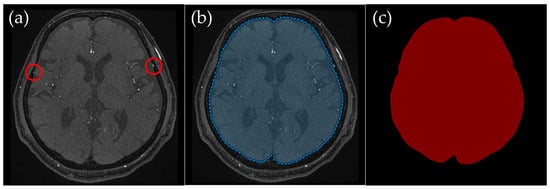
Figure 2.
Example of manually annotated training data at the basal ganglia level. (a) Original TOF-MRA axial image (red circles indicate the superficial temporal artery). (b) Manually defined region of interest (ROI) in blue, excluding the external carotid artery and non-intracranial structures. (c) Corresponding indexPNG mask for semantic segmentation training.
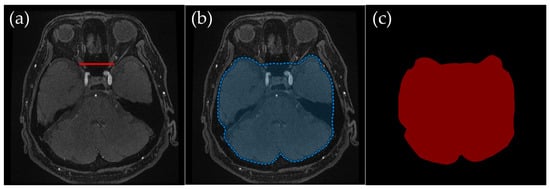
Figure 3.
Example of manually annotated training data at the orbital level. (a) Original TOF-MRA axial image (red line indicates the line connecting both optic nerves’ endpoints). (b) Manually defined region of interest (ROI) in blue, excluding the external carotid artery and non-intracranial structures. (c) Corresponding indexPNG mask for semantic segmentation training.
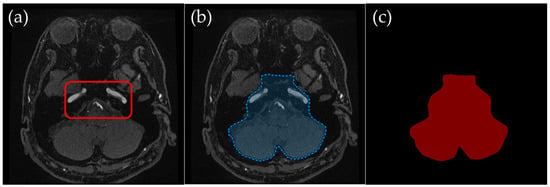
Figure 4.
Example of manually annotated training data at the temporal–occipital junction level. (a) Original TOF-MRA axial image (inside red line indicates internal carotid arteries and vertebrobasilar arteries). (b) Manually defined region of interest (ROI) in blue, excluding the external carotid artery and non-intracranial structures. (c) Corresponding indexPNG mask for semantic segmentation training.
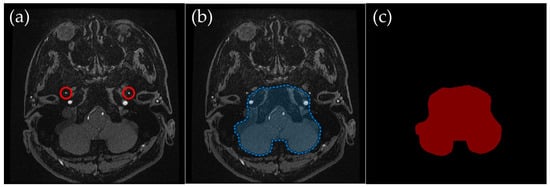
Figure 5.
Example of manually annotated training data at the skull base level. (a) Original TOF-MRA axial image (red circles indicate the middle meningeal artery). (b) Manually defined region of interest (ROI) in blue, excluding the external carotid artery and non-intracranial structures. (c) Corresponding indexPNG mask for semantic segmentation training.
2.3.1. Basal Ganglia Level: Preserving Intracranial Vessels and Removing External Carotid Arteries
Basal Ganglia Level (Figure 2): The external carotid artery branches and other extracranial tissues were excluded, leaving only the brain parenchyma and relevant intracranial vessels. The external carotid artery is particularly problematic in lateral projections, where it can overlap with middle cerebral artery branches, complicating MIP interpretation. This exclusion was crucial to ensuring clear visualization of the middle cerebral arteries in MIP images.
2.3.2. Orbital Level: Excluding Bright Fat Tissue near the Optic Nerves
Orbital Level (Figure 3): An ROI was defined dorsal to the line connecting both optic nerves’ endpoints, thus omitting the orbital region. Bright fat tissue in the orbit can interfere with segmentation due to its high signal intensity in TOF-MRA, leading to potential misclassification as vascular structures. This issue is especially pronounced in elderly patients or those with vascular flow impairments, as a lower flow signal in the arteries can become nearly indistinguishable from bright orbital fat. By removing fat signal contamination, the segmentation process ensures more accurate vessel delineation.
2.3.3. Temporal–Occipital Junction Level: Retaining Internal Carotid and Vertebrobasilar Arteries
Temporal–Occipital Junction Level (Figure 4): Only the internal carotid artery and the occipital lobe region were retained in the ROI. This slice range typically represents a section just inferior to the Circle of Willis, where the internal carotid arteries and the vertebrobasilar system begin to converge near the center of the brain. This level was particularly important for ensuring clear visualization of vertebrobasilar arteries, as overlapping signals from extracranial vessels can obscure diagnostically relevant structures in MIP images.
2.3.4. Skull Base Level: Excluding High-Intensity External Carotid Artery Branches
Skull Base Level (Figure 5): The ROI was drawn to retain the internal carotid artery and occipital regions, excluding the middle meningeal artery and other external carotid branches such as the superficial temporal artery. These external carotid branches exhibit high-intensity signals similar to intracranial arteries in TOF-MRA, making them a frequent source of misinterpretation. By carefully excluding these structures, the segmentation model reduces interference from non-diagnostic vascular signals in MIP images.
Figure 2 through Figure 4 illustrate how external carotid arteries and extracranial tissues were removed on each axial slice. These masks were then loaded into the in-house software, which combines all 2D masks into a 3D volume. In doing so, the software effectively eliminates unwanted structures and retains only the relevant intracranial vessels across all slices. The resulting ROIs were saved as indexed PNG files (indexPNG), each corresponding to a specific slice. The original JPEG images and their corresponding ROI masks thus formed paired data suitable for supervised segmentation tasks. These manually defined ROIs served as the ground truth for training the deep learning model, ensuring that segmentation decisions were anatomically accurate. These data pairs were then divided into training and testing sets at a ratio of 4:1. To address the limited dataset size (20 total cases) and improve generalizability, a 5-fold cross-validation strategy was adopted to make the best use of the limited dataset (20 total cases). Each fold consisted of a unique partition of the dataset, labeled fold 1 through fold 5. To enhance model generalization, we applied data augmentation techniques, including rotation (−25° to +25° in 5° increments), scaling (0.7× to 1.0× in 0.1 steps), and horizontal flipping. While these operations do not increase the absolute number of unique cases, they introduce variability that improves the model’s robustness across different imaging conditions. These augmentations helped the model learn a robust representation of intracranial structures under varying patient anatomies and imaging conditions.
2.4. DeepLab v3+ Training and Evaluation
The DeepLab v3+ network [31], which employs atrous convolution and pyramid pooling to capture features at multiple scales, was trained separately for each of the five folds. Training involved three epochs, a batch size of 64, and an initial learning rate that was reduced by a factor of 0.3 after each epoch. Although the number of epochs was small, data augmentation mitigated underfitting risks. Each model’s performance was then tested on the data held out for that particular fold. Segmentation accuracy was evaluated using the Dice Similarity Coefficient (DSC) and Intersection over Union (IoU). Given a predicted set of pixels S and a ground-truth set G, these metrics were computed as
Both DSC and IoU measure how closely the segmentation output aligns with the manually annotated ground truth. For each test slice, the metrics were calculated on a pixel-by-pixel basis, and mean values were reported. In addition, the inference speed, defined as the number of images processed per second (fps), was recorded to assess potential clinical utility. No further postprocessing was applied, allowing the results to reflect the raw predictive ability of the network.
2.5. MIP Image Assessment
Figure 6 and Figure 7 present the MIP images generated by applying the slice-wise masks shown in Figure 2, Figure 3 and Figure 4 to the 3D volume. After the masks are applied, a MIP is performed on the resulting 3D dataset, providing a final MIP that excludes non-intracranial arteries. When the masks (Figure 2, Figure 3 and Figure 4) accurately preserve only the intracranial arteries, the MIP images (Figure 6 and Figure 7) clearly show reduced interference from external carotid branches and highlight the internal carotid and vertebrobasilar systems. In order to evaluate images produced by the segmentation step, a dedicated software application was developed (Figure 6). This software can load a folder containing DICOM files for axial images acquired by MRA, and upon pressing the “start processing” button, it automatically converts each DICOM file into a JPEG image, applies the segmentation model, and incorporates the resulting mask back into the DICOM data. It then saves a three-dimensional matrix reflecting the applied segmentation, effectively providing an automated workflow from raw images to a fully segmented 3D volume. As a preview, the software displays MIP images viewed in axial, sagittal, and coronal orientations side by side, enabling a direct visual comparison of MIP images without segmentation versus those with segmentation applied. This interface helps verify how effectively undesired regions have been removed.
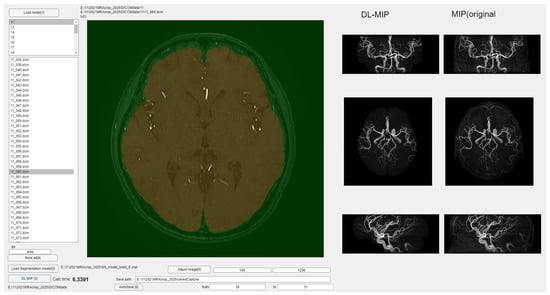
Figure 6.
Overview of the developed software for automated segmentation and MIP image generation.
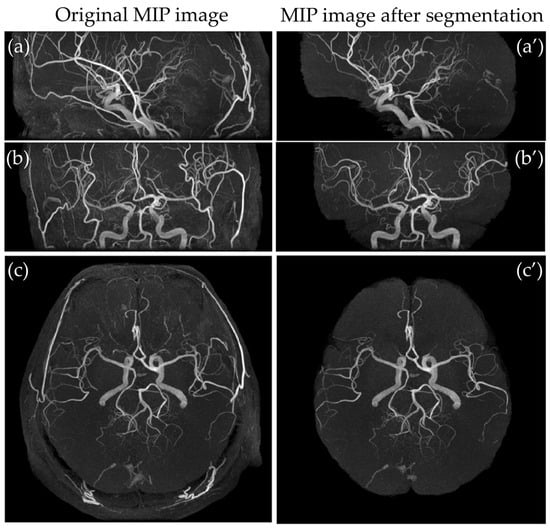
Figure 7.
Comparison of MIP images from different views. (a) Sagittal MIP image, (b) coronal MIP image, and (c) axial MIP image, and corresponding segmented MIP images (a’–c’).
Using this software environment, MIP images were generated for all 20 subjects under two conditions: one without segmentation and one with the segmentation masks. The degree of removal of key external carotid branches, including the superficial temporal, middle meningeal, and occipital arteries, was assessed qualitatively. Specifically, complete absence of removal was assigned a score of 0, removal of approximately 50% of the original depiction was scored as 1, removal of about 75% was scored as 2, removal of about 90% was scored as 3, and complete removal was scored as 4. The scores were determined by consensus among four experienced radiological technologists. Each test case was evaluated according to this scoring system, and for each fold in the five-fold cross-validation, the mean removal score and its 95% confidence interval were calculated.
3. Results
In this section, we present both quantitative and qualitative findings from our studies to demonstrate the effectiveness of our automated MIP image generation approach. Section 3.1 evaluates the semantic segmentation performance through objective metrics (DSC, IoU, and fps), establishing the technical accuracy and efficiency of our model across the five-fold cross-validation. These metrics directly address our objective of creating an accurate and computationally efficient segmentation solution. Section 3.2 then analyzes the impact of this segmentation on actual MIP image quality using a clinical scoring system for external carotid artery removal, which connects our technical achievements to the practical goal of improving diagnostic image clarity for clinical evaluation.
3.1. Evaluation of Semantic Segmentation
The performance of the semantic segmentation model was assessed based on DSC, IoU, and fps across five-fold cross-validation. The results for each fold, along with their mean and standard deviation (SD), are presented in Table 2. The model demonstrated a high segmentation accuracy, with an average DSC of 0.9615 ± 0.0112 and an IoU of 0.9261 ± 0.0206 across the five folds. These values indicate a strong agreement between the predicted segmentation masks and the ground-truth labels.

Table 2.
Segmentation evaluation metrics.
Additionally, the processing speed was evaluated by measuring fps during inference. The average fps across all folds was 16.61 ± 0.92, with individual fold values ranging from 15.59 to 17.60. This suggests that the model is capable of performing segmentation efficiently, maintaining a high processing speed while achieving high segmentation accuracy. Table 2 provides a detailed breakdown of the DSC, IoU, and fps values for each fold.
3.2. Evaluation of MIP Images After Segmentation
To evaluate the impact of the segmentation model on MIP image quality, the degree of removal of non-intracranial arteries was assessed using a scoring system. The evaluation focused on three external carotid artery branches: the superficial temporal artery, middle meningeal artery, and occipital artery. The mean scores and their respective 95% confidence intervals (95% CIs) were calculated for each artery. The superficial temporal artery had a mean removal score of 3.75, with a 95% CI of 3.54 to 3.96. The middle meningeal artery had a mean removal score of 3.80, with a 95% CI of 3.61 to 3.99. The occipital artery had a mean removal score of 3.60, with a 95% CI of 3.36 to 3.84. These scores suggest that the segmentation model was highly effective in suppressing undesired vascular structures, minimizing interference from non-intracranial arteries in MIP images.
Figure 7 illustrates a qualitative comparison between the MIP images generated from the original TOF-MRA dataset and those produced after applying the segmentation model. The images processed through the developed software show a significant reduction in non-essential vascular structures, particularly in the external carotid branches. The results indicate that the segmentation model successfully isolates the intracranial arteries while preserving their anatomical integrity, thereby enhancing the clarity of MIP images for diagnostic purposes.
4. Discussion
The proposed method for automatic MIP image generation using semantic segmentation demonstrated high segmentation accuracy, with a mean DSC exceeding 0.95 and a mean IoU surpassing 0.90. Compared to other studies [38,39,40] that employed semantic segmentation for brain images, these results indicate superior performance in delineating relevant anatomical structures while effectively removing unnecessary regions. When specifically comparing our quantitative metrics with recent state-of-the-art models, our approach shows competitive performance. For instance, Qu et al. [38] reported a median DSC of 0.87 for intracranial aneurysms using a UNet architecture, and Shahzad et al. [39] achieved a median DSC of 0.87 for intracranial aneurysms segmentation. Isensee et al. [40], while focused on skull removal rather than vessel segmentation, reported a DSC of 96.1–97.6 for brain extraction tasks. Although our method and these approaches target different structures, our performance (DSC > 0.95, IoU > 0.90) is comparable to the latest reports. This performance advantage is particularly significant considering the complexity of distinguishing intracranial from extracranial vessels, which have similar intensity profiles, unlike the more distinct intensity differences utilized in traditional skull stripping tasks. The high accuracy achieved in this study can be attributed to the choice of DeepLab v3+ [31] as the network model. DeepLab v3+ incorporates atrous convolution and an improved spatial pyramid pooling approach, enhancing its ability to capture fine details at multiple scales. This characteristic is particularly advantageous for segmenting complex anatomical structures, such as intracranial arteries and surrounding tissues, where precise boundary delineation is crucial. The model’s ability to distinguish brain parenchyma from high-intensity regions such as fat tissues contributed significantly to the overall segmentation accuracy. The DeepLab v3+ network, a 2D CNN-based model, was employed for semantic segmentation. A 2D approach was chosen to optimize computational efficiency and facilitate data augmentation, including rotation and scaling, which would be more complex in a 3D CNN framework. While 3D CNNs can capture spatial dependencies more effectively, the increased computational cost and the relatively small dataset size made a 2D approach more practical for this study. Unlike studies that concentrate solely on refining the architecture of deep segmentation networks, our work focuses on the clinical workflow surrounding MRA scans. By using DeepLab v3+ to remove non-intracranial arteries, we highlight how an established segmentation method can be leveraged in a software pipeline specifically designed for MRA preprocessing. This includes the DICOM-based import and export of data, as well as precise ROI annotation guidelines for consistent operator-independent tissue removal. Through these contributions, we aim to demonstrate a practical solution that integrates seamlessly into standard neurovascular imaging protocols.
To further evaluate the effectiveness of the trained segmentation model, MIP images were generated based on the segmented outputs. As shown in Figure 6, the automatically processed MIP images exhibited clear suppression of external carotid branches, including the superficial temporal artery and middle meningeal artery, which are commonly misrepresented in conventional TOF-MRA MIP images. However, despite the overall success in removing non-intracranial vessels, certain venous structures, such as the occipital venous sinus, were not entirely excluded in some cases. One potential reason for this incomplete removal is that these veins travel within the brain parenchyma, making their signal intensity distribution more variable and less distinguishable from arteries based on conventional segmentation features. Additionally, the anatomical variability in venous structures across individuals may have introduced inconsistencies in the training dataset, thereby reducing the model’s ability to generalize effectively to all cases.
Currently, the developed tool allows for a single ROI per axial slice, meaning that entire unwanted structures must be included or excluded as a whole. Future improvements could involve allowing multiple ROIs per slice, enabling finer control over segmentation and potentially improving the accuracy of venous structure exclusion. By refining the model to handle such detailed segmentations, it may be possible to enhance its ability to discriminate between veins and arteries based on spatial consistency and flow characteristics.
In addition to improvements in segmentation precision, the integration of DICOM-based processing represents a significant advancement. While this study primarily relied on JPEG images for training, a dedicated software tool was developed to directly process DICOM images, apply the segmentation model, and generate MIP images in an automated fashion. The software can take a folder containing DICOM files, convert them into a format suitable for segmentation, apply the trained model, and then integrate the segmentation results back into the DICOM format to produce MIP images. The comparison between MIP images generated from JPEG-based segmentation and those obtained directly from DICOM processing showed strong similarity, indicating that the approach can be feasibly implemented in real-world clinical workflows. Since medical imaging data are natively stored in DICOM format, having the capability to process images without requiring conversion to alternative formats is a crucial advantage for seamless integration into clinical environments.
The efficiency of the developed segmentation pipeline was also evaluated in terms of processing speed. The model achieved an average processing speed of 16.61 fps, meaning that approximately 16 axial slices could be segmented per second. Given that a typical brain TOF-MRA scan consists of around 160 slices, it is estimated that the entire set of axial images can be processed and a segmentation-enhanced MIP image can be displayed within approximately 10 s. This significantly reduces the time required compared to manual editing, which typically takes several minutes per case. This reduction in processing time is particularly beneficial in clinical settings where rapid assessment of cerebrovascular structures is essential, such as in emergency cases involving suspected aneurysms or vascular malformations. The ability to generate MIP images with automatically refined segmentation in near real time presents a potential improvement over conventional MIP image review, where non-intracranial structures can obscure diagnostically relevant features. The proposed method enables rapid MIP image generation, significantly reducing the time required for manual editing. While speed improvement is inherently expected with deep learning-based segmentation, our study emphasizes the clinical advantage of automating a traditionally manual preprocessing step.
Our segmentation task, while seemingly similar to skull removal, differs in several key aspects. Skull removal typically involves extracting the brain by removing the surrounding skull and scalp, which are structurally distinct from the brain and exhibit different intensity profiles in T1-weighted and T2-weighted MRI. In contrast, our approach focuses on selectively removing external carotid arteries, which have high signal intensities similar to intracranial arteries in TOF-MRA images. This makes segmentation more complex, as conventional thresholding-based methods used in skull stripping are not directly applicable. Instead, our model must differentiate between relevant and irrelevant vascular structures based on anatomical context rather than simple intensity differences. Prior research in skull removal, such as the work by Isensee et al. [40], demonstrates the effectiveness of deep learning-based approaches for brain extraction, and similar methodologies can be adapted for vascular segmentation. However, due to the fundamental differences in anatomical structures and segmentation challenges, our task requires a different approach, emphasizing precise vessel boundary delineation rather than complete organ extraction. Future work may explore integrating vessel topology information to further refine segmentation performance.
Despite these promising results, several limitations must be acknowledged. One notable limitation is related to the consistency of intensity variations in the training dataset. During the acquisition of TOF-MRA images, variations in signal intensity across slices were observed, likely due to differences in scanner parameters, patient movement, and inherent variations in flow dynamics. These inconsistencies may have affected segmentation performance, particularly in cases where signal intensity discontinuities led to misclassification of vessel structures. One potential approach to mitigating this issue is to incorporate data augmentation strategies that introduce controlled intensity variations during training. By exposing the model to a wider range of intensity profiles, it may become more robust to natural variations present in clinical datasets.
Another limitation observed in the evaluation was the variance in segmentation accuracy across different anatomical levels. When observing DSC and IoU scores on a per-slice basis, segmentation accuracy was found to be consistently higher at the basal ganglia and orbital levels but lower at the skull base level. This discrepancy can be explained by differences in anatomical complexity between these regions. The basal ganglia level primarily consists of relatively large intracranial arteries with clear boundaries, whereas the skull base region features more intricate vessel networks, including smaller arterial branches and venous structures that may have been misclassified. These findings suggest that a single segmentation model may not be optimal for all anatomical regions. Future work could involve training multiple models specialized for different anatomical levels, with tailored preprocessing and feature extraction techniques for each region. By employing region-specific segmentation models, it may be possible to further enhance segmentation accuracy, particularly in challenging areas such as the skull base.
A basic limitation of this study is the relatively small dataset size (n = 20). To mitigate this issue, we employed extensive data augmentation strategies, including rotation (−25° to +25°) and scaling (0.7× to 1.0×), as well as a 5-fold cross-validation approach. While these measures enhance generalizability, future studies should incorporate larger and more diverse datasets to further validate the robustness of our model. Additionally, expanding the dataset to include images acquired with different scanners and imaging protocols will be crucial to assessing the generalization ability of the proposed segmentation method.
Another aspect that warrants further investigation is the generalizability of the developed segmentation model to datasets acquired from different scanners and imaging protocols. Although the model performed well on the dataset used in this study, variations in MRI acquisition parameters, such as slice thickness, magnetic field strength, and sequence settings, could potentially affect segmentation performance. To address this limitation, additional datasets should be incorporated into the training process, covering a diverse range of imaging conditions. This would allow the model to learn more robust representations and improve its ability to generalize across different imaging environments.
Moreover, while the current study focused on vessel segmentation, the method could potentially be extended to other applications within neurovascular imaging. For example, deep learning-based segmentation could be applied to detect and classify vascular abnormalities such as aneurysms, stenoses, or arteriovenous malformations. By integrating automated segmentation with lesion detection models, a comprehensive AI-assisted workflow could be developed for cerebrovascular diagnosis. Such an approach could not only enhance diagnostic efficiency but also reduce inter-operator variability in image interpretation, leading to more standardized assessments across institutions.
5. Conclusions
In this study, we developed a deep learning-based semantic segmentation approach to automate the removal of non-essential vascular structures in MIP images, improving visualization while reducing manual workload. The proposed system demonstrated high segmentation accuracy, with an average DSC exceeding 0.95 and IoU surpassing 0.90, while maintaining computational efficiency, with a processing speed of approximately 16.61 fps. These results suggest the feasibility of real-time application in clinical settings.
By leveraging DeepLab v3+, the model successfully eliminated unwanted external carotid branches while preserving intracranial arteries crucial for cerebrovascular assessment. The developed software provides an automated pipeline that processes DICOM images, applies segmentation, and generates refined MIP images, facilitating seamless integration into clinical workflows. While venous structure removal remains a challenge, future work should focus on improving segmentation robustness, optimizing region-specific performance, and validating the method across diverse imaging protocols. Deep learning-based MIP image processing has the potential to enhance cerebrovascular imaging workflows and improve diagnostic precision in neurovascular assessments.
Author Contributions
T.Y. (Tomonari Yamada) contributed to the data analysis, algorithm construction, and writing and editing of the manuscript. T.Y. (Takaaki Yoshimura) and S.I. reviewed and edited the manuscript. H.S. proposed the idea and contributed to the data acquisition, performed supervision and project administration, and reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was conducted according to the principles of the Declaration of Helsinki, and was approved by the Institutional Review Board of Hokkaido University Hospital (No. 016-0495: May 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The created models in this study are available on request from the corresponding author. The source code of this study is available at https://github.com/MIA-laboratory/brainMRA_segmentation/ (accessed on 31 January 2025).
Acknowledgments
The authors would like to thank the laboratory members of the Medical Image Analysis Laboratory for their help.
Conflicts of Interest
The authors declare that no conflicts of interest exist.
Abbreviations
The following abbreviations are used in this manuscript:
| MRA | Magnetic Resonance Angiography |
| TOF | Time of Flight |
| MIP | Maximum Intensity Projection |
| DSC | Dice Similarity Coefficient |
| IoU | Intersection over Union |
| DSA | Digital Subtraction Angiography |
| CTA | Computed Tomography Angiography |
| CNN | Convolutional Neural Network |
| ROI | Region of Interest |
| PACS | Picture Archiving and Communication System |
| TR | Time of Repetition |
| TE | Time of Echo |
| FA | Flip Angle |
| FOV | Field of View |
| fps | Frames Per Second |
| SD | Standard Deviation |
| DICOM | Digital Imaging and Communications in Medicine |
References
- Baharoglu, M.I.; Lauric, A.; Gao, B.L.; Malek, A.M. Identification of a Dichotomy in Morphological Predictors of Rupture Status between Sidewall- and Bifurcation-Type Intracranial Aneurysms: Clinical Article. J. Neurosurg. 2012, 116, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Takahashi, S.; Hayakawa, A.; Saito, K.; Takada, A.; Fukunaga, T. Fatal Intracranial Aneurysms and Dissections Causing Subarachnoid Hemorrhage: An Epidemiological and Pathological Analysis of 607 Legal Autopsy Cases. J. Stroke Cerebrovasc. Dis. 2018, 27, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Inagawa, T. Trends in Incidence and Case Fatality Rates of Aneurysmal Subarachnoid Hemorrhage in Izumo City, Japan, between 1980–1989 and 1990–1998. Stroke 2001, 32, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Nakano, M.; Sasanuma, J.; Asari, J.; Watanabe, K. Diagnosis and Treatment of Ruptured Cerebral Aneurysm by 3D-MRA. Surg. Cereb. Stroke 2004, 32, 42–48. [Google Scholar] [CrossRef]
- Matsui, T.; Sugimori, H.; Koseki, S.; Koyama, K. Postharvest Biology and Technology Automated Detection of Internal Fruit Rot in Hass Avocado via Deep Learning-Based Semantic Segmentation of X-Ray Images. Postharvest Biol. Technol. 2023, 203, 112390. [Google Scholar] [CrossRef]
- Yoshimura, T.; Hasegawa, A.; Kogame, S.; Magota, K.; Kimura, R.; Watanabe, S.; Hirata, K.; Sugimori, H. Medical Radiation Exposure Reduction in PET via Super-Resolution Deep Learning Model. Diagnostics 2022, 12, 872. [Google Scholar] [CrossRef]
- Yaqub, M.; Jinchao, F.; Arshid, K.; Ahmed, S.; Zhang, W.; Nawaz, M.Z.; Mahmood, T. Deep Learning-Based Image Reconstruction for Different Medical Imaging Modalities. Comput. Math. Methods Med. 2022, 2022, 8750648. [Google Scholar] [CrossRef]
- Usui, K.; Yoshimura, T.; Tang, M.; Sugimori, H. Age Estimation from Brain Magnetic Resonance Images Using Deep Learning Techniques in Extensive Age Range. Appl. Sci. 2023, 13, 1753. [Google Scholar] [CrossRef]
- Li, M.; Jiang, Y.; Zhang, Y.; Zhu, H. Medical Image Analysis Using Deep Learning Algorithms. Front. Public Health 2023, 11, 1273253. [Google Scholar] [CrossRef]
- Inomata, S.; Yoshimura, T.; Tang, M.; Ichikawa, S.; Sugimori, H. Estimation of Left and Right Ventricular Ejection Fractions from Cine-MRI Using 3D-CNN. Sensors 2023, 23, 6580. [Google Scholar] [CrossRef]
- Ichikawa, S.; Itadani, H.; Sugimori, H. Prediction of Body Weight from Chest Radiographs Using Deep Learning with a Convolutional Neural Network. Radiol. Phys. Technol. 2023, 16, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Fourcade, A.; Khonsari, R.H. Deep Learning in Medical Image Analysis: A Third Eye for Doctors. J. Stomatol. Oral Maxillofac. Surg. 2019, 120, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent Advances and Clinical Applications of Deep Learning in Medical Image Analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Ito, M.; Sugimori, H.; Tang, M.; Nakamura, T.; Ogasawara, K.; Matsuzawa, H.; Nakayama, N.; Lama, S.; Sutherland, G.R.; et al. Tissue Acceleration as a Novel Metric for Surgical Performance During Carotid Endarterectomy. Oper. Neurosurg. 2023, 25, 343–352. [Google Scholar] [CrossRef]
- Asami, Y.; Yoshimura, T.; Manabe, K.; Yamada, T.; Sugimori, H. Development of Detection and Volumetric Methods for the Triceps of the Lower Leg Using Magnetic Resonance Images with Deep Learning. Appl. Sci. 2021, 11, 12006. [Google Scholar] [CrossRef]
- Faron, A.; Sichtermann, T.; Teichert, N.; Luetkens, J.A.; Keulers, A.; Nikoubashman, O.; Freiherr, J.; Mpotsaris, A.; Wiesmann, M. Performance of a Deep-Learning Neural Network to Detect Intracranial Aneurysms from 3D TOF-MRA Compared to Human Readers. Clin. Neuroradiol. 2020, 30, 591–598. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Sun, Y.; Fan, Q.; Wang, L.; Ji, C.; Gu, H.; Chen, B.; Zhao, S.; Wang, D.; et al. Deep Learning-Based Platform Performs High Detection Sensitivity of Intracranial Aneurysms in 3D Brain TOF-MRA: An External Clinical Validation Study. Int. J. Med. Inform. 2024, 188, 105487. [Google Scholar] [CrossRef]
- Claux, F.; Baudouin, M.; Bogey, C.; Rouchaud, A. Dense, Deep Learning-Based Intracranial Aneurysm Detection on TOF MRI Using Two-Stage Regularized U-Net. J. Neuroradiol. 2023, 50, 9–15. [Google Scholar] [CrossRef]
- Sohn, B.; Park, K.Y.; Choi, J.; Koo, J.H.; Han, K.; Joo, B.; Won, S.Y.; Cha, J.; Choi, H.S.; Lee, S.K. Deep Learning-Based Software Improves Clinicians’ Detection Sensitivity of Aneurysms on Brain TOF-MRA. Am. J. Neuroradiol. 2021, 42, 1769–1775. [Google Scholar] [CrossRef]
- Faron, A.; Sijben, R.; Teichert, N.; Freiherr, J.; Wiesmann, M.; Sichtermann, T. Deep Learning-Based Detection of Intracranial Aneurysms in 3D TOF-MRA. Am. J. Neuroradiol. 2019, 40, 25–32. [Google Scholar] [CrossRef]
- Chatterjee, S.; Prabhu, K.; Pattadkal, M.; Bortsova, G.; Sarasaen, C.; Dubost, F.; Mattern, H.; de Bruijne, M.; Speck, O.; Nürnberger, A. DS6, Deformation-Aware Semi-Supervised Learning: Application to Small Vessel Segmentation with Noisy Training Data. J. Imaging 2022, 8, 259. [Google Scholar] [CrossRef] [PubMed]
- Quon, J.L.; Chen, L.C.; Kim, L.; Grant, G.A.; Edwards, M.S.B.; Cheshier, S.H.; Yeom, K.W. Deep Learning for Automated Delineation of Pediatric Cerebral Arteries on Pre-Operative Brain Magnetic Resonance Imaging. Front. Surg. 2020, 7, 517375. [Google Scholar] [CrossRef]
- Hilbert, A.; Madai, V.I.; Akay, E.M.; Aydin, O.U.; Behland, J.; Sobesky, J.; Galinovic, I.; Khalil, A.A.; Taha, A.A.; Wuerfel, J.; et al. BRAVE-NET: Fully Automated Arterial Brain Vessel Segmentation in Patients With Cerebrovascular Disease. Front. Artif. Intell. 2020, 3, 552258. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wu, H.; Luo, G.; Zhou, H. Deep Learning-Based 3D Cerebrovascular Segmentation Workflow on Bright and Black Blood Sequences Magnetic Resonance Angiography. Insights Imaging 2024, 15, 81. [Google Scholar] [CrossRef] [PubMed]
- Hokamura, M.; Uetani, H.; Nakaura, T.; Matsuo, K.; Morita, K.; Nagayama, Y.; Kidoh, M.; Yamashita, Y.; Ueda, M.; Mukasa, A.; et al. Exploring the Impact of Super-Resolution Deep Learning on MR Angiography Image Quality. Neuroradiology 2024, 66, 217–226. [Google Scholar] [CrossRef]
- Huang, K.; Das, P.; Olanrewaju, A.M.; Cardenas, C.; Fuentes, D.; Zhang, L.; Hancock, D.; Simonds, H.; Rhee, D.J.; Beddar, S.; et al. Automation of Radiation Treatment Planning for Rectal Cancer. J. Appl. Clin. Med. Phys. 2022, 23, e13712. [Google Scholar] [CrossRef]
- Aslan, M.F. A Robust Semantic Lung Segmentation Study for CNN-Based COVID-19 Diagnosis. Chemom. Intell. Lab. Syst. 2022, 231, 104695. [Google Scholar] [CrossRef]
- Khodadadi Shoushtari, F.; Sina, S.; Dehkordi, A.N.V. Automatic Segmentation of Glioblastoma Multiform Brain Tumor in MRI Images: Using Deeplabv3+ with Pre-Trained Resnet18 Weights. Phys. Medica 2022, 100, 51–63. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, J.; Gong, Y.; Pu, Z.; Jiang, J.; Chu, Y.; Xia, L. Semantic Segmentation Method for Myocardial Contrast Echocardiogram Based on DeepLabV3+ Deep Learning Architecture. Math. Biosci. Eng. 2023, 20, 2081–2093. [Google Scholar] [CrossRef]
- Nguyen, T.T.U.; Nguyen, A.T.; Kim, H.; Jung, Y.J.; Park, W.; Kim, K.M.; Park, I.; Kim, W. Deep-Learning Model for Evaluating Histopathology of Acute Renal Tubular Injury. Sci. Rep. 2024, 14, 9010. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin, Germany, 2018; Volume 11211, pp. 833–851. [Google Scholar] [CrossRef]
- Li, Z.; Zeng, C.M.; Dong, Y.G.; Cao, Y.; Yu, L.Y.; Liu, H.Y.; Tian, X.; Tian, R.; Zhong, C.Y.; Zhao, T.T.; et al. A Segmentation Model to Detect Cevical Lesions Based on Machine Learning of Colposcopic Images. Heliyon 2023, 9, e21043. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin, Germany, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin, Germany, 2016; Volume 9901, pp. 424–432. [Google Scholar] [CrossRef]
- Zheng, B.; Shen, Y.; Luo, Y.; Fang, X.; Zhu, S.; Zhang, J.; Wu, M.; Jin, L.; Yang, W.; Wang, C. Automated Measurement of the Disc-Fovea Angle Based on DeepLabv3. Front. Neurol. 2022, 13, 949805. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Christian, M.; Chang, D.H.; Lai, F.; Liu, T.J.; Chen, Y.S.; Chen, W.J. Deep Learning Approach Based on Superpixel Segmentation Assisted Labeling for Automatic Pressure Ulcer Diagnosis. PLoS ONE 2022, 17, e0264139. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Li, J.; Shi, L.; Jiang, Q.; Yan, B.; Wang, Z. DME-DeepLabV3+: A Lightweight Model for Diabetic Macular Edema Extraction Based on DeepLabV3+ Architecture. Front. Med. 2023, 10, 1150295. [Google Scholar] [CrossRef]
- Qu, J.; Niu, H.; Li, Y.; Chen, T.; Peng, F.; Xia, J.; He, X.; Xu, B.; Chen, X.; Li, R.; et al. A Deep Learning Framework for Intracranial Aneurysms Automatic Segmentation and Detection on Magnetic Resonance T1 Images. Eur. Radiol. 2024, 34, 2838–2848. [Google Scholar] [CrossRef]
- Shahzad, R.; Pennig, L.; Goertz, L.; Thiele, F.; Kabbasch, C.; Schlamann, M.; Krischek, B.; Maintz, D.; Perkuhn, M.; Borggrefe, J. Fully Automated Detection and Segmentation of Intracranial Aneurysms in Subarachnoid Hemorrhage on CTA Using Deep Learning. Sci. Rep. 2020, 10, 21799. [Google Scholar] [CrossRef]
- Isensee, F.; Schell, M.; Pflueger, I.; Brugnara, G.; Bonekamp, D.; Neuberger, U.; Wick, A.; Schlemmer, H.P.; Heiland, S.; Wick, W.; et al. Automated Brain Extraction of Multisequence MRI Using Artificial Neural Networks. Hum. Brain Mapp. 2019, 40, 4952–4964. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).