Abstract
The search for new biologically active compounds with prospective pharmaceutical applications has motivated the investigation of alternative synthesis pathways. One such approach involves the development of compounds with established biological activity as lead compounds. The focus on compounds of natural origin is gaining prominence, with steroids and alkaloids representing notable examples. Our research aimed to synthesize novel steroid–alkaloid bioconjugates with potential biological activity. The structure of all new compounds was determined using spectroscopic methods. The final heats of formation (HOF) for all bioconjugates were also calculated. In silico methods demonstrated that most obtained compounds, especially caffeine derivatives, exhibited potential biological activity. These compounds act as cholesterol antagonists, analeptics, antihypercholesterolemic, and respiratory analeptic compounds. The molecular docking results for the 1HWK and 6RZ4 protein domains indicate that the selected bioconjugates exhibit affinities comparable to or lower than those of atorvastatin (−9.6 kcal/mol), the reference ligand in cholesterol-lowering. Conversely, the affinities of the selected bioconjugates are higher than those of caffeine (−6.2 kcal/mol), which is used as the reference ligand for analeptic drugs.
Keywords:
natural compounds; alkaloids; steroids; caffeine; gramine; bioconjugates; molecular docking 1. Introduction
One strategy for the synthesis of compounds with potential pharmacological applications is to use biologically active compounds as starting materials. Modifications of naturally occurring compounds with established biological activity, such as steroids and alkaloids, exemplified this approach.
Steroids are one of the most important classes of natural compounds. These include hormones, pheromones, sterols, and bile acids. Bile acids, e.g., lithocholic acid, deoxycholic acid as well as cholic acid, are the primary metabolites of cholesterol in the form of polyhydroxylated steroids. The specific skeleton geometry and hydroxyl groups of the bile acids (3α or 3α, 12α, and also 3α, 7α, 12α) play a significant role in their amphiphilic properties. These compounds and their derivatives have led to their application in many fields of chemistry, such as molecular recognition, biomimetic chemistry, host–guest chemistry, and pharmacology [1,2,3]. Moreover, bile acids have found applications as building blocks in the synthesis of molecular receptors; their dimers are infused in synthesizing macrocyclic artificial receptors with good organogelling properties [4,5,6,7,8,9,10]. At the same time, cholesterol and cholestanol as sterols have a secondary hydroxyl group in the steroid skeletons at the C-3 position. Within this class of compounds, they also differ in modifying the side chains or the presence of double bonds [11,12,13].
Among the alkaloids, indole and purine compounds are particularly noteworthy. Indole compounds display a range of biological activities. They have been shown to have anti-inflammatory [14], antioxidant [15] and anticancer [15,16] effects. Of particular interest is the naturally occurring indole alkaloid gramine. Its broad spectrum of biological activity and ease of modification make it the starting point for many syntheses [17]. Gramine and its derivatives have various properties ranging from antiviral [18] and anticancer [19] to insecticidal [20] and anticorrosive [21]. They also reduce the toxicity of cholic acid to human erythrocytes [22,23].
Another alkaloid that has attracted the attention of scientists is caffeine. It has stimulating [24], antibacterial [25] and antifungal [26] effects. Caffeine derivatives in which the substituent is located at the C-8 position are particularly interesting for their antioxidant activity [27,28]. They are also monoamine oxidase (MAO) inhibitors, which significantly increase their potential in the fight against neurodegenerative diseases such as Alzheimer’s and Parkinson’s [29,30]. Sulfur derivatives of caffeine are potential anticancer agents [31]. These compounds also exhibit significant antibacterial activity, making them promising candidates for the development of new antibiotics, especially in the face of increasing resistance to traditional antibiotics [32].
Both caffeine and gramine have been used to synthesize hybrid compounds. An interesting example of a caffeine hybrid is the combination of caffeine with Eddystone D. This unique combination results in a compound with a higher affinity for adenosine receptors than caffeine alone [33]. Gramine–uracil and gramine–imidazole hybrids show cytoprotective effects against AAPH-induced oxidative hemolysis [34,35]. Steroid–gramine bioconjugates with a triazole linker are examples of hybrids with potential antibacterial and antifungal activity [36]. In view of the above, we plan to synthesize the unique steroid–gramine bioconjugates and evaluate their pharmacological activities using the in silico program PASS (Pharma Expert Predictive Services ©2011–2013, Version 2.0.), a widely accepted tool for predicting biological activities. The lack of literature reports on the synthesis of caffeine–steroid hybrids prompted us to extend our studies to the caffeine molecule. Considering the promising potential of the new compounds as biologically active agents, their selected biological activity was evaluated using molecular docking methodology. Furthermore, to ensure the comprehensive nature of our research, PM5 calculations were performed for all compounds.
2. Materials and Methods
2.1. Chemicals and Reagents
Lithocholic acid, deoxycholic acid, cholic acid, acetic anhydride, pyridine, propiolic acid, sodium azide, sodium ascorbate, sodium hydrosulfate hydrate, 1,2-dihydro-2H-1,3-benzimidazole-2-thione, gramine, and caffeine were acquired from Sigma-Aldrich (Schnelldorf, Germany). DMF, chloroform, dichloromethane, t-butanol, and methanol solvents were obtained from standard commercial sources such as Merck (Darmstadt, Germany) and were used without purification.
The characterization methods used were as follows: IR Spectra: FT/IR Nicolet iS5 (Thermo Scientific, Waltham, MA, USA) (KBr pellet, oil, cm−1). 1H and 13C NMR spectra: Varian Mercury 300 MHz spectrometer (Oxford, UK) operating at 300.07 MHz and 75.4614 MHz for 1H and 13C, respectively. Chemical shifts are reported in ppm relative to Me4Si used as the internal standard and coupling constants (J values) in Hz. Typical conditions for 1H spectra include a pulse width of 32°, acquisition time of 5 s, FT size of 32 K, digital resolution of 0.3 Hz per point, and scans ranging from 1200 to 10,000 per spectrum. For 13C spectra, typical conditions include a pulse width of 60°, FT size of 60 K, digital resolution of 0.6 Hz per point, and the number of scans varied accordingly. ESI-MS was conducted with a Waters/Micromass (Manchester, UK) ZQ mass spectrometer equipped with a Harvard Apparatus (Saint Laurent, QC, Canada) syringe pump, with mass-to-charge ratio (m/z) reported. Sample solutions were prepared in MeOH at a concentration of approximately 10–5 M. Standard ESI-MS mass spectra were recorded at a cone voltage of 90 V. The melting points were measured using the Stuart Melting point apparatus, Model SMP30 (Bibby Scientific Ltd. Stone Staffordshire, UK). TLC analysis was conducted using silica gel 60 plates with a fluorescent indicator at 254 nm.
2.2. Synthesis
Thione derivatives of caffeine and gramine (1-((1H-indol-3-yl)methyl)-1,3-dihydro-2H-benzo[d]imidazole-2-thione and 1,3,7-trimethyl-8-sulfanylidene-9H-purine-2,6-dione) were synthesized according to the procedure of [37] and [38], respectively. The bromoacetate compounds were obtained according to [39].
A typical procedure for the synthesis of compounds 1–10 is reported to the following.
In a round-bottom flask, 0.15 mmol of bromoacetate (methyl lithocholate, methyl deoxycholate, methyl cholate, cholesteryl, dihydrocholesteryl) was dissolved in 2 mL of DMF (dimethylformamide). Two eq (0.30 mmol) of K2CO3 (44 mg) and 0.15 mmol of gramine or caffeine derivatives were added and stirred at room temperature for 2–3 h. Five mL of water was added to the reaction mixture, and the reaction mixture was transferred to a separatory funnel. It was extracted with ethyl acetate. It was washed with brine and water. The organic layer was dried over anhydrous Na2SO4. The resulting oil was purified on a chromatography column using chloroform as eluent.
3β-[2-thio-(1-((1H-indol-3-yl)methyl)-1,3-dihydro-2H-benzo[d]imidazole]acetate-cholest-5-ene (1)
Oil, 86%, 1H NMR (400 MHz, CDCl3) δ: 8.31 (s, 1H, NH), 7.65–7.56 (m, 2H, aromatic), 7.36–7.30 (m, 2H, aromatic), 7.20–7.07 (m, 4H, aromatic), 6.99 (d, J = 2.5 Hz, 1H, aromatic), 5.47 (s, 2H, CH2-10′), 5.33 (d, J = 5.0 Hz, 1H, 6-H), 4.69–4.61 (m 1H, 3α-H), 4.19 (s, 2H, CH2-29), 0.99 (s, 3H, CH3-19), 0.91 (d, J = 6.5 Hz, 3H, CH3-21), 0.86 (dd, J = 6.6, 1.8 Hz, 6H, CH3-26,27), 0.67 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ: 168.16 (C-28), 150.48 (C-2″), 143.44 (C-8″), 139.43 (C-5), 136.47 (C-7″), 136.34 (C-9′), 125.96 (C-8′), 123.37 (C-5″), 122.82 (C-6), 122.55 (C-4″), 122.02 (C-2′), 121.87 (C-6′), 120.10 (C-5′), 118.60 (C-3″), 118.39 (C-4′), 111.43 (C-3′), 110.76 (C-7′), 109.44 (C-6″), 75.77 (C-3), 56.68, 56.16, 50.00, 42.31, 40.32, 39.73, 39.52, 37.89, 36.91, 36.56, 36.19, 35.77, 35.24, 31.89, 31.85, 29.68, 28.20, 28.00, 27.58, 24.27, 23.83, 22.80, 22.55, 21.02, 19.28 (C-19), 18.72 (C-21), 11.85 (C-18). FT-IR (KBr) νmax (cm−1): 3410, 2935, 2867, 1732, 1447, 1379, 1349, 1319, 1296, 1241, 1168. ESIMS (MeOH): C45H59N3O2S m/z 707 [M+H]+, 729 [M+Na]+, 745 [M+K]+.
3β-[2-thio-(1-((1H-indol-3-yl)methyl)-1,3-dihydro-2H-benzo[d]imidazole]acetate-5β-cholestan (2)
Solid, m.p. 187.5-189.5 °C, 76%, 1H NMR (400 MHzCDCl3) δ: 8.20 (s, 1H, NH), 7.64–7.60 (m, 2H, aromatic), 7.40–7.29 (m, 2H, aromatic), 7.22–7.10 (m, 4H, aromatic), 7.03 (d, J = 2.5 Hz, 1H, aromatic), 5.50 (s, 2H, CH2-10′), 4.77–4.69 (m, 1H, 3α-H), 4.18 (s, 2H, CH2-29), 0.89 (d, J = 6.5 Hz, 3H, CH3-21), 0.86 (dd, J = 6.6, 1.9 Hz, 6H, CH3-26,27), 0.79 (s, 3H, CH3-19), 0.64 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ: 168.28 (C-28), 150.49 (C-2″), 143.43 (C-8″), 136.41 (C-7″), 136.27 (C-9′), 125.90 (C-8′), 123.30 (C-5″), 122.58 (C-4″), 121.98 (C-2′), 121.83 (C-6′), 120.11 (C-5′), 118.63 (C-3″), 118.39 (C-4′), 111.39 (C-3′), 110.83 (C-7′), 109.41 (C-6″), 75.59 (C-3), 56.37, 56.22, 54.13, 44.58, 42.55, 40.29, 39.93, 39.49, 36.65, 36.13, 35.77, 35.42, 35.39, 35.27, 33.76, 31.93, 29.69, 28.52, 28.22, 27.99, 27.26, 24.18, 23.81, 22.81, 22.55, 21.16 (C-21), 18.64 (C-19), 12.04 (C-18). FT-IR (KBr) νmax (cm−1): 3418, 2929, 2865, 1737, 1731, 1716, 1447, 1378, 1281, 1172. ESIMS (MeOH): C45H61N3O2S m/z 710 [M+H]+, 731 [M+Na]+, 747 [M+K]+.
3β-[8-thio-1,3,7-trimethyl-1H-purine-2,6(3H,7H)-dione]acetate-cholest-5-ene (3)
Solid, m.p. 181–182 °C, 82%, 1H NMR (401 MHz, CDCl3) δ: 5.37 (d, J = 5.1 Hz, 1H, 6-H), 4.71–4.62 (m, 1H, 3α-H), 4.02 (s, 2H, CH2-29), 3.90 (s, 3H, CH3-14′), 3.53 (s, 3H, CH3-12′), 3.39 (s, 3H, CH3-10′), 1.01 (s, 3H, CH3-19), 0.91 (d, J = 6.5 Hz, 3H, CH3-21), 0.86 (dd, J = 6.6, 1.9 Hz, 6H, CH3-26,27), 0.68 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ: 167.54 (C-28), 154.61 (C-6′), 151.46 (C-2′), 149.32 (C-4′), 148.28 (C-8′), 139.11 (C-5), 123.12 (C-6), 108.88 (C-5′), 76.00 (C-3), 56.63, 56.09, 49.94, 42.28, 39.66, 39.48, 37.94, 36.84, 36.53, 36.15, 35.76, 34.91, 32.35 (C-14′), 31.85, 31.79, 29.70 (C-12′), 28.19 (C-10′), 27.99, 27.86, 27.63, 24.25, 23.80, 22.80, 22.54, 20.99, 19.26 (C-19), 18.69 (C-21), 11.83 (C-18). FT-IR (KBr) νmax (cm−1): 2946, 2867, 1733, 1703, 1659, 1538, 1454, 1367, 1176. ESI-MS (MeOH): C37H56N4O4S m/z 675 [M+Na]+.
3β-[8-thio-1,3,7-trimethyl-1H-purine-2,6(3H,7H)-dione]acetate-5β-cholestan (4)
Oil, 91%,1H NMR (401 MHz, CDCl3) δ: 4.80–4.71 (m, 1H, 3α-H), 4.01 (s, 2H, CH2-29), 3.89 (s, 3H, CH3-14′), 3.52 (s, 3H, CH3-12′), 3.39 (s, 3H, CH3-10′), 0.89 (d, J = 6.5 Hz, 3H, 10′), 0.86 (dd, J = 6.6, 1.9 Hz, 6H, CH3-26,27), 0.81 (s, 3H, CH3-19), 0.64 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ: 167.6 (C-28), 154.61 (C-6′), 151.46 (C-2′), 149.31 (C-4′), 148.30 (C-8′), 108.88 (C-5′), 75.83 (C-3), 56.38, 56.26, 54.16, 44.59, 42.57, 39.94, 39.49, 36.64, 36.15, 35.76, 35.44, 35.42, 35.00, 33.88, 32.35 (C-14′), 31.91, 29.69 (C-12′), 28.57, 28.20 (C-10′), 27.98, 27.84, 27.34, 24.17, 23.81, 22.78, 22.53, 21.18 (C-19), 18.65 (C-21), 12.04 (C-18). FT-IR (KBr) νmax(cm−1): 2935, 2867, 1732, 1704, 1659, 1538, 1452, 1367, 1179. ESI-MS (MeOH): C37H58N4O4S m/z 678 [M+Na]+, 694 [M+K]+.
Methyl 3α-[2-thio-(1-((1H-indol-3-yl)methyl)-1,3-dihydro-2H-benzo[d]imidazole]-acetoxy-5β-cholan-24-oate (5)
Oil. 90%, 1H NMR (400 MHz, CDCl3) δ: 8.24 (s, 1H, NH), 7.65–7.60 (m, 2H, aromatic), 7.35–7.30 (m, 2H, aromatic), 7.22–7.10 (m, 4H, aromatic), 7.02 (d, J = 2.6 Hz, 1H, aromatic), 5.50 (s, 2H, CH2-10′), 4.80–4.72 (m, 1H, 3β-H), 4.18 (s, 2H, CH2-27), 3.67 (s, 3H, CH3-25), 0.92 (d, J = 6.4 Hz, 3H, CH3-21), 0.90 (s, 3H, CH3-19), 0.63 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ: 174.81 (C-24), 168.29 (C-26), 150.44 (C-2″), 143.44 (C-8″), 136.41 (C-7″), 136.28 (C-9′), 125.91 (C-8′), 123.32 (C-5″), 122.57 (C-4″), 122.01 (C-2′), 121.85 (C-6′), 120.10 (C-5′), 118.61 (C-3″), 118.39 (C-4′), 111.40 (C-3′), 110.79 (C-7′), 109.43 (C-6″), 76.22 (C-3), 56.38, 55.92, 51.49 (C-25), 42.69, 41.83, 40.35, 40.07, 35.72, 35.34, 35.24, 34.93, 34.51, 31.96, 31.05, 30.99, 28.17, 26.93, 26.40, 26.23, 24.15, 23.25, 20.79 (C-21), 18.26 (C-19), 12.00 (C-18). FT-IR (KBr) νmax (cm−1): 3378, 2932, 2865, 1731, 1448, 1379, 1349, 1294, 1281, 1171. ESI-MS (MeOH): C43H55N3O4S m/z 711 [M+H]+, 733 [M+Na]+, 749 [M+K]+.
Methyl 3α-[2-thio-(1-((1H-indol-3-yl)methyl)-1,3-dihydro-2H-benzo[d]imidazole]-acetoxy-12α-hydroxy-5β-cholan-24-oate (6)
Oil, 61%, 1H NMR (400 MHz, CDCl3) δ: 8.43 (s, 1H, NH), 7.63–7.58 (m, 2H, aromatic), 7.34–7.29 (m, 2H, aromatic), 7.20–7.08 (m, 4H, aromatic), 7.00 (d, J = 2.3 Hz, 1H, aromatic), 5.47 (s, 2H, CH2-10′), 5.08 (d, J = 2.9 Hz, 1H, 12β-H), 4.80–4.71 (m 1H, 3β-H), 4.20 (s, 2H, CH2-27), 3.66 (s, 3H, CH3-25), 2.06 (s, 3H, 12-OAc, CH3-29), 0.88 (s, 3H, CH3-19), 0.80 (d, J = 6.3 Hz, 3H, CH3-21), 0.72 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ:174.57 (C-24), 170.53 (C-28), 168.28 (C-26), 150.42 (C-2″), 143.40 (C-8″), 136.41 (7″), 136.31 (9′), 125.90 (C-8′), 123.36 (C-5″), 122.49 (C-4″), 121.99 (C-2′), 121.84 (C-6′), 120.03 (C-5′), 118.54 (C-3″), 118.29 (C-4′), 111.42 (C-3′), 110.62 (C-7′), 109.44 (C-6″), 75.96 (C-12), 75.86 (C-3), 51.46 (C-25), 49.37, 47.53, 44.98, 41.74, 40.29, 35.61, 35.18, 34.66, 34.34, 33.96, 31.97, 30.95, 30.80, 27.30, 26.77, 26.40, 25.79, 25.57, 23.39, 22.95 (C-19), 21.31, 17.47 (C-21), 12.36 (C-18). FT-IR (KBr) νmax (cm−1): 3379, 2948, 2867, 1735, 1448, 1376, 1247, 1192, 1169. ESI-MS (MeOH): C45H57N3O6S m/z 769 [M+H]+, 791 [M+Na]+, 807 [M+K]+.
Methyl 3α-[2-thio-(1-((1H-indol-3-yl)methyl)-1,3-dihydro-2H-benzo[d]imidazole]-acetoxy-7α,12α-dihydroxy-5β-cholan-24-oate (7)
Oil, 89%, 1H NMR (400 MHz, CDCl3) δ: 8.36 (s, 1H, NH), 7.62–7.59 (m, 2H, aromatic), 7.36–7.30 (m, 2H, aromatic), 7.24–7.09 (m, 4H, aromatic), 7.05 (d, J = 2.6 Hz, 1H, aromatic), 5.49 (d, J = 1.0 Hz, 2H, CH2-10′), 5.09 (s, 1H, 12β-H), 4.89 (s, 1H, 7β-H), 4.63–4.59 (m, 1H, 3β-H), 4.22 (s, 2H, CH2-27), 3.66 (s, 3H, CH3-25), 2.12 (s, 3H, s, 3H, 7-OAc), 1.98 (s, 2H, s, 3H, 12-OAc), 0.90 (s, 3H, CH3-19), 0.81 (d, J = 6.4 Hz, 3H, CH3-21), 0.72 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ: 174.50 (C-24), 170.57 (C-28), 170.43 (C-30), 168.28 (C-26), 150.41 (C-2″), 143.36 (C-8″), 136.42 (7″), 136.30 (C-9′), 125.92 (C-8′), 123.33 (C-5″), 122.59 (C-4″), 122.06 (C-2′), 121.93 (C-6′), 120.13 (C-5′), 118.57 (C-3″), 118.24 (C-4′), 111.41 (C-3′), 110.70 (C-7′) 109.46 (C-6″), 75.85 (C-12), 75.38 (C-3), 70.63 (C-7), 51.49 (C-25), 47.37, 45.07, 43.40, 40.88, 40.31, 37.73, 35.16, 34.59, 34.51, 34.43, 34.29, 31.18, 30.88, 30.76, 28.90, 27.15, 26.66, 25.54, 22.79 (C-19), 22.46, 21.48, 21.37, 17.49 (C-21), 12.19 (C-18). FT-IR (KBr) νmax (cm−1): 3380, 2948, 2870, 1732, 1437, 1377, 1249, 1169. ESIMS (MeOH): C47H59N3O8S m/z 828 [M+H]+, 849 [M+Na]+, 865 [M+K]+.
Methyl 3α-[8-thio-1,3,7-trimethyl-1H-purine-2,6(3H,7H)-dione]acetoxy-5β-cholan-24-oate (8)
Solid, m.p. 139–140 °C, 78%, 1H NMR (400 MHz, CDCl3) δ: 4.83–4.73 (m, 1H, 3β-H), 4.02 (s, 2H, CH2-27), 3.90 (s, 3H, CH3-14′), 3.67 (s, 3H, CH3-25), 3.53 (s, 3H, CH3-12′), 3.39 (s, 3H, CH3-10′), 0.93 (s, 3H, CH3-19), 0.92 (d, J = 6.52 Hz, 3H, CH3-21), 0.64 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ: 174.73 (C-24), 167.60 (C-26), 154.58 (C-6′), 151.44 (C-2′), 149.31 (C-4′), 148.27 (C-8′), 108.85 (C-5′), 76.46 (C-3), 56.46, 55.96, 51.46 (C-25), 42.69, 41.82, 40.44, 40.09, 35.72, 35.33, 34.96, 34.88, 34.54, 32.35 (C-14′), 32.11, 31.03, 30.96, 29.68 (C-12′), 28.14 (C-10′), 27.84, 26.96, 26.50, 26.28, 24.14, 23.25, 20.79 (C-19), 18.24 (C-21), 12.00 (C-18). FT-IR (KBr) νmax (cm−1): 2944, 2866, 1739, 1705, 1665, 1538, 1452, 1366, 1293, 1157. ESI-MS (MeOH): C35H52N4O6S m/z 680 [M+K]+, 696 [M+K]+,
Methyl 3α-[8-thio-1,3,7-trimethyl-1H-purine-2,6(3H,7H)-dione]acetoxy-12α-hydroxy-5β-cholan-24-oate (9)
Oil, 94%, 1H NMR (400 MHz, CDCl3) δ: 5.08 (t, J = 2.9 Hz, 1H, 12β-H), 4.82–4.74 (m, 1H, 3β-H), 4.03 (d, J = 3.9 Hz, 2H, CH2-27), 3.90 (s, 3H, CH3-14′), 3.67 (s, 3H, CH3-25), 3.53 (s, 3H, CH3-12′), 3.39 (s, 3H, CH3-10′), 2.10 (s, 3H, 12-OAc), 0.91 (s, 3H, CH3-19), 0.81 (d, J = 6.3 Hz, 3H, CH3-21), 0.73 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ: 174.49 (C-24), 170.28 (C-28), 167.52 (C-26), 154.57 (C-6′), 151.41 (C-2′), 149.20 (C-4′), 148.27 (C-8′), 108.87 (C-5′), 76.24 (C-3*), 75.88 (C-12), 51.44 (C-25), 49.42, 47.59, 45.00, 41.77, 35.63, 34.98, 34.67, 34.60, 34.43, 34.00, 32.32 (C-14′), 32.16, 30.96, 30.81, 29.64 (C-12′), 27.81 (C-10′), 27.29, 26.83, 26.51, 25.83, 25.59, 23.39, 22.98, 21.28 (C-19), 20.97, 17.48 (C-21), 12.37 (C-18). FT-IR (KBr) νmax (cm−1): 2949, 2869, 1737, 1704, 1665, 1538, 1454, 1367, 1246, 1168. ESI-MS (MeOH): C37H54N4O8S m/z 737 [M+Na]+.
Methyl 3α-[8-thio-1,3,7-trimethyl-1H-purine-2,6(3H,7H)-dione]acetoxy-7α,12α-dihydroxy-5β-cholan-24-oate (10)
Oil, 87%, 1H NMR (400 MHz, CDCl3) δ: 5.09 (t, J = 2.9 Hz, 1H,12β-H), 4.91 (d, J = 3.1 Hz, 1H, 7β-H), 4.69–4.61 (m, 1H, 3β-H), 4.07 (d, J = 5.0 Hz, 2H, CH2-27), 3.90 (s, 3H, CH3-14′), 3.66 (s, 3H, CH3-25), 3.52 (s, 3H, CH3-12′), 3.39 (s, 3H, CH3-10′), 2.14 (s, 3H, 7-OAc), 2.08 (s, 3H, 12-OAc), 0.92 (s, 3H, CH3-19), 0.82 (d, J = 6.3 Hz, 3H, CH3-21), 0.73 (s, 3H, CH3-18). 13C NMR (101 MHz, CDCl3) δ 174.49 (C-24), 170.40 (C-30), 170.17 (C-28), 167.59 (C-26), 154.57 (C-6′), 151.40 (C-2′), 149.05 (C-4′), 148.24 (C-8′), 108.89 (C-5′), 76.16 (C-3), 75.37 (C-12), 70.66 (C-7), 51.51 (C-25), 47.34, 45.02, 43.34, 40.90, 37.70, 35.09, 34.57, 34.54, 34.47, 34.30, 32.35 (C-14′), 31.19, 30.85, 30.72, 29.63 (C-12′), 28.85 (C-10′), 27.85, 27.14, 26.82, 25.51, 22.77, 22.49 (C-19), 21.55 (C-31), 21.37, 17.47 (C-21),12.18 (C-18). FT-IR (KBr) νmax(cm−1): 2950, 2871, 1735, 1704, 1663, 1538, 1453, 1368, 1248, 1166. ESI-MS (MeOH): C39H56N4O10S m/z 795 [M+Na]+, 812 [M+K]+.
2.3. PM5 Calculation
The PM5 semiempirical calculations were performed using CAChe Fujitsu (Chiba, Japan), the WinMopac 2003 program.
2.4. PASS Analysis
The potential pharmacological activities were determined using the PASS website: www.way2drug.com/passonline/ (accessed on 2 October 2024).
2.5. Docking Study
The molecular docking process, conducted on Windows 11 version 23H2 and Anaconda environment, commenced by converting the SMILES representation of given chemical structures into 3D structures, and this was accomplished through the application of OpenBabel pythonic version (pybel, version 3.1.0) [40,41] and biopandas [42]. Subsequently, the protein domains corresponding to PDB [43], IDs 1HWK [44,45], and 6RZ4 [46,47] were prepared following the standard AutoDock tool 1.5.7 scheme [48]. Molecular dockings were then carried out using AutoDock Vina v.1.1.2 [49], with the specific parameters outlined in Table 1 for each docking search, which were determined based on native ligand coordinates, namely atorvastatin (117 A 2) and pranlukast (KNT A 2001), respectively, for 1HWK and 6RZ4 protein domains.

Table 1.
The search spaces of the analyzed binding sites of the protein domains.
The depictions were performed using UCSF Chimera 1.16 software [50], and the 2D diagrams, maps of interactions, were prepared by the ProteinsPlus algorithms [51,52], namely PoseView [53] and PoseEdit [54]. In the case of an inability to obtain results with the ProteinsPlus algorithm, the contacts and clashes visualized via Chimera are given.
3. Results and Discussion
3.1. Synthesis and Spectroscopic Characterization
C3-substituted indole compounds have attracted significant attention due to their widespread pharmaceutical application [34,35,37]. Among the various C8 caffeine derivatives, 8-mercaptocaffeine derivatives are particularly noteworthy because of their considerable pharmaceutical properties [28,29]. Furthermore, bromosubstituted steroid derivatives serve as valuable substrates in the synthesis of new molecules [39].
In this study, we synthesized a series of bioconjugates derived from thione alkaloids and bromosubstituted steroid derivatives to develop novel bioactive compounds.
Since thione compounds can exist as tautomeric forms of thione-thiol, we carried out the reaction of gramine and caffeine thione derivatives with bromoacetate of methyl lithocholate, methyl deoxycholate, methyl cholate, cholesteryl, and dihydrocholesteryl (see Scheme 1). Yields ranged from 61% to 90% (for gramine derivatives) and 82% to 97% (for caffeine derivatives).
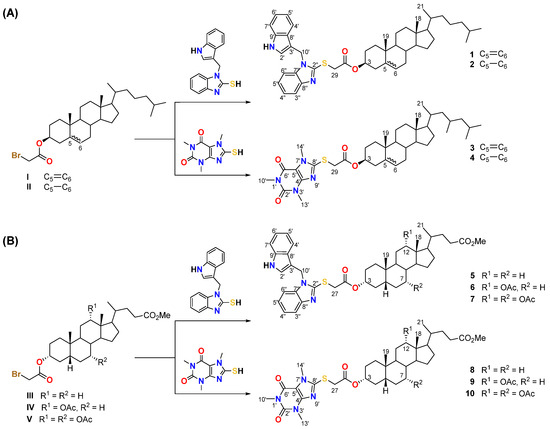
Scheme 1.
Synthesis of new sterol–alkaloid bioconjugates 1–4 (A) and bile acid–alkaloid bioconjugates 5–10 (B).
All new compounds were characterized by the spectroscopic method. The 1H NMR spectra of all new compounds showed signals characteristic of the cholic acid and alkaloid derivatives skeletons. The characteristic signals from axial C3β-H protons were observed for all derivatives at 4.59–4.83 ppm. The presence of signals from C7β-H protons at 4.89 ppm and 4.91 ppm for derivatives 7 and 10, and signals from C12β-H at 5.08 ppm for compounds 6 and 9 and at 5.09 ppm for derivatives 7 and 10, provided further confirmation of the structure of the newly synthesized bioconjugates. The signals assigned to CH3-18 were present in the range of 0.63–0.73 ppm, those assigned to CH3-19 in the range of 0.79–1.01 ppm, and those assigned to CH3-21 in the range of 0.80–0.92 ppm. These signals were observed in all compounds studied.
For compounds 1, 2, and 5–7 (gramine derivatives), aromatic ring signals were present in the range of 6.99–7.65 ppm, while signals from the protons of the C10′ -methylene group were observed at about 5.50 ppm. For the caffeine bioconjugates (compounds 3, 4, 8–10), the three N-methyl group diagnostic signals were observed at about 3.39, 3.53, and 3.90 ppm. The diagnostic signals for the CH2-27 protons in derivatives 5–7 and 8–10 and the CH2-29 protons in compounds 1, 2 and 3, 4 were observed in the range 4.01–4.22 ppm.
In the 13C NMR spectra, diagnostic signals from the C-18, C-19, and C-21 carbon atoms were present at 11.83–12.37 ppm, 18.26–22.95 ppm, and 17.47–21.16 ppm, respectively, for all derivatives. The signal from the C-3 carbon atom was present in the range of 75.38–76.46 ppm for all derivatives, while the signal from the C-5 carbon atom was observed at 139.43 ppm and 139.11 ppm, and C-6 carbon atom at 122.82 ppm and 123.12 ppm for compounds 1 and 3, respectively. The signal from the C-7 carbon atom was observed at 70.63 ppm and 70.66 ppm for compounds 7 and 10, respectively. The indole ring signals for the gramine derivatives (compounds 1, 2, 5–7) were observed in the 111-137 ppm range. The carbonyl carbon atoms of caffeine (derivatives 3, 4, 8–10) exhibited signals at 150 ppm, whereas the carbonyl group of steroid derivatives displayed signals near 170 ppm.
The FT-IR spectra of all new derivatives exhibited absorption bands between 2800 and 3000 cm−1, originating from the stretching vibration of the C-H bonds in aromatic rings. In compounds 1, 2, 5, and 7, characteristic wide bands at about 3400 cm−1 corresponding to the N-H bond in unsubstituted indole rings were found. The C=O bonds from the caffeine ring (3, 4, 8–10) were observed as two bands in the 1663–1706 cm−1 range. Bands originating from the C-N bonds in benzimidazole–indole and caffeine rings were found in the 1280–1380 cm−1 range. The most characteristic feature of the FT-IR spectra present in the structure of the steroid part is two strong representative bands at 17535–1704 cm−1 and 1297–1157 cm−1 region, appropriately assigned, respectively, to the symmetric groups ν(C=O) and ν(C–O). In addition, characteristic stretching vibrations of C–H bonds were present in the 2950–2865 cm−1.
In the ESI MS spectra of newly obtained bioconjugates, the positive ion mode showed signals from the m/z value corresponding to [M+H]+ (for 1), [M+Na]+ (for 2–4, 5–7, 10) and [M+K]+ (for 5, 6).
The NMR (1H and 13C), ESI-MS, and FT-IR spectra of the investigated compounds are provided in the Supplementary Materials (Figures S1–S30).
3.2. PM5 Calculation
PM5 semiempirical calculations were performed using the WinMopac 2003 program. The molecular models of representative conjugates are shown in Figure 1. The final heats of formation (HOF) for bioconjugates 1–10 are presented in Table 2.
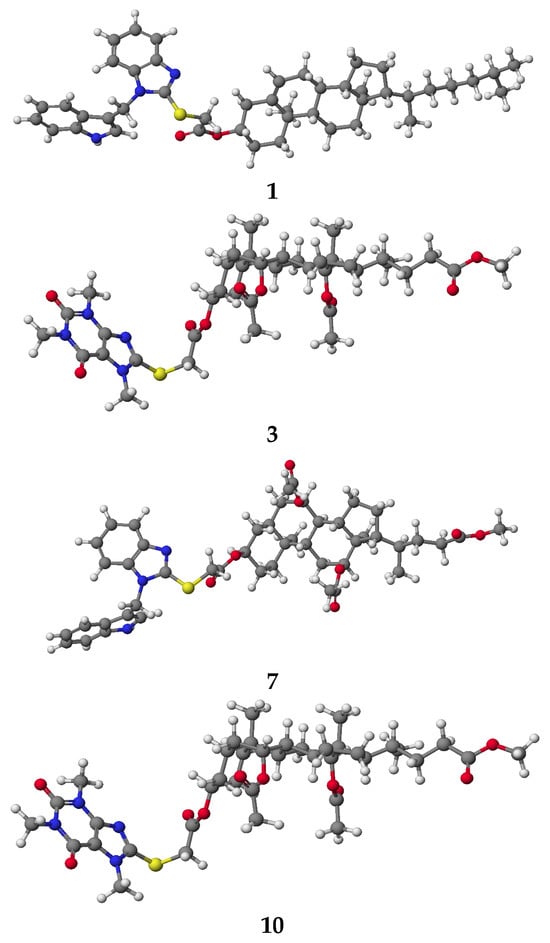
Figure 1.
Molecular models of selected bioconjugates 1, 3, 7 and 10 calculated by PM5 method.

Table 2.
Heat of formation (HOF) of bioconjugates 1–10 from steroids and alkaloids.
The lowest HOF values for conjugates 1–10, comprising bile acids and sterols, were observed for cholic acid bioconjugates with gramine and caffeine, compounds 7 and 10, respectively. A reduction in the number of hydroxyl groups in the bile acid skeleton resulted in a corresponding decrease in the HOF value. A comparable correlation was identified in the hydroxyl groups blocked by acetate groups.
The OAc groups facilitate the formation of intramolecular H–H bonds and stable host–guest complexes. These complexes may be stabilized by H–H bonding or electrostatic interactions resulting from the OAc groups in the bile acid molecule. In the case of sterol derivatives, the lowest HOF values appear for cholestanol derivatives, which is a consequence of greater stability resulting from the lack of multiple bonds in the B ring of the steroid skeleton.
The spatial arrangement and interaction of the bioconjugates 7 and 10 are shown in Figure 2. The final heat of formation is −1655.7660 kcal/mol for 7 and –2335.0571 kcal/mol for 10, respectively. The molecules of bioconjugates are arranged to form a specific kind of channel through which ions or neutral molecules can be transferred. Compensation charges occur only through intermolecular electrostatic interaction. This is a very good confirmation of the conclusion that interactions reduce HOF.
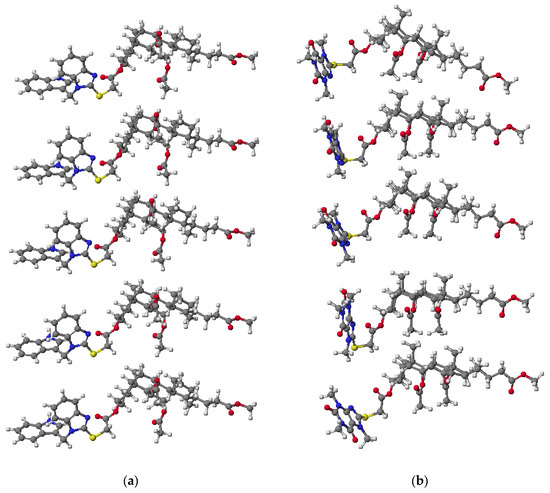
Figure 2.
Molecular models of derivatives 7 (a) and 10 (b) calculated by PM5 method for five molecules.
3.3. PASS Analysis
The potential pharmacological activities of new bioconjugates were determined using a computer-aided drug discovery approach, using the in silico prediction of activity spectra for substances (PASS) program [55,56,57]. The biological activity predicted for a potential compound with the highest probability (focal activities) has also been selected and is presented in Table 3.

Table 3.
Probability “to be active” (PA) values for the predicted biological activity of bioconjugates 1–10.
Table 3 demonstrates that the most active compounds belong to the caffeine derivatives (compounds 3, 4, 8–10). The most frequently predicted types of biological activity are cholesterol antagonist activity (with a probability of 90%), analeptic activity (with a probability of 73%), respiratory analeptic activity (with a probability of 70%), and antihypercholesterolemic activity. Furthermore, the caffeine bioconjugates demonstrated notably inhibitory activity against lipoprotein lipase (Pa > 75% for 4, 8, and 9) and acylcarnitine hydrolase (Pa > 71% for 4 and 8–10). Bioconjugate 6 did not display any discernible activity. Among the remaining gramine derivatives (1, 2, 5, and 7), compounds 1, 2, and 5 showed the most significant cholesterol antagonist activity. In particular, derivative 5 demonstrated cytoprotective and antieczema properties. It is estimated that over 80 percent of eczema cases are of the atopic type. Atopic eczema is a chronic inflammatory skin condition associated with sleep disorders and psychological issues. Treatment options for eczema typically include corticosteroids, antihistamines, and immunosuppressants. However, these medications can lead to numerous side effects. Natural products are currently being researched as potential alternatives for treating dermatitis, as they may offer effective and safer treatment options [58]. Therefore, new gramine bioconjugates could be a promising solution.
Glyceryl ether monooxygenase is an enzyme that facilitates the breakdown of lipid ethers into glycerol and their corresponding aldehydes. Several substances can inhibit this enzyme, including N5-methyl tetrahydrobiopterin and certain metal ions [59]. As indicated in Table 3, derivative 7 demonstrated cytoprotective activity and showed the ability to inhibit glyceryl ether monooxygenase.
Cholesterol is a substrate for the biosynthesis of vitamin D, bile acids, and steroid hormones and is an essential structural component of cell membranes [60]. However, its accumulation in the body is associated with the development of atherosclerosis, hypertension, and, ultimately, cardiovascular disease, leading to increased mortality and morbidity worldwide. Statins are widely recognized as effective cholesterol-lowering drugs. Unfortunately, these drugs have been associated with many adverse effects. However, the safety profile of natural compounds offers a promising alternative. These compounds have a safer use profile, fewer side effects, and potential health benefits, including cholesterol-lowering properties [61]. As shown in Table 3, an in silico analysis of compounds 1–10 revealed that all caffeine and steroid derivative bioconjugates demonstrated significant potential for cholesterol reduction.
Analeptics are stimulants that act on the central nervous system, particularly the respiratory and vasomotor centers. A notable example of a compound with analeptic activity is caffeine. Caffeine, a naturally occurring stimulant, has a long history of use in the treatment of barbiturate overdose [62]. This alkaloid acts on the cerebral cortex and has also been demonstrated to stimulate the respiratory, metabolic, and thermoregulatory centers. As a pharmaceutical agent, it is used to treat cardiovascular insufficiency, as a psychostimulant, and as a vasoconstrictor to relieve headaches.
3.4. Docking Study
3.4.1. Selected Bioconjugates and Protein Domains
As cholesterol antagonists, three caffeine derivatives (3, 4, and 8) were selected for molecular docking. Additionally, derivative 1, a gramine compound with a high probability of being an effective cholesterol antagonist (Pa = 82%) was also used for comparison purposes. As potential respiratory analeptic compounds, derivatives 4 and 8 were selected for molecular docking. In addition, caffeine was molecularly docked as a representative analeptic drug (CFN).
The selection of protein domains was guided by their specific biological functions within the physiological system:
- −
- HMG-CoA reductase activity (PDB ID: 1HWK): This enzyme plays a pivotal role in the mevalonate pathway, catalyzing HMG-CoA conversion to mevalonate, a key precursor in cholesterol biosynthesis. The structure of HMG-CoA reductase in complex with statins reveals their mechanism of inhibition and provides valuable insights into the design of drugs for treating hypercholesterolemia and cardiovascular disease. Understanding the binding interactions at the active site can lead to developing more effective and selective inhibitors that minimize the side effects associated with cholesterol-lowering therapies [44,45].
- −
- Cysteinyl leukotriene receptor 1 (CysLT1R) activity (PDB ID: 6RZ4): This G protein-coupled receptor is crucial for mediating inflammatory responses, particularly in asthma and allergic reactions. The crystal structure of CysLT1R complexed with the antagonists zafirlukast and pranlukast elucidates the binding mechanisms and conformational changes associated with receptor inhibition. These insights are essential for the design of new therapeutic agents targeting CysLT1R, potentially leading to more effective treatments for asthma and other inflammatory diseases [46,47].
These proteins are integral to various physiological processes, and their modulation can have significant implications for treating various diseases. Both HMG-CoA reductase and CysLT1R are critical targets in pharmacology, with their structural insights providing a foundation for designing specific inhibitors that could enhance therapeutic efficacy while reducing adverse effects associated with existing treatments.
3.4.2. Similarities and Differences Between Novel and Endogenous Ligands
To visualize the differences between native ligands and new ligands, some of the QED (Quantitative Estimate of Druglikeness) [63] molecular descriptors were calculated with the RDKit Python library [64]. The resulting data are shown in Table 4.

Table 4.
Druglikeness properties of selected ligands.
Both the native and new ligands exhibit a wide range of molecular weights. The new ligands have a higher molecular weight than the reference ligands. The number of hydrogen bond acceptors in new ligands is generally higher than the number of hydrogen bond acceptors in reference ligands. In the case of hydrogen bond donors, the new ligands have none except compound 1. The polar surface area is generally lower for the new ligands compared to the atorvastatin and higher when compared to the caffeine. The lipophilicity differs for the new ligands exposed by higher alogP values. This indicates that new ligands exhibit much higher lipophilicity than native ligands, which may lead to low solubility and poor absorption [65]. There is also variability in the number of rotatable bonds, meaning that the flexibility of the new ligands is higher than that of the caffeine, and it is similar to atorvastatin.
3.4.3. Docking Analysis
The RMSD (root mean square deviation) measures the average distance between the atoms of two superimposed molecules. It is commonly used to quantify the structural similarity between two molecules in space. In the context of molecular docking, RMSD describes how far the predicted pose of a ligand differs from the native pose. Lower RMSD values indicate higher accuracy in pose prediction. The RMSD, given in Å, was 0.67 and 0.83 for 1HWK and 6RZ4, respectively. This means that the redocking procedure was successful [66].
The molecular docking data revealed that the newly acquired derivatives exhibited affinity for the investigated protein domains. In Table 5, their affinity for the protein domains is comparable to the binding energies of the reference ligands.

Table 5.
The results of molecular docking to the 1HWK and 6RZ4 protein domains of all the compounds analyzed. Atorvastatin and caffeine were used as reference molecules. STD of binding energy was calculated based on the nine best poses.
The conducted studies indicate that the analyzed ligands exhibited affinity profiles comparable to or lower than the atorvastatin as a reference ligand and higher than the caffeine as a reference ligand, thus indicating higher strength of binding to the 6RZ4 protein domain and lower affinities to 1HWK. The best poses for the 1HWK protein domain were 6 and 3, respectively, for compounds 4 and 8. The poses with the lowest energy have binding energies of −9.3 and −9.6 kcal/mol, respectively [66]. The interactions between the ligands and the 1HWK protein domain can be found in Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7, whereas the 6RZ4 can be found in Figure 8, Figure 9 and Figure 10.
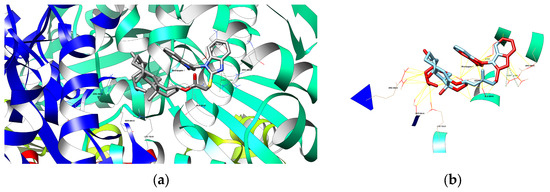
Figure 3.
(a) The depiction of possible hydrogen bond formation between bioconjugate 1 and 1HWK protein domain. (b) The depiction of interactions (contacts/clashes) between bioconjugate 1 and 1HWK protein domain.
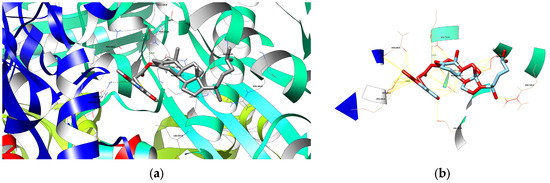
Figure 4.
(a) The depiction of possible hydrogen bond formation between bioconjugate 3 and 1HWK protein domain. (b) The depiction of interactions (contacts/clashes) between bioconjugate 3 and 1HWK protein domain.
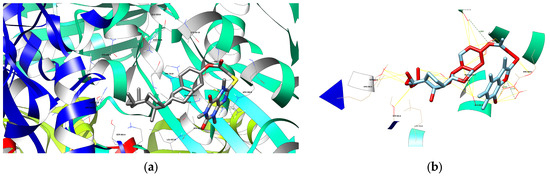
Figure 5.
(a) The depiction of possible hydrogen bond formation between bioconjugate 4 and 1HWK protein domain. (b) The depiction of interactions (contacts/clashes) between bioconjugate 4 and 1HWK protein domain.
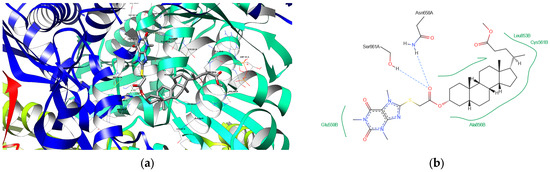
Figure 6.
(a) The depiction of possible hydrogen bond formation between bioconjugate 8 and 1HWK protein domain. (b) The 2D depiction of interactions between bioconjugate 8 and 1HWK protein domain. Blue dashed lines—hydrogen bonds, green solid lines—hydrophobic contacts.
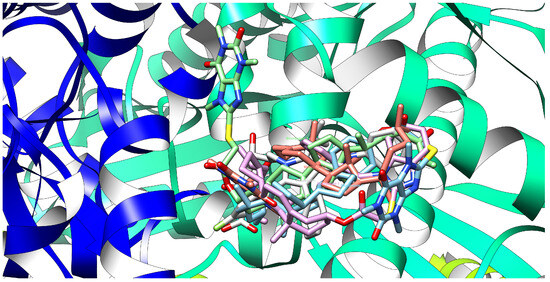
Figure 7.
Native ligand (atorvastatin, grey-colored) and new ligands: bioconjugates 1 (pink-colored), 3 (bittersweet-colored), 4 (cyan-colored), and 8 (green-colored) at once in the binding site of the 1HWK protein domain. The new ligands occupy the same space as the native ligands.
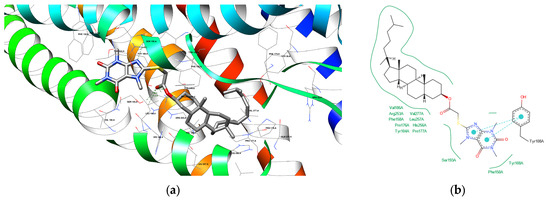
Figure 8.
(a) The depiction of possible hydrogen bond formation between bioconjugate 4 and 6RZ4 protein domain. (b) The 2D depiction of interactions between bioconjugate 4 and 6RZ4 protein domain. Cyan lines with big dots—pi–pi interactions, green solid lines—hydrophobic contacts.
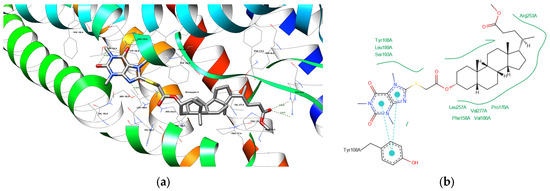
Figure 9.
(a) The depiction of possible hydrogen bond formation between bioconjugate 8 and 6RZ4 protein domain. (b) The 2D depiction of interactions between bioconjugate 8 and 6RZ4 protein domain. Cyan lines with big dots—pi–pi interactions, green solid lines—hydrophobic contacts.
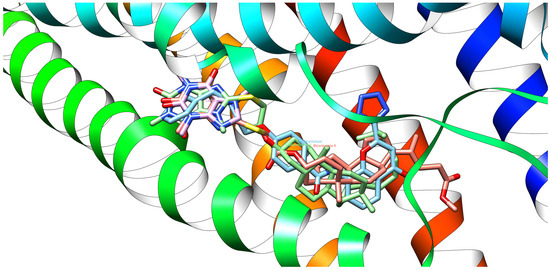
Figure 10.
Native ligand (pranlukast, cyan-colored), reference ligand (caffeine, pink-colored), and new ligands: bioconjugates 4 (green-colored) and 8 (bittersweet-colored) at once in the binding site of a 6RZ4 protein domain.
All the predicted possible hydrogen bonds formed between the protein domains and the ligands are gathered in Table 6. This table contains only the highest probable hydrogen bonds if there are a few possible between certain atoms of a ligand and a protein domain. The potential hydrogen bonds whose length is higher than 3.5 Å are not listed, as their probability of formation is very low [67].

Table 6.
The predicted hydrogen bonds between investigated ligands and protein domains.
A potential hydrogen bond between compound 4 and SER 155 A residue of the 6RZ4 protein domain (Figure 8) can indeed be formed as the hydrogens are added automatically and can be placed at different angles, shortening the distance. The same applies to the potential hydrogen bond between compound 8 and THR 154 A residue of the 6RZ4 protein domain (Figure 9).
Analyzing bond distances in molecular docking studies is important for understanding how stable and biologically relevant ligand–protein interactions are. Generally, shorter bond distances suggest stronger interactions, which help make the ligand–protein complex more stable. For instance, compound 8 has a short bond distance of 2.20 Å with GLY 560 B, indicating a strong interaction that could lead to better binding affinity. In comparison, atorvastatin shows competitive distances, such as 1.91 Å with LYS 735 B, highlighting its effectiveness as a ligand. The biological significance of these distances is considerable; interactions with amino acids like ARG and SER are crucial for protein function. When comparing these compounds to reference ligands like atorvastatin and caffeine—where caffeine has a distance of 2.80 Å with SER 155 A—it becomes clear that some analyzed compounds may have advantages in binding efficiency. This detailed look at bond distances not only helps interpret molecular docking results but also improves our understanding of how effective ligands can be, guiding the design of better ligands for specific protein interactions.
4. Conclusions
The use of compounds of natural origin, such as alkaloids or steroids, as substrates for the synthesis of new bioactive compounds is becoming increasingly popular, particularly in the context of drug discovery. The possibility of modifying the starting compounds allows the synthesis of a wide range of derivatives with potential biological activity. Using caffeine and gramine derivatives as well as steroid derivatives as starting compounds, we obtained ten new steroid–alkaloid bioconjugates. All compounds were characterized by spectroscopic methods. The lowest HOF values for gramine and caffeine bioconjugates with cholic acid were observed. The preliminary results based on in silico data indicated that the obtained compounds, particularly caffeine bioconjugates, can function as cholesterol antagonists, analeptic and respiratory agents, and antihypercholesterolemic compounds. The results of molecular docking to selected protein domains (1HWK and 6RZ4) highlight the potential of the newly developed bioconjugates in pharmacology and give hope for future drug discovery.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/app15020591/s1, Figures S1–10: NMR spectra of compounds 1–10; Figures S11–S20: FT-IR spectra of compounds 1–10; Figures S21–S30: ESI-MS spectra of compounds 1–10.
Author Contributions
Conceptualization, T.P. and B.J.; methodology, K.O., K.B., H.K. and D.N.; software, D.N. and T.P.; validation, D.N. and T.P.; formal analysis, D.N.; investigation, B.J. and T.P.; resources, B.J. and T.P.; data curation, D.N. and T.P.; writing—original draft preparation, H.K., D.N. and B.J.; writing—review and editing, B.J. and T.P.; visualization, K.B., K.O., D.N. and T.P.; supervision, B.J. and T.P.; project administration, B.J. and T.P.; funding acquisition, B.J. and T.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lednicer, D. Strategies for Organic Drug Synthesis and Design; John Wiley & Sons Ltd.: Chichester, UK, 2011. [Google Scholar]
- Santiago-Sampedro, G.I.; Aguilar-Granda, A.; Torres-Huerta, A.; Flores-Álamo, M.; Maldonado-Domínguez, M.; Rodríguez-Molina, B.; Iglesias-Arteaga, A. Self-assembly of an amphiphilic bile acid dimer: A combined experimental and theoretical study of its medium-responsive fluorescence. J. Org. Chem. 2022, 87, 2255–2266. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Bariya, D.; Mishra, R.; Mishra, B. Bile acid-based receptors and their applications in recognition. Steroids 2022, 179, 108981. [Google Scholar] [CrossRef]
- Jain, M.; Nowak, B.P.; Ravoo, B.J. Supramolecular hydrogels based on cyclodextrins: Progress and perspectives. Chem. Nano Mat. 2022, 8, e202200077. [Google Scholar] [CrossRef]
- Kovacevic, B.; Jones, M.; Ionsecu, C.; Walker, D.; Wagle, S.; Chester, J.; Foster, T.; Brown, D.; Mikov, M.; Mooranian, A.; et al. The effect of deoxycholic acid on chitosan-enabled matrices for tissue scaffolding and injectable nanogels. Biomaterials 2022, 8, 358. [Google Scholar] [CrossRef]
- Gao, H.; Dias, J.R. Synthesis and characterization of dimeric bile acid ester derivatives. J. Für Prakt. Chem. Chem.-Ztg. 1997, 339, 187–190. [Google Scholar] [CrossRef]
- Li, Y.; Dias, J.R. Syntheses of α-and β-dimers of lithocholic acid esters. Org. Prep. Proced. Int. 1996, 28, 203–209. [Google Scholar] [CrossRef]
- Paryzek, Z.; Joachimiak, R.; Piasecka, M.; Pospieszny, T. A new approach to steroid dimers and macrocycles by the reaction of 3-chlorocarbonyl derivatives of bile acids with O,O-, N,N-, and S,S-dinucleophiles. Tetrahedron Lett. 2012, 53, 6212–6215. [Google Scholar] [CrossRef]
- Wallimann, P.; Marti, T.; Fürer, A.; Diederich, F. Steroids in molecular recognition. Chem. Rev. 1997, 97, 1567–1608. [Google Scholar] [CrossRef]
- Tamminen, J.; Kolehmainen, E. Bile acids as building blocks of supramolecular hosts. Molecules 2001, 6, 21–46. [Google Scholar] [CrossRef]
- Nagrady, T.; Weaver, D.F. Medicinal Chemistry: A Molecular and Biochemical Approach, 3rd ed.; Oxford University Press: New York, NY, USA, 2005; pp. 316–320. [Google Scholar]
- Parish, E.J.; Nes, W.D. Biochemistry and Function of Sterols; CRC Press: Boca Raton, FL, USA, 1997. [Google Scholar]
- Schaller, H. The role of sterols in plant growth and development. Prog. Lipid Res. 2003, 42, 163–175. [Google Scholar] [CrossRef]
- Abdellatif, K.R.A.; Abdelall, E.K.A.; Elshemy, H.A.H.; El-Nahass, E.S.; Abdel-Fattah, M.M.; Abdelgawad, Y.Y.M. New indomethacin analogs as selective COX-2 inhibitors: Synthesis, COX-1/2 inhibitory activity, anti-inflammatory, ulcerogenicity, histopathological, and docking studies. Arch. Der Pharm. 2021, 354, 2000328. [Google Scholar] [CrossRef]
- Jagadeesan, S.; Karpagam, S. Novel series of N-acyl substituted indole-based piperazine, thiazole, and tetrazoles as potential antibacterial, antifungal, antioxidant, and cytotoxic agents, and their docking investigation as potential Mcl-1 inhibitors. J. Mol. Struct. 2023, 1271, 134013. [Google Scholar] [CrossRef]
- Berdeja, J.G.; Laubach, J.P.; Richter, J.; Stricker, S.; Spencer, A.; Richardson, P.G.; Chari, A. Panobinostat from bench to bedside: Rethinking the treatment paradigm for multiple myeloma. Clin. Lymphoma Myeloma Leuk. 2021, 21, 752–765. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, Q.; Li, N.; Gu, L.; Dan, W.; Dai, J. Recent developments of gramine: Chemistry and biological activity. Molecules 2023, 28, 5695. [Google Scholar] [CrossRef] [PubMed]
- Centofanti, F.; Alonzi, T.; Latini, A.; Spitalieri, P.; Murdocca, M.; Chen, X.; Cui, W.; Shang, Q.; Goletti, D.; Shi, Y.; et al. Indole-3-carbinol In Vitro antiviral activity against SARS-CoV-2 virus and in vivo toxicity. Cell Death Discov. 2022, 8, 491. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.R.; Tsai, C.H.; Kulp, S.K.; Chen, C.S. Indole-3-carbinol as a chemopreventive and anti-cancer agent. Cancer Lett. 2008, 262, 153–163. [Google Scholar] [CrossRef]
- Cai, Q.N.; Han, Y.; Cao, Y.Z.; Hu, Y.; Zhao, X.; Bi, J.L. Detoxification of gramine by the cereal aphid Sitobion avenae. J. Chem. Ecol. 2009, 35, 320–325. [Google Scholar] [CrossRef]
- Quartarone, G.; Ronchin, L.; Vavasori, A.; Tortato, C.; Bonaldo, L. Inhibitive action of gramine towards corrosion of mild steel in deaerated 1.0 M hydrochloric acid solutions. Corros. Sci. 2012, 64, 82–89. [Google Scholar] [CrossRef]
- Kozanecka-Okupnik, W.; Jasiewicz, B.; Pospieszny, T.; Matuszak, M.; Mrówczyńska, L. Haemolytic activity of formyl- and acetyl-derivatives of bile acids and their gramine salts. Steroids 2017, 126, 50–56. [Google Scholar] [CrossRef]
- Kozanecka, W.; Mrówczyńska, L.; Pospieszny, T.; Jasiewicz, B.; Gierszewski, M. Synthesis, spectroscopy, theoretical and biological studies of new gramine-steroids salts and conjugates. Steroids 2015, 98, 92–99. [Google Scholar] [CrossRef]
- Fiani, B.; Zhu, L.; Musch, B.L.; Briceno, S.; Andel, R.; Sadeq, N.; Ansari, A.Z. The neurophysiology of caffeine as a central nervous system stimulant and the resultant effects on cognitive function. Cureus 2021, 13, e15032. [Google Scholar] [CrossRef] [PubMed]
- Woziwodzka, A.; Krychowiak-Maśnicka, M.; Gołuński, G.; Łosiewska, A.; Borowik, A.; Wyrzykowski, D.; Piosik, J. New life of an old drug: Caffeine as a modulator of antibacterial activity of commonly used antibiotics. Pharmaceuticals 2022, 15, 872. [Google Scholar] [CrossRef] [PubMed]
- AlEraky, D.M.; Abuohashish, H.M.; Gad, M.M.; Alshuyukh, M.H.; Bugshan, A.S.; Almulhim, K.S.; Mahmoud, M.M. The antifungal and antibiofilm activities of caffeine against Candida albicans on polymethyl methacrylate denture base material. Biomedicines 2022, 10, 2078. [Google Scholar] [CrossRef]
- Sierakowska, A.; Jasiewicz, B.; Piosik, Ł.; Mrówczyńska, L. New C8-substituted caffeine derivatives as promising antioxidants and cytoprotective agents in human erythrocytes. Sci. Rep. 2023, 13, 1785. [Google Scholar] [CrossRef]
- Jasiewicz, B.; Sierakowska, A.; Wandyszewska, N.; Warżajtis, B.; Rychlewska, U.; Wawrzyniak, R.; Mrówczyńska, L. Antioxidant properties of thio-caffeine derivatives: Identification of the newly synthesized 8-[(pyrrolidin-1-ylcarbonothioyl)sulfanyl] caffeine as antioxidant and highly potent cytoprotective agent. Bioorg. Med. Chem. Lett. 2016, 26, 3994–3998. [Google Scholar] [CrossRef]
- Chen, J.F.; Xu, K.; Petzer, J.P.; Staal, R.; Xy, Y.H.; Beilstein, M.; Sonsalla, P.K.; Castagnoli, K.; Castagnoli, N.; Schwarzschild, M.A. Neuroprotection by caffeine and A(2A) adenosine receptor inactivation in a model of Parkinson’s disease. J. Neurosci. 2001, 15, RC143. [Google Scholar] [CrossRef]
- Booysen, H.P.; Moraal, C.; Terre’Blanche, G.; Petzer, A.; Bergh, J.J.; Petzer, J.P. Thio- and aminocaffeine analogues as inhibitors of human monoamine oxidase. Bioorg. Med. Chem. 2011, 19, 7507–7518. [Google Scholar] [CrossRef]
- Soltani Rad, M.N.; Behrouz, S.; Aghajani, S.; Behrouz, M.; Zarenezhad, E.; Ghanbariasad, A. Design, synthesis, anticancer and in silico assessment of 8-caffeinyl-triazolylmethoxy hybrid conjugates. RSC Adv. 2023, 13, 3056–3070. [Google Scholar] [CrossRef]
- Reshetnikov, D.V.; Burova, L.G.; Rybalova, T.V.; Bondareva, E.A.; Patrushev, S.S.; Evstropov, A.N.; Shults, E.E. Synthesis and Antibacterial Activity of Caffeine Derivatives Containing Amino-Acid Fragments. Chem. Nat. Compd. 2022, 58, 800–806. [Google Scholar] [CrossRef]
- Ohshita, K.; Ishiyama, H.; Oyanagi, K.; Nakata, H.; Kobayashi, J. Synthesis of hybrid molecules of caffeine and eudistomin D and its effects on adenosine receptors. Bioorg. Med. Chem. 2007, 15, 3235–3240. [Google Scholar] [CrossRef]
- Kozanecka-Okupnik, W.; Jasiewicz, B.; Pospieszny, T.; Jastrząb, R.; Skrobańska, M.; Mrówczyńska, L. Spectroscopy, molecular modeling, and antioxidant activity studies on novel conjugates containing indole and uracil moiety. J. Mol. Struct. 2018, 1169, 130–137. [Google Scholar] [CrossRef]
- Jasiewicz, B.; Babijczuk, K.; Warżajtis, B.; Rychlewska, U.; Starzyk, J.; Cofta, G.; Mrówczyńska, L. Indole derivatives bearing imidazole, benzothiazole-2-thione, or benzoxazole-2-thione moieties—Synthesis, structure, and evaluation of their cytoprotective, antioxidant, antibacterial, and fungicidal activities. Molecules 2023, 28, 708. [Google Scholar] [CrossRef]
- Berdzik, N.; Koenig, H.; Mrowczyńska, L.; Nowak, D.; Jasiewicz, B.; Pospieszny, T. Synthesis and hemolytic activity of bile acid-indole bioconjugates linked by triazole. J. Org. Chem. 2023, 88, 16719–16734. [Google Scholar] [CrossRef] [PubMed]
- Babijczuk, K.; Berdzik, N.; Nowak, D.; Warżajtis, B.; Rychlewska, U.; Starzyk, J.; Mrówczyńska, L.; Jasiewicz, B. Novel C3-methylene-bridged indole derivatives with and without substituents at N1: The influence of substituents on their hemolytic, cytoprotective, and antimicrobial activity. Int. J. Mol. Sci. 2024, 25, 5364. [Google Scholar] [CrossRef]
- Yan, B.; Liu, L.; Huang, S.; Ren, Y.; Wang, H.; Yao, Z.; Li, L.; Chen, S.; Wang, X.; Zhang, Z. Discovery of a new class of highly potent necroptosis inhibitors targeting the mixed lineage kinase domain-like protein. Chem. Commun. 2017, 53, 3637. [Google Scholar] [CrossRef]
- Kawka, A.; Hajdaś, G.; Kułaga, D.; Koenig, H.; Kowalczyk, I.; Pospieszny, T. Molecular structure, spectral and theoretical study of new type bile acid–sterol conjugates linked via 1,2,3-triazole ring. J. Mol. Struct. 2023, 1273, 134313. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Morley, C.; Hutchison, G.R. Pybel: A Python wrapper for the OpenBabel cheminformatics toolkit. Chem. Cent. J. 2008, 2, 5. [Google Scholar] [CrossRef]
- Raschka, S. Biopandas: Working with molecular structures in pandas dataframes. J. Open Source Softw. 2017, 2, 14. [Google Scholar] [CrossRef]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. D 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef]
- Istvan, E.S.; Deisenhofer, J. Complex of the catalytic portion of human HMG-CoA reductase with atorvastatin. Protein Data Bank. 2001. Available online: https://www.wwpdb.org/pdb?id=pdb_00001hwk (accessed on 3 December 2024). [CrossRef]
- Luginina, A.; Gusach, A.; Marin, E.; Mishin, A.; Brouillette, R.; Popov, P.; Shiriaeva, A.; Besserer-Offroy, É.; Longpré, J.-M.; Lyapina, E.; et al. Structure-Based Mechanism of Cysteinyl Leukotriene Receptor Inhibition by Antiasthmatic Drugs. Sci. Adv. 2019, 5, eaax2518. [Google Scholar] [CrossRef]
- Luginina, A.; Gusach, A.; Marin, E.; Mishin, A.; Brouillette, R.; Popov, P.; Shiryaeva, A.; Besserer-Offroy, É.; Longpré, J.-M.; Lyapina, E.; et al. Crystal Structure of Cysteinyl Leukotriene Receptor 1 in Complex with Pranlukast. Protein Data Bank. 2019. Available online: https://www.wwpdb.org/pdb?id=pdb_00006rz4 (accessed on 3 December 2024).
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDock-Tools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- ProteinsPlus Development Team. ProteinsPlus. Available online: https://proteins.plus (accessed on 18 November 2024).
- Schöning-Stierand, K.; Diedrich, K.; Ehrt, C.; Flachsenberg, F.; Graef, J.; Sieg, J.; Penner, P.; Poppinga, M.; Ungethüm, A.; Rarey, M. ProteinsPlus: A comprehensive collection of web-based molecular modeling tools. Nucleic Acids Res. 2022, 50, W611–W615. [Google Scholar] [CrossRef]
- Stierand, K.; Maaß, P.C.; Rarey, M. Molecular complexes at a glance: Automated generation of two-dimensional complex diagrams. Bioinformatics 2006, 22, 1710–1716. [Google Scholar] [CrossRef]
- Diedrich, K.; Krause, B.; Berg, O.; Rarey, M. PoseEdit: Enhanced ligand binding mode communication by interactive 2D diagrams. J. Comput. Aided Mol. Des. 2023, 37, 491–503. [Google Scholar] [CrossRef]
- Poroikov, V.V.; Filimonov, D.A. Predictive Toxicology; Helma, C., Ed.; Taylor & Francis: Boca Raton, FL, USA, 2005; pp. 459–478. [Google Scholar]
- Poroikov, V.V.; Filimonov, D.A. How to acquire new biological activities in old compounds by computer prediction. J. Comput. Aided Mol. Des. 2002, 16, 819–824. [Google Scholar] [CrossRef]
- Poroikov, V.V.; Filimonov, D.A.; Borodina, Y.V.; Lagunin, A.A.; Kos, A. Robustness of biological activity spectra predicting by computer program PASS for non-congeneric sets of chemical compounds. J. Chem. Inf. Comput. Sci. 2000, 40, 1349–1355. [Google Scholar] [CrossRef]
- Mohammed, A.I.; Musa, A.; Abu-Bakr, M.S.; Abbass, H.S. Anti-eczematic and molecular modeling of anthraquinones isolated from the seeds of Asphodelus microcarpus salzm. viv. growing in Egypt. Pharmacogn. Mag. 2019, 15, 586–591. [Google Scholar] [CrossRef]
- Watschinger, K.; Keller, M.A.; Hermetter, A.; Golderer, G.; Werner-Felmayer, G.; Werner, E.R. Glyceryl ether monooxygenase resembles aromatic amino acid hydroxylases in metal ion and tetrahydrobiopterin dependence. Biol. Chem. 2009, 390, 3–10. [Google Scholar] [CrossRef]
- Rahmati-Ahmadabad, S.; Broom, D.R.; Ghanbari-Niaki, A.; Shirvani, H. Effects of exercise on reverse cholesterol transport: A systemized narrative review of animal studies. Life Sci. 2019, 224, 139–148. [Google Scholar] [CrossRef]
- Agosto, N.J.; Alambatin, P.B.; Bacalso, J.; Cabisada, J.; Carating, B.D. The evaluation of Citrus bergamia phytochemicals as potential cholesterol-lowering agents against HMG-CoA reductase: An in silico molecular docking study. Biol. Life Sci. Forum 2024, 35, 7. [Google Scholar] [CrossRef]
- Peppin, J.F.; Pergolizzi, J.V.; Fudin, J.; Meyer, T.A.; Raffa, R.B. History of respiratory stimulants. J. Pain Res. 2021, 14, 1043–1049. [Google Scholar] [CrossRef]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef]
- Landrum, G. RDKit: Open-Source Cheminformatics Software; Zenodo: Geneva, Switzerland, 2022. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: An analysis of ALOGP and CLOGP methods. J. Phys. Chem. A 1998, 102, 3762–3772. [Google Scholar] [CrossRef]
- Ramírez, D.; Caballero, J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules 2018, 23, 1038. [Google Scholar] [CrossRef]
- Herschlag, D.; Pinney, M.M. Hydrogen bonds: Simple after all? Biochemistry 2018, 57, 3338–3352. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).