MemGanomaly: Memory-Augmented Ganomaly for Frost- and Heat-Damaged Crop Detection
Abstract
1. Introduction
2. Related Work
2.1. Machine Learning-Based Approaches
2.2. Convolutional Neural Networks
2.3. Autoencoders
2.4. Generative Adversarial Networks
2.5. Transformer-Based Models
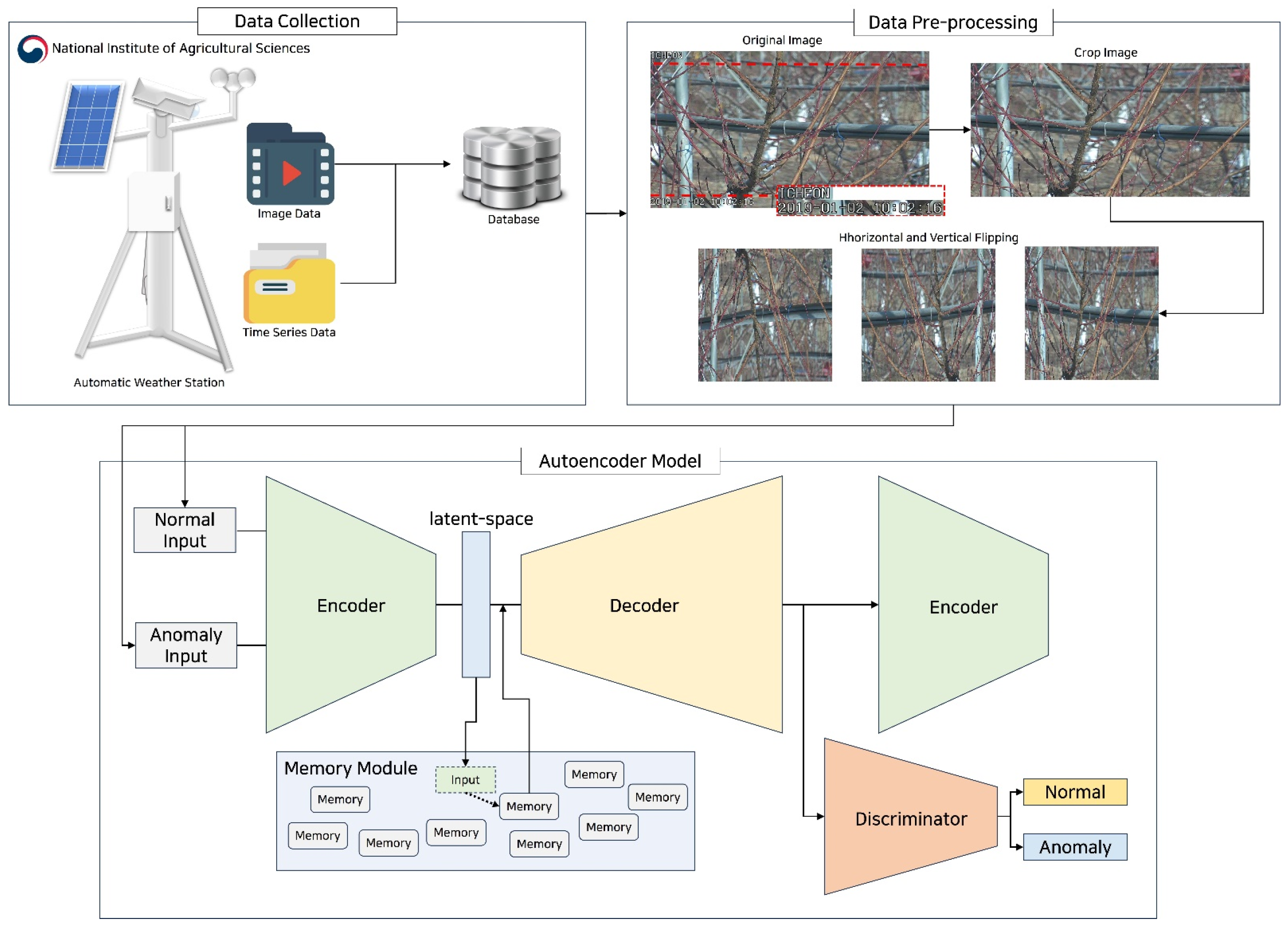
3. Materials and Methods
3.1. Data Preprocessing Module
3.2. Data Collection
3.3. Data Preprocessing
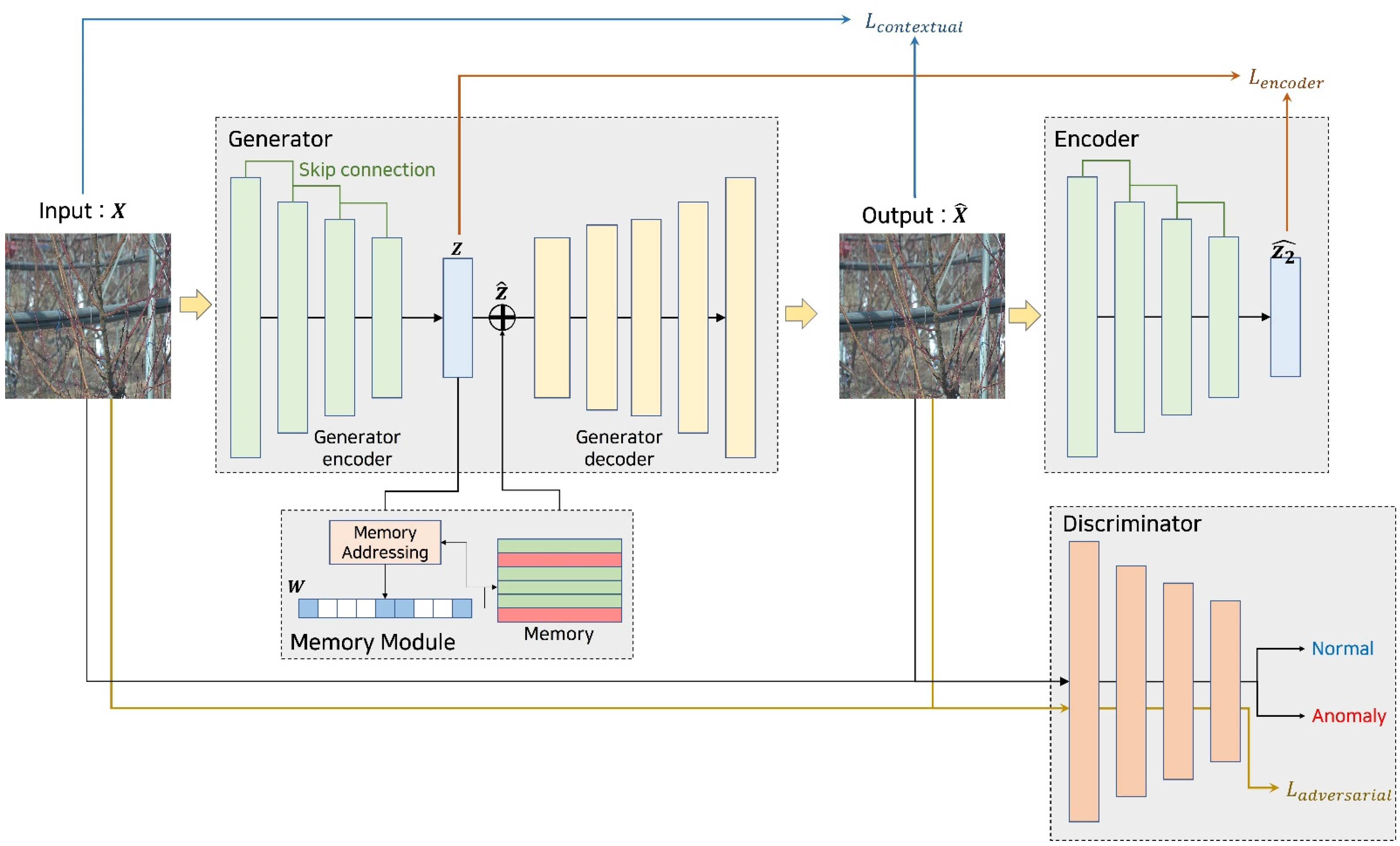
3.4. Autoencoder Model
3.4.1. Generator
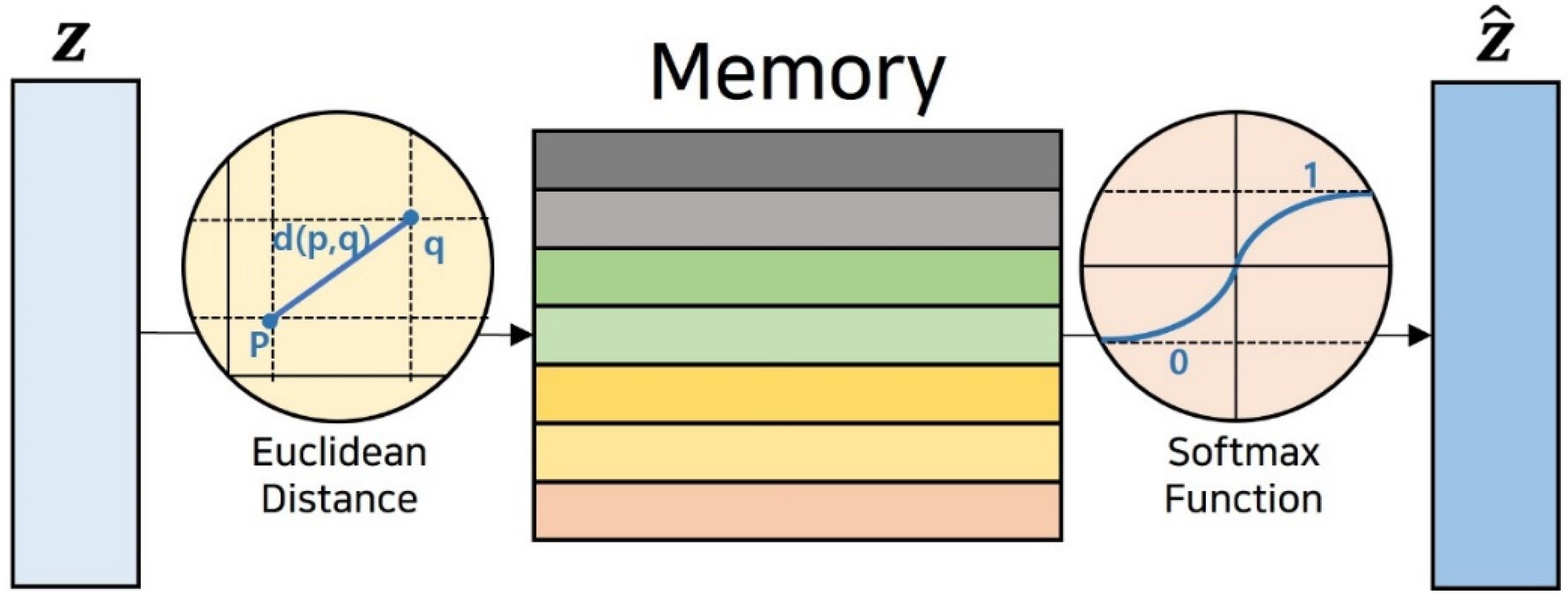
3.4.2. Memory Module
3.4.3. Encoder
3.4.4. Discriminator
3.4.5. Hyperparameter Settings
3.4.6. Programming Environment
3.5. Training Details
4. Loss Function
4.1. Loss Function Overview
4.2. Contextual Loss
4.3. Encoder Loss
4.4. Adversarial Loss
4.5. Total Loss Function
5. Results
5.1. Crop Dataset
5.2. Model Performance
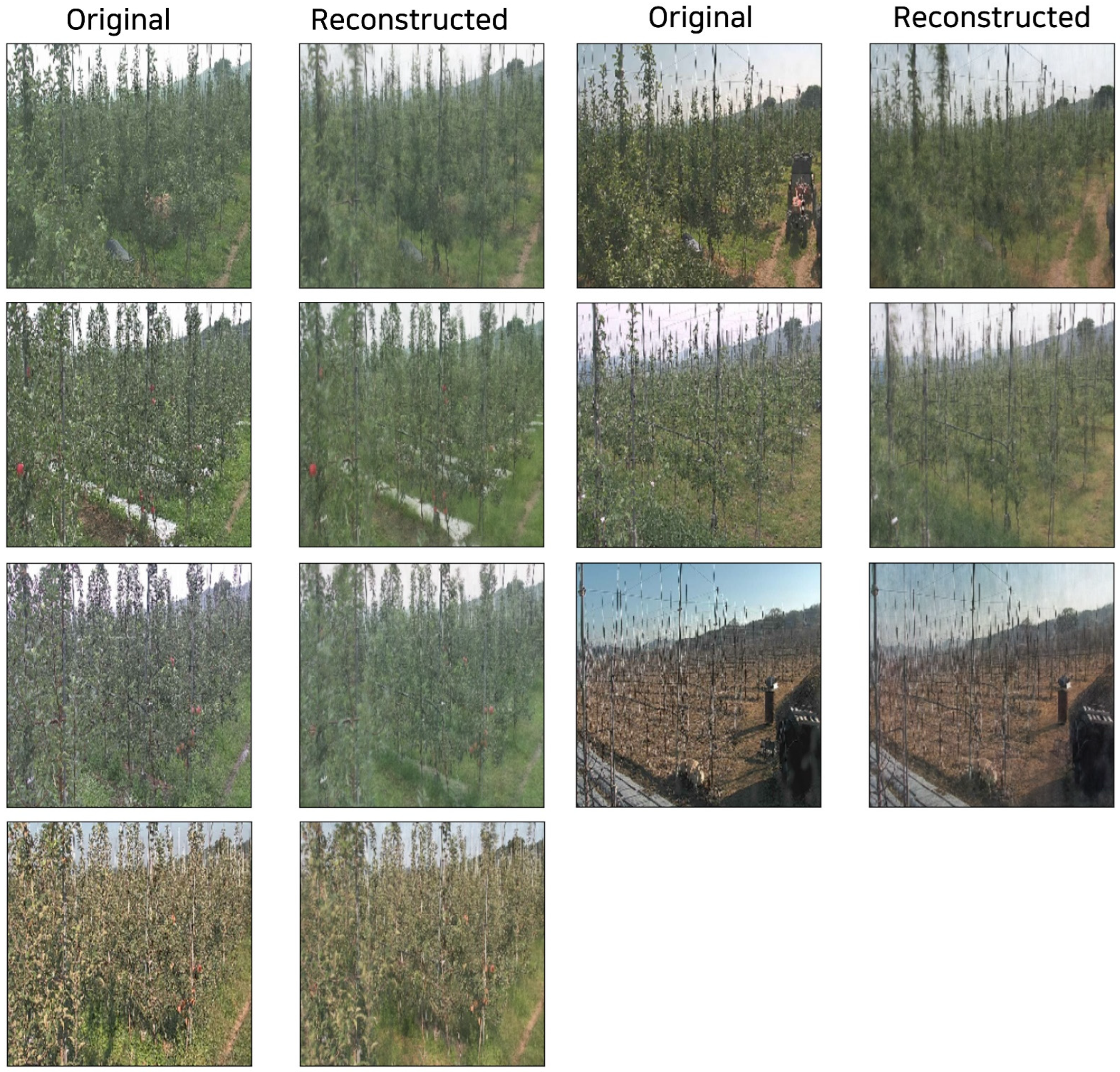
5.3. Reconstruction Results
5.4. Computational Efficiency
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- You, J.; Li, X.; Low, M.; Lobell, D.; Ermon, S. Deep Gaussian Process for Crop Yield Prediction Based on Remote Sensing Data. Proc. AAAI Conf. Artif. Intell. 2017, 31, 4559–4566. [Google Scholar] [CrossRef]
- Verma, S.; Singh, A.; Pradhan, S.S.; Kushuwaha, M. Impact of Climate Change on Agriculture: A Review. Int. J. Environ. Clim. Change 2024, 14, 615–620. [Google Scholar] [CrossRef]
- Arora, N.K. Impact of Climate Change on Agriculture Production and Its Sustainable Solutions. Environ. Sustain. 2019, 2, 95–96. [Google Scholar] [CrossRef]
- Afsar, M.M.; Bakhshi, A.D.; Iqbal, M.S.; Hussain, E.; Iqbal, J. High-Precision Mango Orchard Mapping Using a Deep Learning Pipeline Leveraging Object Detection and Segmentation. Remote Sens. 2024, 16, 3207. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Sharma, A.; Jain, A.; Gupta, P.; Chowdary, V. Machine Learning Applications for Precision Agriculture: A Comprehensive Review. IEEE Access 2020, 9, 4843–4873. [Google Scholar] [CrossRef]
- Nevavuori, P.; Narra, N.; Lipping, T. Crop yield prediction with deep convolutional neural networks. Comput. Electron. Agric. 2019, 163, 104859. [Google Scholar] [CrossRef]
- Dong, S.; Wang, P.; Abbas, K. A Survey on Deep Learning and Its Applications. Comput. Sci. Rev. 2021, 40, 100379. [Google Scholar] [CrossRef]
- Mujkic, E.; Philipsen, M.P.; Moeslund, T.B.; Christiansen, M.P.; Ravn, O. Anomaly Detection for Agricultural Vehicles Using Autoencoders. Sensors 2022, 22, 3608. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep Learning in Agriculture: A Survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Jeyabose, A.; Eunice, J.; Popescu, D.E.; Chowdary, M.K.; Hemanth, J. Deep Learning-Based Leaf Disease Detection in Crops Using Images for Agricultural Applications. Agronomy 2022, 12, 2395. [Google Scholar] [CrossRef]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehn, H.-W.; Plümer, L. Early Detection and Classification of Plant Diseases with Support Vector Machines Based on Hyperspectral Reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Demilie, W.B. Plant disease detection and classification techniques: A comparative study of the performances. J. Big Data 2024, 11, 5. [Google Scholar] [CrossRef]
- Oğuz, A.; Ertuğrul, Ö.F. A survey on applications of machine learning algorithms in water quality assessment and water supply and management. Water Supply 2023, 23, 895–922. [Google Scholar] [CrossRef]
- Meshram, V.; Patil, K.; Meshram, V.; Hanchate, D.; Ramteke, S.D. Machine learning in agriculture domain: A state-of-art survey. Artif. Intell. Life Sci. 2021, 1, 100010. [Google Scholar] [CrossRef]
- Garofalo, S.P.; Ardito, F.; Sanitate, N.; De Carolis, G.; Ruggieri, S.; Giannico, V.; Rana, G.; Ferrara, R.M. Robustness of Actual Evapotranspiration Predicted by Random Forest Model Integrating Remote Sensing and Meteorological Information: Case of Watermelon (Citrullus lanatus, (Thunb.) Matsum. & Nakai, 1916). Water 2025, 17, 323. [Google Scholar] [CrossRef]
- Araújo, S.O.; Peres, R.S.; Ramalho, J.C.; Lidon, F.; Barata, J. Machine Learning Applications in Agriculture: Current Trends, Challenges, and Future Perspectives. Agronomy 2023, 13, 2976. [Google Scholar] [CrossRef]
- Sutaji, D.; Rosyid, H. Convolutional Neural Network (CNN) Models for Crop Diseases Classification. Kinetik 2022, 7. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep Learning Models for Plant Disease Detection and Diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Too, E.C.; Li, Y.; Njuki, S.; Liu, Y. A Comparative Study of Fine-Tuning Deep Learning Models for Plant Disease Identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar] [CrossRef]
- Ramcharan, A.; Baranowski, A.; McCloskey, P.; Ahmed, B.; Legg, J.; Hughes, D.P. Deep Learning for Image-Based Cassava Disease Detection. Front. Plant Sci. 2017, 8, 1852. [Google Scholar] [CrossRef]
- Tokunaga, T.; Katafuchi, R. Image-based Plant Disease Diagnosis with Unsupervised Anomaly Detection Based on Reconstructability of Colors. In Proceedings of the International Conference on Image Processing and Vision Engineering (IMPROVE 2021), Online, 28–30 April 2021; pp. 112–120. [Google Scholar] [CrossRef]
- Garcia-Huerta, R.A.; González-Jiménez, L.E.; Villalon-Turrubiates, L.E. Sensor Fusion Algorithm Using a Model-Based Kalman Filter for the Position and Attitude Estimation of Precision Aerial Delivery Systems. Sensors 2020, 20, 5227. [Google Scholar] [CrossRef] [PubMed]
- Benfenati, A.; Causin, P.; Oberti, R.; Stefanello, G. Unsupervised deep learning techniques for automatic detection of plant diseases: Reducing the need of manual labelling of plant images. J. Math. Ind. 2023, 13, 5. [Google Scholar] [CrossRef]
- Ciniglio, A.; Guiotto, A.; Spolaor, F.; Sawacha, Z. The Design and Simulation of a 16-Sensors Plantar Pressure Insole Layout for Different Applications: From Sports to Clinics, a Pilot Study. Sensors 2021, 21, 1450. [Google Scholar] [CrossRef]
- Schlegl, T.; Seeböck, P.; Waldstein, S.M.; Langs, G.; Schmidt-Erfurth, U. f-AnoGAN: Fast Unsupervised Anomaly Detection with Generative Adversarial Networks. Med. Image Anal. 2019, 54, 30–44. [Google Scholar] [CrossRef]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2016, arXiv:1511.06434. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the 28th International Conference on Neural Information Processing Systems—Volume 2 (NIPS’14), Montreal, QC, Canada, 8–13 December 2014; MIT Press: Cambridge, MA, USA, 2014; pp. 2672–2680. Available online: https://dl.acm.org/doi/10.5555/2969033.2969125 (accessed on 25 September 2025).
- Prosvirin, A.E.; Islam, M.M.M.; Kim, J.-M. An Improved Algorithm for Selecting IMF Components in Ensemble Empirical Mode Decomposition for Domain of Rub-Impact Fault Diagnosis. IEEE Access 2019, 7, 121728–121741. [Google Scholar] [CrossRef]
- Gong, D.; Liu, L.; Le, V.; Saha, B.; Mansour, M.R.; Venkatesh, S. Memorizing Normality to Detect Anomaly: Memory-Augmented Deep Autoencoder for Unsupervised Anomaly Detection. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 16000–16009. [Google Scholar] [CrossRef]
- Akçay, S.; Atapour-Abarghouei, A.; Breckon, T.P. Ganomaly: Semi-Supervised Anomaly Detection via Adversarial Training. Proc. Asian Conf. Comput. Vis. 2018, 11363, 622–637. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar] [CrossRef]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated Residual Transformations for Deep Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5987–5995. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11966–11976. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 9992–10002. [Google Scholar] [CrossRef]
- Koch, G.; Zemel, R.; Salakhutdinov, R. Siamese Neural Networks for One-Shot Image Recognition. Available online: https://www.cs.cmu.edu/~rsalakhu/papers/oneshot1.pdf (accessed on 25 September 2025).
- Snell, J.; Swersky, K.; Zemel, R.S. Prototypical Networks for Few-Shot Learning. Adv. Neural Inf. Process. Syst. 2017, 30, 4077–4087. [Google Scholar] [CrossRef]
- Vinyals, O.; Blundell, C.; Lillicrap, T.; Kavukcuoglu, K.; Wierstra, D. Matching Networks for One-Shot Learning. Adv. Neural Inf. Process. Syst. 2016, 29, 3630–3638. [Google Scholar] [CrossRef]




| State | Apple | Peach | ||
|---|---|---|---|---|
| Original | Used | Original | Used | |
| Dormancy | 4504 | 401 | 10,877 | 1047 |
| Bud stage | 683 | 401 | 2584 | 1047 |
| Flowering | 401 | 401 | 1047 | 1047 |
| Post-flowering | 619 | 401 | 1417 | 1047 |
| Early fruit growth | 1560 | 401 | 2960 | 1047 |
| Fruit growth | 2364 | 401 | 9298 | 1047 |
| Harvest | 544 | 401 | 2120 | 1047 |
| Nutrient accumulation | 3667 | 401 | 4122 | 1047 |
| Total | 14,342 | 3208 | 34,425 | 8376 |
| State | Apple | Peach | ||
|---|---|---|---|---|
| Original | Used | Original | Used | |
| Cold Damage | 31 | 93 | 36 | 108 |
| Heat Damage | 27 | 81 | 32 | 96 |
| Total | 58 | 174 | 68 | 204 |
| Model | Accuracy (%) | Precision (%) | Recall (%) |
|---|---|---|---|
| ResNet-50 | 94.86 | 0 | 0 |
| EfficientNet-B3 | 94.86 | 0 | 0 |
| ResNeXt-50 | 94.86 | 0 | 0 |
| ConvNeXt-Tiny | 94.86 | 0 | 0 |
| Swin Transformer-Tiny | 94.86 | 0 | 0 |
| Siamese Network(10-shot) | 68.12 ± 2.7 | 64.11 ± 2.1 | 70.3 ± 2.8 |
| Prototypical Network(10-shot) | 68.64 ± 3.1 | 65.32 ± 4.4 | 65.6 ± 5.7 |
| Matching Network(10-shot) | 65.81 ± 3.4 | 61.55 ± 3.2 | 64.0 ± 4.7 |
| Siamese Network(20-shot) | 73.11 ± 3.7 | 77.43 ± 2.6 | 75.3 ± 2.8 |
| Prototypical Network(20-shot) | 66.64 ± 6.9 | 61.32 ± 7.4 | 62.6 ± 7.1 |
| Matching Network(20-shot) | 71.81 ± 4.3 | 73.55 ± 5.0 | 72.0 ± 4.7 |
| Ganomaly | 77.29 ± 2.6 | 76.98 ± 3.1 | 77.12 ± 2.9 |
| MemAE | 79.61 ± 1.7 | 78.21 ± 2.5 | 78.0 ± 2.1 |
| Proposed Model | 80.32 ± 1.3 | 79.4 ± 1.6 | 79.1 ± 1.4 |
| Model | Accuracy (%) | Precision (%) | Recall (%) |
|---|---|---|---|
| ResNet-50 | 97.63 | 0 | 0 |
| EfficientNet-B3 | 97.63 | 0 | 0 |
| ResNeXt-50 | 97.63 | 0 | 0 |
| ConvNeXt-Tiny | 97.63 | 0 | 0 |
| Swin Transformer-Tiny | 97.63 | 0 | 0 |
| Siamese Network(10-shot) | 70.64 ± 2.7 | 69.9 ± 3.7 | 71.9 ± 1.1 |
| Prototypical Network(10-shot) | 67.33 ± 3.6 | 69.33 ± 6.7 | 68.31 ± 4.7 |
| Matching Network(10-shot) | 68.17 ± 4.1 | 64.87 ± 3.4 | 57.74 ± 5.4 |
| Siamese Network(20-shot) | 71.43 ± 3.1 | 76.3 ± 4.9 | 75.1 ± 2.8 |
| Prototypical Network(20-shot) | 61.7 ± 5.5 | 63.27 ± 3.3 | 59.6 ± 7.1 |
| Matching Network(20-shot) | 73.94 ± 5.2 | 69.64 ± 4.6 | 71.0 ± 3.7 |
| Ganomaly | 81.42 ± 3.1 | 81.08 ± 2.7 | 76.9 ± 3.2 |
| MemAE | 80.96 ± 2.1 | 79.01 ± 2.9 | 77.8 ± 2.5 |
| Proposed Model | 81.06 ± 1.7 | 83.23 ± 1.6 | 80.3 ± 1.7 |
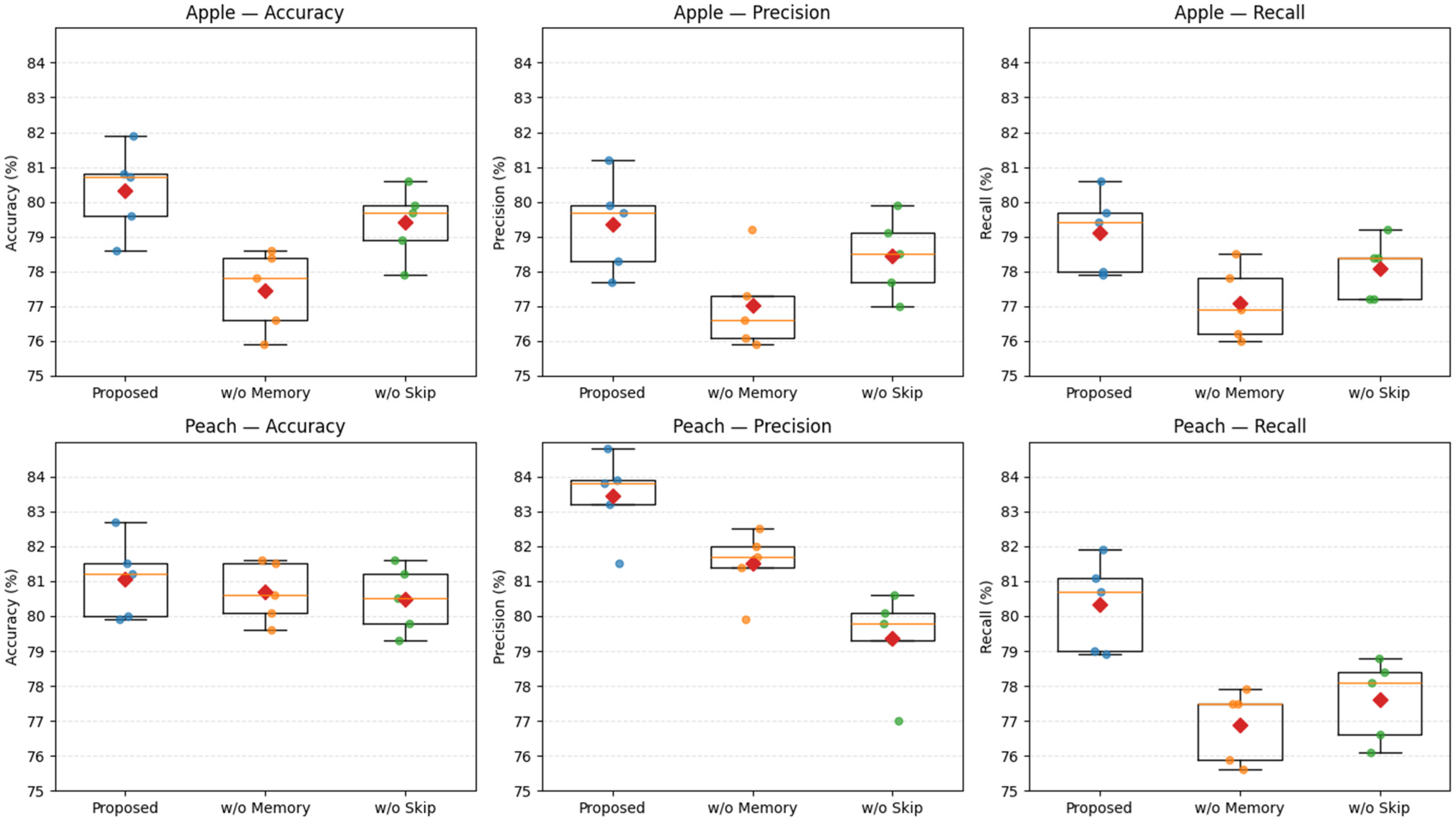
| Dataset | Metric | Proposed Model | w/o Memory Module (Ganomaly) | w/o Skip Connections (MemAE) |
|---|---|---|---|---|
| Apple | Accuracy | 80.32 ± 1.3 | 77.29 ± 2.6 | 79.61 ± 1.7 |
| Precision | 79.4 ± 1.6 | 76.98 ± 3.1 | 78.21 ± 2.5 | |
| Recall | 79.1 ± 1.4 | 77.12 ± 2.9 | 78.0 ± 2.1 | |
| Peach | Accuracy | 81.06 ± 1.7 | 81.42 ± 3.1 | 80.96 ± 2.1 |
| Precision | 83.23 ± 1.6 | 81.08 ± 2.7 | 79.01 ± 2.9 | |
| Recall | 80.3 ± 1.7 | 76.9 ± 3.2 | 77.8 ± 2.5 |
| Dataset | Condition | SSIM (↑) | PSNR (dB, ↑) |
|---|---|---|---|
| Apple | Undamaged | 0.87 ± 0.02 | 28.5 ± 1.1 |
| Apple | Damaged | 0.62 ± 0.03 | 21.7 ± 1.4 |
| Peach | Undamaged | 0.89 ± 0.01 | 29.2 ± 1.0 |
| Peach | Damaged | 0.64 ± 0.02 | 22.1 ± 1.2 |
| Model | Component | Params (M) | FLOPs (G) |
|---|---|---|---|
| GANomaly | Generator | 6.07 | 120.1 |
| Discriminator | 0.37 | 127.8 | |
| Total | 6.45 | 247.9 | |
| MemAE | AE (Total) | 5.24 | 769.2 |
| MemGanomaly (Proposed) | Generator | 34.18 | 2022.6 |
| Discriminator | 3.91 | 218.5 | |
| Total | 38.09 | 2241.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Park, S.-W.; Kim, Y.-S.; Jung, S.-H.; Sim, C.-B. MemGanomaly: Memory-Augmented Ganomaly for Frost- and Heat-Damaged Crop Detection. Appl. Sci. 2025, 15, 10503. https://doi.org/10.3390/app151910503
Park J, Park S-W, Kim Y-S, Jung S-H, Sim C-B. MemGanomaly: Memory-Augmented Ganomaly for Frost- and Heat-Damaged Crop Detection. Applied Sciences. 2025; 15(19):10503. https://doi.org/10.3390/app151910503
Chicago/Turabian StylePark, Jun, Sung-Wook Park, Yong-Seok Kim, Se-Hoon Jung, and Chun-Bo Sim. 2025. "MemGanomaly: Memory-Augmented Ganomaly for Frost- and Heat-Damaged Crop Detection" Applied Sciences 15, no. 19: 10503. https://doi.org/10.3390/app151910503
APA StylePark, J., Park, S.-W., Kim, Y.-S., Jung, S.-H., & Sim, C.-B. (2025). MemGanomaly: Memory-Augmented Ganomaly for Frost- and Heat-Damaged Crop Detection. Applied Sciences, 15(19), 10503. https://doi.org/10.3390/app151910503







