Abstract
Super-resolution microscopy has been recently applied to understand the 3D topology of chromatin at an intermediated genomic scale (kilobases to a few megabases), as this corresponds to a sub-diffraction spatial scale crucial for the regulation of gene transcription. In this context, polycomb proteins are very renowned gene repressors that organize into the multiprotein complexes Polycomb Repressor Complex 1 (PRC1) and 2 (PRC2). PRC1 and PRC2 operate onto the chromatin according to a complex mechanism, which was recently recapitulated into a working model. Here, we present a functional colocalization study at 100–140 nm spatial resolution targeting PRC1 and PRC2 as well as the histone mark H3K27me3 by Image Scanning Microscopy (ISM). ISM offers a more flexible alternative to diffraction-unlimited SRMs such as STORM and STED, and it is perfectly suited to investigate the mesoscale of PRC assembly. Our data suggest a partially simultaneous effort of PRC1 and PRC2 in locally shaping the chromatin topology.
1. Introduction
The genomes of metazoans are organized at multiple spatial scales, which range from the double helix of DNA to whole chromosomes [1]. These scales hint at different levels of functional organization, which are essential to transferring the genome information to cell machinery yielding the actual cellular phenotype.
The intermediate genomic scale of kilobases to megabases, corresponding to 50–300 nm spatial scale, is fascinating as the 3D arrangement of DNA, proteins and other biomolecules—commonly referred to as chromatin—is implicated in multiple regulatory mechanisms [2]. Here, polycomb group (PcG) proteins stand as major epigenetic modulators of chromatin function, acting mostly as repressors of gene transcription by a synergistic combination of chemical histone modifications with the physical crosslinking of distal genomic regions [3]. PcG proteins mainly assemble in two multiprotein complexes, which are known as polycomb repressive complexes 1 and 2 (PRC1 and PRC2) [4,5]. PRC1, through its catalytic subunit RING1B (or its less diffused isoform RING1A) and in association with one of the six PCGF paralog genes (PCGF1-6), monoubiquitinylates histone H2A at the lysine 119 (H2AK119ub1). The nature of PCGF further classifies PRC1 into “variant” PRC1 (vPRC1, containing PCGF1,3,5,6) and “canonical” PRC1 (cPRC1, containing PCGF2,4). PCGF4, known as BMI1, is a crucial molecular player in cell differentiation and cancers [6,7,8]. vPRC1 has been demonstrated to possess an enhanced ubiquitination activity with respect to cPRC1 [9]. Notably, PRC1 complexes are endowed with multiple functions by the presence of ancillary scaffold proteins [10,11].
PRC2 trimethylates histone 3 on lysine 27 (H3K27me3) via its enzymatic unit EZH2 (or its paralog EZH1, more abundant in resting cells) in combination with the PRC2 scaffold proteins SUZ2 and EED [5]. A further subdivision of PRC2 into PRC2.1 and PRC2.2 (with PRC2.2 endowed with higher catalytic activity) stems from the different ancillary proteins that participate in the complexes [12].
The working model of PcG repressing activity assumes a coordinated interplay of PRC1 and PRC2 along the genomic regions that must be silenced [3]. At first, vPRC1.1 (containing PCGF1) is tethered at non-methylated CG-rich sequences (CGIs) by the ancillary protein KDM2B. PRC1.1 deposits the H2AK119ub mark, which in turn recruits PRC2, which activates H3 methylation. The latter modification allosterically activates PRC2 to expand the methylation region, and a similar mechanism also operates for vPRC1 from ubiquitinated H2A foci. Finally, the H3K27me3 mark is used for docking cPRC1 by the ancillary protein CBXs. cPRC1, through the Sterile Alpha Motif (SAM) of PHC1-3 proteins, activates the physical crosslinking of distal chromatin sequences [13]. Of note, a few details on PcG repressing activities came from super-resolution microscopy (SRM) studies, mostly by STORM imaging combined with multilabel Fluorescence In Situ Hybridization [14,15,16]. These studies complemented chromosome capture studies in recapitulating PcG-modulated 3D topologies of chromatin molecules at nanoscale resolution [16].
Yet, some aspects of PRC1 and PRC2 activities are still obscure. For instance, it is unclear whether the activity of vPRC1, PRC2, and cPRC1 is temporally sequential or, vice-versa, some of these protein complexes operate on the same DNA strands at the same time. We recently questioned this aspect by investigating the functional colocalization of cPRC1 component PCGF4 (aka: BMI1) with H2AK119ub by confocal microscopy in a model epithelial (i.e., differentiated) cell line [17]. Functional colocalization was expressed by the Pearson’s coefficient R, which quantifies the degree of correlated variability between two acquisition channels and thereby samples the functional association between the two fluorescent partners through their stoichiometric correlation [18]. Remarkably, statistically significant positive R values were found between H2AK119ub and BMI1, albeit the concentrations of these two proteins were highly correlated at the whole nuclear level [17]. This was interpreted according to a dynamically “lagged” docking of cPRC1 and histone ubiquitination. Another explanation may invoke the prevalent E3-ligase activity of vPRC1 [9], which does not contain BMI1.
On account of our long-standing interest in SRM and its applications, in this work, we extended our previous colocalization approach by replacing traditional confocal with ISM imaging, as the gain in spatial resolution is expected to increase the significance of positive channel correlation in terms of functional biochemical relationships. Indeed, ISM resolution (100–200 nm) fits perfectly the mesoscale dimension of most PcG foci reported by Wani et al. [15]. Of note, we recently adopted the ISM colocalization approach in a study targeting the endosomal entry in cells of SARS-Cov-2 virus [19].
Owing to the pivotal role of PRC2 as “mediator” between vPRC1 and cPRC1, we focused our attention on EZH2, whose housekeeping role makes it the best proxy of PRC2 localization in the nucleus. Accordingly, we set out to determine the functional colocalization of EZH2 with RING1b, BMI1, and H3K27me3, to infer the stoichiometric relationship at mesoscale of PRC2 with (vPRC1 + cPRC2), cPRC1, and the product of its enzymatic activity, respectively. As these experiments represented a preliminary step within a larger project on PRCs in the context of differentiated and neoplastic cells, our cell model was represented by HCC827, a Non-Small Cell Lung Cancer (NSCLC) epithelial cell line bearing the classic oncogenic 746–750 deletion on the EGFR gene [20].
2. Materials and Methods
Reagents were acquired from Merck (Milan, Italy). Cell Signaling antibodies were purchased from EuroClone (Milan, Italy). AbCam antibodies were acquired from Prodotti Gianni (Milan, Italy). Life Technologies antibodies were purchased from Life Technologies Italy (Monza, Italy). Merck antibodies were acquired from Merck Italia (Milan, Italy)
2.1. Primary Antibodies
- Mouse anti-human EZH2 monoclonal antibody (3147S, Cell Signaling Technologies): 1/100 dilution.
- Rabbit anti-human BMI1 monoclonal antibody (6964S, Cell Signaling Technologies): 1/600 dilution.
- Rabbit anti-human RING1b monoclonal antibody (5694S, Cell Signaling Technologies): 1/250 dilution.
- Rabbit anti-human H3K27me3 monoclonal antibody (ab192985, AbCam): 1/1000 dilution.
2.2. Secondary Antibodies
- Donkey anti-mouse IgG AlexaFluor 488 (A32766, Life Technologies): 1/400 dilution.
- Donkey anti-rabbit IgG AlexaFluor 647 (A32795, Life Technologies): 1/400 dilution.
- Goat anti-rabbit IgG Abberior StarRed (41699, Merck Italia): 1/400 dilution.
2.3. Cell Culture
HCC827 NSCLC cells were grown at 37 °C and with 5% CO2 in a RPMI-1640 medium supplied with phenol red and enriched with NaHCO3, L-glutamine, 1% of sodium pyruvate (Merck, Milan, Italy), 1% of penstreptomycin (Merck, Milan, Italy), and 10% of fetal bovine serum (Merck, Milan, Italy). For immunofluorescence, we seeded HCC827 cells (1–3 × 105) in 35 mm glass-bottom dishes (Willco, Amsterdam, The Netherlands) with 2 ml of medium, and we maintained this culture for 1 day at 37 °C and 5% CO2.
2.4. Cell Indirect Immunofluorescence Protocol
After rinsing (3 times) with phosphate buffer saline 1× (PBS), HCC827 cells were fixed with paraformaldehyde (Merck, Milan, Italy) 2% in PBS for 15 min. Cells were washed again (3 times) with PBS and then permeabilized for 15 min with 0.1% Triton X-100 (Merck, Milan, Italy). After rinsing with PBS (3 times) and 0.5% Bovine Serum Albumin (BSA, Merck, Milan, Italy) in PBS (PBB, 4 times), cells were maintained in the blocking solution (2% BSA in PBS) for 40 min. Then, cells were incubated with the primary antibody (dilution in PBB) for 12 h at 4 °C. After rinsing with PBB (4 times), cells were incubated with the secondary antibody (dilution in PBB) for 1 h at room temperature in the dark. Finally, cells were washed with PBB (4 times) and PBS (3 times), chromatin was stained with 0.01 mg/mL Hoechst 33342 (Merck, Milan, Italy) for 5 min, and it was washed again with PBS (three times). Cells were kept at 4 °C in the dark up to the imaging session.
2.5. Confocal Fluorescence Microscopy
Measurements were carried out by a confocal Zeiss LSM 900 with Airyscan (Carl Zeiss, Jena, Germany). Samples were viewed with a 63× Apochromat oil-immersion objective (NA = 1.4). The pinhole size was set to 1 airy unit (AU) for the green acquisition channel. Then, 512 × 512 pixel images were acquired line by line in the three channels (blue, green, and far-red, acquisition in this sequence) by setting the pixel dwell time to 1.52 ms and taking 4 averages per line. Cells were imaged at the focal depth maximizing the nuclear section on the image plane. The spectral windows of the acquisition channels were: (i) blue (Hoechst 33342): λex = 405 λem = 420–500 nm; (ii) green: λex = 488 (Alexa488), λem = 500–560 nm; and (iii) far-red (Alexa647 and Abberior StarRed): λex = 640, λem = 650–700 nm. Image processing was carried out by the open-source software Fiji (NIH, Bethesda, USA, 2022) [21]. Intensity detection of green and far-red nuclear fluorescence in multicellular images was performed by applying a preliminary segmentation of single nuclei from the blue (Hoechst) channel, according to the following algorithm:
- Blurring (Gaussian, 1 pixel standard deviation) of the blue channel image.
- Background subtraction routine of Fiji (rolling ball radius: 50 pixels) of the blue channel image.
- Thresholding [22], filling holes and watershed separation of the blue channel image to yield an 8-bit mask of single nuclei.
- Particle analysis routine of Fiji of the green and red channels by using the mask of single nuclei.
2.6. Image Scanning Microscopy Setup
For this work, we used a custom ISM setup, based on what previously described in [23] and coupled with an off-the-shelf Nikon Eclipse Ti microscope body. Two triggerable pulsed (pulse-width: <100 ps) diode lasers emitting at 488 nm and 642 nm afforded the excitation beams (QuixX 488-200 PS, QuixX 642-140 PS; Omicron-laserage Laserprodukte GmbH, Rodgau, Germany). The power of the excitation beams was controlled through the analog input provided by their drivers. The excitation laser beams were combined by a long/short-pass dichroic mirror (F43-491; AHF Analysentechnik, Tübingen, Germany) and reflected by a multi-band dichroic mirror (quad-line beamsplitter ZT 405-488-561-640; Chroma, Bellows Falls, VT, USA); they were then deflected by two galvanometric scanning mirrors (GVS002; Thorlabs, Newton, NJ, USA) and directed toward the objective lens (CFI Plan Apo VC 60x 1.49NA, oil; Nikon Instruments, Yokohama, Japan) by a scan lens (SL50-CLS2; Thorlabs), and the tube lens was included in the Nikon Ti microscope body. The emission was collected by the same objective lens, descanned, and filtered via the aforementioned multi-band dichroic mirror and finally through a multi-bandpass fluorescence filter (quad-line laser rejection band ZET 405-488-561-640; Chroma). A 500 mm aspheric lens (Thorlabs) focused the fluorescent light at the image plane (no-pinhole) into the SPAD array. This gave a final 600x magnification on the SPAD array plane, yielding a ∼1.1 AU size of the SPAD array projected on the specimen at the wavelength of far-red emission. Pixel size was set to about 40 nm and pixel dwell time was set to 0.2 ms, collecting 1024 × 1024 raster images. In detection, we used the PRISM-Light kit for Image Scanning Microscopy (Genoa Instruments Srl, Genova, Italy), which includes a 25-element SPAD detector array and FPGA-based acquisition electronics. Briefly, each photon detected by each element of the SPAD array generated a digital signal that was delivered to the FPGA-based data-acquisition unit through a dedicated channel. The entire custom microscope was controlled by Carma software (Genoa Instruments Srl, Genova, Italy), enabling the control and synchronization of all the microscope parts and devices used during image acquisition (i.e., the galvo mirrors, the laser sources and the SPAD detector array).
2.7. Image Scanning Microscopy Data Analysis
Data were saved in a standard binary format and then analyzed and processed using Python or Fiji. In particular, pixel reassignment and image deconvolution were implemented exactly as described in [23]. In short, the 25 independent images (i.e., the scanned images) were phase-correlated—with respect to the image of the array central element—to estimate the so-called shift vectors. The scanned images were registered using the shift-vectors and summed all together to obtain the final super-resolved image (adaptive pixel reassignment). Given the shift vectors, the emission and excitation wavelengths and the numerical aperture of the objective lens, it was possible to estimate the Gaussian point-spread functions of the system (one for each scanned images) and reconstruct the final super-resolved image also through a multi-image Richardson-Lucy deconvolution approach [23,24].
2.8. Determination of Image Resolution
The Fourier Ring Correlation (FRC) method was applied to determine the effective lateral (x, y) image spatial resolution [25]. In short, diffraction makes a microscope analogous to a short-pass filter with a fixed cutoff frequency. Only the sample’s frequencies below this (diffraction) limit are transmitted to the image, identifying the maximum resolution of the microscope. This actually holds for an ideal, i.e., noise-free microscope, as noise decreases the effective cutoff frequency of the system. The FRC methods determine this effective cutoff frequency by measuring the correlation degree of two statistically-independent images across a range of different spatial frequencies. Below the effective cutoff, the two images must be strongly correlated at the spatial level, which means that these frequencies contain structural information of the sample. At spatial frequencies above the effective cutoff, non-correlated noise dominates. FRC recovers the effective cutoff frequency by setting a threshold (usually 1/7 of the maximum value, which is obtained at zero frequency) to the correlation curve. For confocal measurements, two independent images were obtained as two sequential frames. For ISM images, we followed a different approach that did not require a specific set of sequential measurements. Remembering that every measurement results in a dataset of 25 scanned images, we split every dataset into two groups, respectively, with 13 and 12 images (belonging to the even and odd detectors of the array). Then, we independently calculated the adaptive pixel reassignment image using the partial images of the two groups, obtaining two independent reconstructed images, which were used to calculate the FRC. It is important to highlight that the final super-resolved image can always be retrieved by summing these two “partial” reconstructions. In both confocal and ISM acquisition, care was taken to sample at pixel size fulfilling the Nyquist’s criterium (50 nm for confocal, 40 nm for ISM).
2.9. Colocalization Analysis
Pearson’s colocalization (R) of the green and far-red images of HCC827 was quantified after background subtraction (50 pixel rolling ball) for both channels, according to the colocalization method by Costes et al. [26]. The quantitative colocalization analysis was performed by the JACoP localization plugin [27]. Of note, before the analysis, the two images were equally cropped to isolate the minimum rectangular area containing the nucleus of interest and avoid spurious colocalization signals from the surrounding cytoplasmic regions.
2.10. Statistical Data Analysis and Fitting
Statistical analyses of data and fitting were carried out by using GraphPad Prism 7 (GraphPad, San Diego, CA, USA, 2016).
3. Results and Discussion
3.1. Immunolabeling and Confocal Imaging of PcG Proteins and H3K27me3
EZH2 and its partners were immunolabeled by an indirect immunocytochemistry approach which minimizes spectral cross-talk, as it leverages green-emitting (Alexa488) and far-red-emitting (Abberior StarRed or Alexa647) dyes. Chromatin was labeled by blue-emitting Hoechst 33342. At first, we imaged the cells by confocal microscopy. The fluorescence of EZH2, RING1b, BMI1, and H3K27me3 was observable predominantly in cell nuclei, consistently with the nuclear localization of these proteins (Figure 1). There, the classical organization in submicron foci (“polycomb bodies”) was visible for polycombs EZH2, RING1b and BMI1. Of note, also, H3K27me3 assembles in sub-micron nuclear structures that correspond to chromatin domains enriched in H3 methylation. This pattern is analogous to the other epigenetic mark of PcGs, H2AK119ub [17].
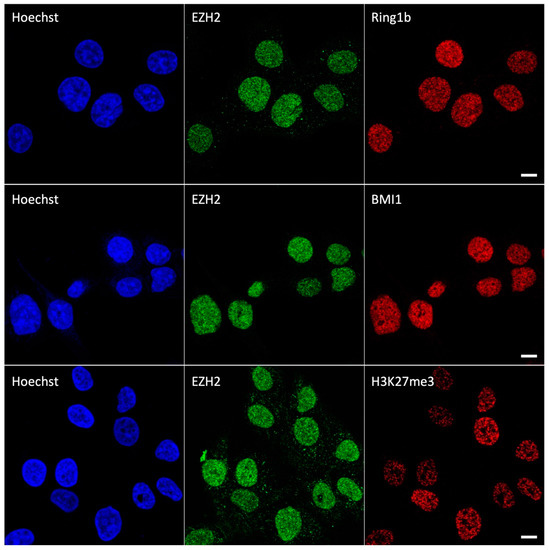
Figure 1.
Confocal images of HCC827 nuclei immunolabeled by EZH2/RING1b (upper row), EZH2/BMI1 (central row), and EZH2/H3K27me3 (lower row). Scale bar: 10 µm.
3.2. Spatial Resolution Improvement by Image Scanning Microscopy
The optical sectioning ability of a confocal microscope is mainly due to the pinhole spatial filter placed in a conjugate image plane in front of the detector along the fluorescence detection path. Our preliminary measurements (Figure 1) were obtained by setting the pinhole aperture of the confocal system at 1 Airy unit (AU), as commonly performed. In these conditions, the radial (i.e., on the object xy plane) resolution dxy was 240 nm for the green channel and dxy = 310 nm for the far-red channel, as judged by the Fourier Ring Correlation (FRC) method [25]. Ideally, by closing the pinhole below 0.2 AU, dxy would be improved by a factor compared to 1 AU at the price of a dramatic decrease in signal reaching the detector (95%) and severe degradation of the signal to noise ratio (SNR) of the final image. Image Scanning Microscopy (ISM) overcomes this limitation by replacing the single point detector of a confocal microscope with a detector array [28]. Each element of the array generates a “confocal” image of the sample, differing in information content as they map different spatial regions of the point spread function generated by the light coming from the objective focus.
The most transparent way to form an ISM image is represented by the so-called pixel-reassignment (PR) method. To generate the PR-ISM image, all the scanned images are summed together after shifting each image by a vector (shift-vector) proportional to the relative position of the corresponding detector element and properly scaled by a factor. We implemented an adaptive version of the PR method (APR) in which the shift vectors are straightforwardly obtained from the data, and there is no need for calibrations or prior information. APR is potentially able to correct misalignment of the system or aberrations introduced by the sample. To extend the resolution further, we also developed alternative approaches fusing together the scanned images by more advanced methods considering also prior knowledge (e.g., the point-spread-functions) of the system and based on inverse filtering or multi-image deconvolution (mode ISM++). ISM can be considered the laser-scanning microscopy alternative to wide-field structured illumination microscopy (SIM). In SIM, the sample is illuminated with a series of grating-like light patterns, which encode into the resulting image’s normally inaccessible high-frequency information. It is worth noting that the single-photon timing ability of the SPAD array allows the combination of ISM with FLIM, thereby enabling super-resolution FLIM: fluorescence lifetime image-scanning microscopy (FLISM).
In our study, we used a custom ISM setup whose core is a single-photon avalanche diode (SPAD) array composed of 25 detection elements (organized in a 5 × 5 square area configuration) and FPGA-based acquisition electronics. PR-ISM images of PcG proteins and H3K27me3 appeared qualitatively endowed with higher resolution as compared to their confocal counterparts (Figure 2). Accordingly, FRC analysis showed dxy = 145 nm (green channel) and dxy = 210 nm (far-red channel), indicating a resolution improvement of 1.5–1.7× over the confocal acquisition. A further significant increase in resolution was obtained by applying the Lucy–Richardson deconvolution to PR-ISM images (Figure 2): ISM++ images were characterized by dxy = 95 nm (green channel) and dxy = 160 nm (far-red channel). These data indicate that ISM may conveniently fit the spatial mesoscale of polycomb domains without the need of special fluorescent labels, sample preparation, or excitation patterns typical of more challenging SRM techniques (e.g., STORM and STED).
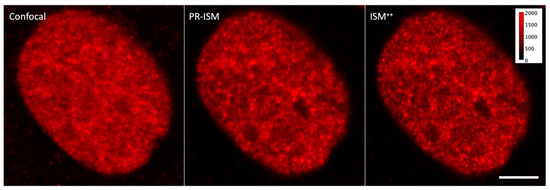
Figure 2.
Confocal, PR-ISM, and ISM++ images of an HCC827 nuclei immunolabeled by RING1b. All images were rescaled to the same digital level interval (0–2000) for better comparison. Scale bar: 10 µm.
3.3. Colocalization by ISM
Colocalization studies were carried out in PR-ISM and ISM++ modes targeting single nuclei. Notably, color-merged images already posited the existence of actual colocalization between EZH2 and all its partners (Figure 3).
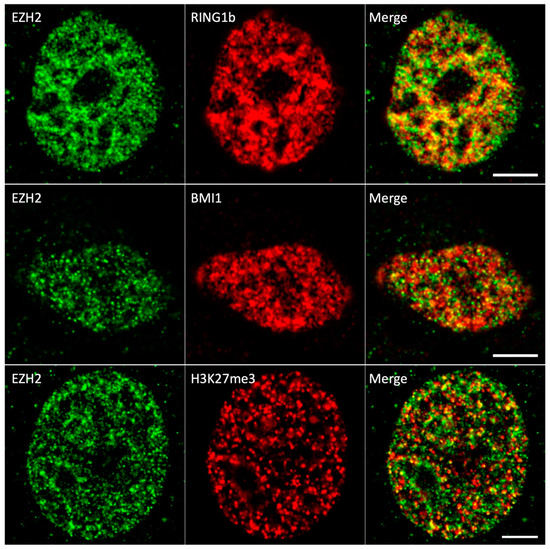
Figure 3.
PR-ISM images of HCC827 nuclei immunolabeled by EZH2/RING1b (upper row), EZH2/BMI1 (central row), and EZH2/H3K27me3 (lower row). For all images, the background was subtracted by a rolling ball algorithm. Scale bar: 10 µm.
This colocalization was quantified by the Pearson coefficient R, which measures the correlation between the intensities of the two fluorescent partners in each pixel as a proxy of their functional association. As complexes are composed by an integer number of associated subunits, we shall refer to R as a measure of the strength of “stoichiometric relationship (or correlation)” between the two partners. Ideally, two partners that engage always in the same complex have a perfect stoichiometric relationship; i.e., their amounts are always found in a fixed ratio and R = 1. Incomplete engagement in the complex (for instance due to the presence of isoforms) leads to R < 1.
R was calculated by applying the thresholding method of Costes [26], which is known to remove the removes the bias of visual interpretation, and it was moderately positive for all protein couples (Figure 4). The statistical significance (p > 0.05) of these values with respect to the random spatial overlapping of the two signals was assessed by the Costes’ statistical test on colocalization [18]. Notably, ISM++ super-resolved images were characterized by slightly lower R values than PR-ISM. This could be attributed to the colocalization overestimation effect due to the lower resolution of PR-ISM [29]. Alternatively, ISM++ could underestimate colocalization owing to a working region’s size of the protein complexes over chromatin, to which the spacing effect of antibody labels (20–30 nm) must be added, exceeding the resolution of the technique (~100 nm).
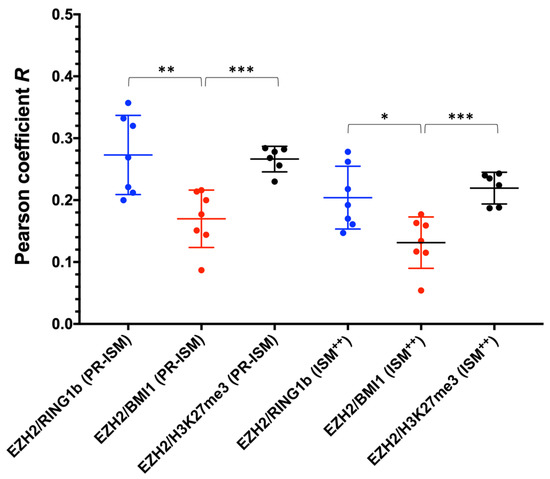
Figure 4.
Pearson colocalization coefficients (R) of EZH2/RING1b (blue), EZH2/BMI1 (red), and EZH2/H3K27Me3(black) in HCC827 cells as determined by ISM. EZH2/RING1b: R = 0.27 ± 0.02 (PR-ISM), R = 0.20 ± 0.02 (ISM++); EZH2/BMI1: R = 0.17 ± 0.02 (PR-ISM), R = 0.13 ± 0.02 (ISM++); EZH2/H3K27me3: R = 0.27 ± 0.01 (PR-ISM), R = 0.22 ± 0.01 (ISM++); values expressed as average ± SEM, * p < 0.05, ** p < 0.01, *** p < 0.001.
Overall, these data support a model where PRC1 and PRC2 have a moderate stoichiometric relationship at mesoscale, i.e., suggesting that the activity of PRC1 and PRC2 is partially concomitant in space and time. The slightly higher colocalization values found for EZH2/RING1b over EZH2/BMI1 (Figure 4) might suggest a stronger stoichiometric correlation (and spatio/temporal overlap) between vPRC1 and PRC2 as compared to cPRC1. Interestingly, a recent elegant study addressing PRC1 dynamics by single-particle tracking has revealed that only a minor (20%) fraction of PRC1 is stably bound to chromatin [30]. This highly dynamic nature of PRC1 could be at the basis of the observed incomplete functional colocalization between PRC1 and PRC2.
3.4. Single-cell Analysis of EZH2/H3K27me3 Correlation
The moderately positive EZH2/H3K27me3 colocalization prompted us to verify whether a stoichiometric correlation between the methyltransferase enzyme and its product would be visible at the whole nuclear level, taking advantage of the intrinsically heterogeneous protein expression across different cells. Accordingly, we measured the fluorescence given by both proteins for a large number of nuclei (n = 186). Figure 5 shows the H3K27me3 vs. EZH2 plot. Qualitatively, the trend of H3K27me3 with increasing EZH2 expression posited a positive stoichiometric correlation between the two partners, as quantitatively demonstrated by the Spearman non-parametric correlation test (r = 0.435, p < 0.0001). In addition to a minor effect due to the immunolabeling procedure quantified in ref. [19], the presence of scattered values that reduce the monotonicity of the H3K27me3 vs. EZH2 reflects the cell heterogeneity with respect to the extent of methylated H3 per EZH2 molecule. Albeit some genomic heterogeneity of the cell population could be envisaged (due, for instance, to a variable number of chromosomes), a realistic contribution to the observed dispersion should be attributed to the non-synchronous cell cycle within the cell population. Indeed, a perfect stoichiometric correlation between the concentrations of an enzyme and its product in an ensemble of independent units occurs only if they are synchronized in time, owing to the monotonic dependence of the catalyzed reaction rate from the enzyme concentration.
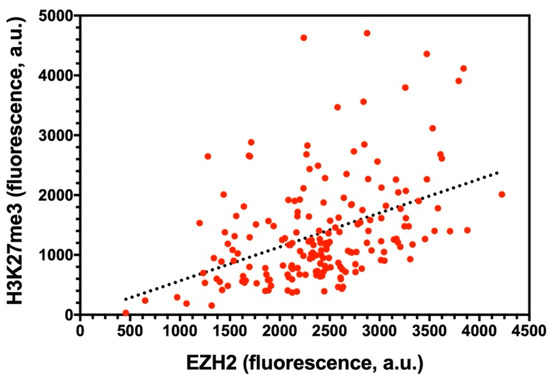
Figure 5.
H3K27me3 vs. EZH2 nuclear fluorescence plot. Each red circle stands for a single nucleus. The black line represents the linear fitting of data and was drawn only to highlight the presence of a stoichiometric correlation between the two partners.
Overall, our findings highlight a partial spatiotemporal overlap between EZH2 and its enzymatic product H3K27me3 at target genomic regions, whereas the functional correlation at the whole nuclear level is well established. This may hint to a mechanism where some PRC2 complexes remain stably bound to chromatin after deposition of the H3K27me3 mark, whereas another pool of PRC2 acts more according to a hit-and-run methylation mechanism, similarly to what was observed for PRC1 [17,30].
5. Conclusions
A crucial hallmark of cell life is the widespread ordering of many biological processes in nano/mesoscopic domains, which now may be revealed by an imaging toolbox referred to as super-resolution microscopy (SRM). Recently, this approach has been applied to visualize chromatin topology. In this context, polycomb group proteins stand as major modulators of chromatin function, mostly by silencing genes by a combination of chemical modifications and physical interactions, which has been recently recapitulated in a dynamic working model by gathering results from several approaches, including SRM. Nonetheless, some aspects of the PcG gene-silencing mechanism are still obscure, including the possible spatiotemporal co-presence of the PcG complexes and their histone marks. Here, we present a functional colocalization study at 100–140 nm resolution targeting the PcG complexes PRC1 and PRC2 as well as the histone mark H3K27me3 by Image Scanning Microscopy (ISM). ISM represents a more flexible SRM technique as compared to STORM and STED microscopy, and its resolution fits the spatial scale of PRC assembly. It is worth noting that ISM allows using the well-established sample labeling protocols adopted for conventional confocal and wide-field imaging. Our data suggest that a fraction of EZH2 is stoichiometrically linked to BMI1, RING1b, and H3K27me3 at mesoscale, suggesting that PRC1 and PRC2 are partially concomitant in shaping the chromatin topology at the PcG foci.
Author Contributions
Conceptualization: I.N., S.P., P.B., A.D., G.V., M.C., B.S. and R.B.; methodology, formal analysis and data curation: I.N., S.P., M.C., B.S. and R.B.; experimental investigation: I.N., S.P., M.C., M.R., S.C., E.B., T.S., B.S. and R.B.; manuscript draft preparation: all authors; manuscript review and editing, supervision of research: M.C., B.S. and R.B. All authors have read and agreed to the published version of the manuscript.
Funding
R.B. acknowledges support from the University of Pisa under the framework of project PRA_2020_77.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data contained within the article are available on request from the authors.
Acknowledgments
The authors gratefully acknowledge Vittoria Carnicelli and Michele Oneto for precious technical assistance. We thank the center of instrument sharing of the University of Pisa (CISUP) for the access to the confocal-Airyscan laboratory facility.
Conflicts of Interest
S.P., M.C., P.B., G.V., and A.D. have personal financial interest (co-founders) in Genoa Instruments. The remaining authors declare no competing interests. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Bonev, B.; Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016, 17, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Szabo, Q.; Donjon, A.; Jerkovic, I.; Papadopoulos, G.L.; Cheutin, T.; Bonev, B.; Nora, E.P.; Bruneau, B.G.; Bantignies, F.; Cavalli, G. Regulation of single-cell genome organization into TADs and chromatin nanodomains. Nat. Genet. 2020, 52, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Klose, R.J. The molecular principles of gene regulation by Polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 815–833. [Google Scholar] [CrossRef] [PubMed]
- Giner-Laguarda, N.; Vidal, M. Functions of Polycomb Proteins on Active Targets. Epigenomes 2020, 4, 17. [Google Scholar] [CrossRef]
- Loubiere, V.; Martinez, A.M.; Cavalli, G. Cell Fate and Developmental Regulation Dynamics by Polycomb Proteins and 3D Genome Architecture. BioEssays News Rev. Mol. Cell Dev. Biol. 2019, 41, 15. [Google Scholar] [CrossRef]
- Yong, K.J.; Basseres, D.S.; Welner, R.S.; Zhang, W.C.; Yang, H.; Yan, B.; Alberich-Jorda, M.; Zhang, J.Y.; de Figueiredo-Pontes, L.L.; Battelli, C.; et al. Targeted BMI1 inhibition impairs tumor growth in lung adenocarcinomas with low CEBP alpha expression. Sci. Transl. Med. 2016, 8, 350ra104. [Google Scholar] [CrossRef]
- Maroni, G.; Bassal, M.A.; Krishnan, I.; Fhu, C.W.; Savova, V.; Zilionis, R.; Maymi, V.A.; Pandell, N.; Csizmadia, E.; Zhang, J.Y.; et al. Identification of a targetable KRAS-mutant epithelial population in non-small cell lung cancer. Commun. Biol. 2021, 4, 370. [Google Scholar] [CrossRef]
- Cao, L.X.; Bombard, J.; Cintron, K.; Sheedy, J.; Weetall, M.L.; Davis, T.W. BMI1 As a Novel Target for Drug Discovery in Cancer. J. Cell. Biochem. 2011, 112, 2729–2741. [Google Scholar] [CrossRef]
- Rose, N.R.; King, H.W.; Blckledge, N.P.; Fursova, N.A.; Ember, K.J.I.; Fischer, R.; Kessler, B.M.; Klose, R.J. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. Elife 2016, 5, e18591. [Google Scholar] [CrossRef]
- Pachano, T.; Crispatzu, G.; Rada-Iglesias, A. Polycomb proteins as organizers of 3D genome architecture in embryonic stem cells. Brief. Funct. Genom. 2019, 18, 358–366. [Google Scholar] [CrossRef]
- Gil, J.; O’Loghlen, A. PRC1 complex diversity: Where is it taking us? Trends Cell Biol. 2014, 24, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.H.; Veenstra, G.J.C. The Role of Polycomb Proteins in Cell Lineage Commitment and Embryonic Development. Epigenomes 2022, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Illingworth, R.S. Chromatin folding and nuclear architecture: PRC1 function in 3D. Curr. Opin. Genet. Dev. 2019, 55, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Boettiger, A.N.; Bintu, B.; Moffitt, J.R.; Wang, S.Y.; Beliveau, B.J.; Fudenberg, G.; Imakaev, M.; Mirny, L.A.; Wu, C.T.; Zhuang, X.W. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016, 529, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Wani, A.H.; Boettiger, A.N.; Schorderet, P.; Ergun, A.; Munger, C.; Sadreyev, R.I.; Zhuang, X.; Kingston, R.E.; Francis, N.J. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat. Commmun. 2016, 7, 10291. [Google Scholar] [CrossRef]
- Boettiger, A.; Murphy, S. Advances in Chromatin Imaging at Kilobase-Scale Resolution. Trends Genet. 2020, 36, 273–287. [Google Scholar] [CrossRef]
- Storti, B.; Civita, S.; Faraci, P.; Maroni, G.; Krishnan, I.; Levantini, E.; Bizzarri, R. Fluorescence imaging of biochemical relationship between ubiquitinated histone 2A and Polycomb complex protein BMI1. Biophys. Chem. 2019, 253, 12. [Google Scholar] [CrossRef]
- Adler, J.; Parmryd, I. Quantifying Colocalization by Correlation: The Pearson Correlation Coefficient is Superior to the Mander’s Overlap Coefficient. Cytom. Part A 2010, 77A, 733–742. [Google Scholar] [CrossRef]
- Storti, B.; Quaranta, P.; Di Primio, C.; Clementi, N.; Mancini, N.; Criscuolo, E.; Spezia, P.G.; Carnicelli, V.; Lottini, G.; Paolini, E.; et al. A spatial multi-scale fluorescence microscopy toolbox discloses entry checkpoints of SARS-CoV-2 variants in Vero E6 cells. Comput. Struct. Biotechnol. J. 2021, 19, 6140–6156. [Google Scholar] [CrossRef]
- Choi, K.; Creighton, C.J.; Stivers, D.; Fujimoto, N.; Kurie, J.M. Transcriptional Profiling of Non-Small Cell Lung Cancer Cells with Activating EGFR Somatic Mutations. PLoS ONE 2007, 2, e1226. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.K.; Wang, M.J.J. Image thresholding by minimizing the measures of fuzziness. Pattern Recognit. 1995, 28, 41–51. [Google Scholar] [CrossRef]
- Castello, M.; Tortarolo, G.; Buttafava, M.; Deguchi, T.; Villa, F.; Koho, S.; Pesce, L.; Oneto, M.; Pelicci, S.; Lanzano, L.; et al. A robust and versatile platform for image scanning microscopy enabling super-resolution FLIM. Nat. Methods 2019, 16, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Zunino, A.; Castello, M.; Vicidomini, G. Reconstructing the Image Scanning Microscopy Dataset: An Inverse Problem. arXiv 2022, arXiv:2211.12510. [Google Scholar]
- Tortarolo, G.; Castello, M.; Diaspro, A.; Koho, S.; Vicidomini, G. Evaluating image resolution in stimulated emission depletion microscopy. Optica 2018, 5, 32–35. [Google Scholar] [CrossRef]
- Costes, S.V.; Daelemans, D.; Cho, E.H.; Dobbin, Z.; Pavlakis, G.; Lockett, S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004, 86, 3993–4003. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelieres, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- Sheppard, C.J.R. Super-Resolution in Confocal Imaging. Optik 1988, 80, 53–54. [Google Scholar]
- Xu, L.; Ronnlund, D.; Aspenstrom, P.; Braun, L.J.; Gad, A.K.B.; Widengren, J. Resolution, target density and labeling effects in colocalization studies—Suppression of false positives by nanoscopy and modified algorithms. Febs. J. 2016, 283, 882–898. [Google Scholar] [CrossRef]
- Huseyin, M.K.; Klose, R.J. Live-cell single particle tracking of PRC1 reveals a highly dynamic system with low target site occupancy. Nat. Commmun. 2021, 12, 887. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).