Abstract
The major challenges for medical image segmentation tasks are complex backgrounds and fuzzy boundaries. In order to reduce their negative impacts on medical image segmentation tasks, we propose an enhanced feature extraction network (EFEN), which is based on U-Net. Our network is designed with the structure of feature re-extraction to strengthen the feature extraction ability. In the process of decoding, we use improved skip-connection, which includes positional encoding and a cross-attention mechanism. By embedding positional information, absolute information and relative information between organs can be captured. Meanwhile, useful information will be strengthened and useless information will be weakened by using the cross-attention mechanism. Our network can finely identify the features of each skip-connection and cause the features in the process of decoding to have less noise in order to reduce the effect of fuzzy object boundaries in medical images. Experiments on the CVC-ClinicDB, the task1 from ISIC-2018, and the 2018 Data Science Bowl challenge dataset demonstrate that EFEN outperforms U-Net and some recent networks. For example, our method obtains 5.23% and 2.46% DSC improvements compared to U-Net on CVC-ClinicDB and ISIC-2018, respectively. Compared with recent works, such as DoubleU-Net, we obtain 0.65% and 0.3% DSC improvements on CVC-ClinicDB and ISIC-2018, respectively.
1. Introduction
Medical image segmentation is an important task in medical image processing and analysis. It has great application and research value in medical research [1], clinical diagnosis, pathological analysis, surgical planning, computer-assisted surgery [2], and so on. The purpose of the medical image segmentation task is to extract and segment special features, such as lesions, and to provide a reliable basis for clinical diagnosis and pathological research. The main challenges of medical image segmentation are as follows: first, it is very difficult to construct a database of medical images, because the medical images themselves are extremely unbalanced, with many normal samples and few and variable lesion samples, which leads to insufficient well-labeled training samples [3]. There are still some problems, such as limited image quality, an absence of universally adopted segmentation protocols, and significant inter-patient variations in image characteristics [4]. In addition, medical images generally have a lot of noise and artifacts in imaging. The quantification of segmentation accuracy and uncertainty is critical for estimating its performance in other applications [5]. There are many types of medical images, including computed tomography, X-ray, magnetic resonance imaging, and positron emission computed tomography images. Medical image segmentation mainly includes methods based on thresholds, regions, deformation models, and fuzzy and neural networks.
In recent years, deep learning technology has played an important role in the field of computer vision, and it is rapidly being applied to other fields, especially in the field of medicine. FCN [6] stands out as one of the initial deep learning architectures specifically designed for end-to-end training, which enables pixel-wise predictions in semantic segmentation tasks. U-Net [7] is a network with an end-to-end encoder–decoder structure, and it overcomes the shortcoming that FCN cannot keep some pixel spatial position information and context information, which leads to the loss of local features and global features. Liu et al. [8] proposed that the U-shaped end-to-end network is the best medical image segmentation architecture. The U-Net structure includes two stages: the first stage is the down-sampling process, which is mainly responsible for feature extraction, and the second stage is the up-sampling process based on the first stage, which is responsible for outputting the feature map with the same size as the original sample. Because the features obtained in the first stage comprise highly compressed feature information, and the task of image segmentation needs to classify each pixel in the image, it cannot completely rely on up-sampling features, and it needs to be supplemented with the information in the down-sampling process. The skip-connection operation is used in U-Net. On the basis of U-Net, many people have improved this network. For example, Zhou et al. [9] proposed U-Net++. This network changes the original U-Net layers, redesigns the skip-connection in the original network, and adds depth supervision to different segmentation tasks to achieve a good segmentation result. Because of the excellent performance of U-Net in medical image segmentation tasks, this framework is widely used for lesion segmentation in organs, such as the eyes, heart, liver, brain, skin, prostate, and breast. Li et al. [10] proposed ANU-Net for the organ cancer segmentation task. The network has achieved good segmentation results on the LiTS (liver tumor segmentation) dataset and the CHAOS (combined healthy abdominal organ segmentation) dataset with dense skip-connection and an attention mechanism. Zhang et al. [11] proposed DIU-Net, which integrates the inception module and the dense skip-connection module into U-Net to increase its width and depth. This network is used to segment the brain tumors of MRI images and computed tomography images of lungs and retinal blood vessels, and it achieves good results. Jha et al. [12] proposed DoubleU-Net, which is a combination of two U-Net structures. The first U-Net uses pre-trained VGG-19 as the encoding network, and the second U-Net uses ASPP to capture context information. It achieves better segmentation results than U-Net in colonoscopy, dermatoscope, and microscope images.
In addition, due to the good performance of U-Net in the field of medical image analysis, many scholars combine it with other models to further improve its performance. Li Jianfei et al. [13] proposed an image fusion algorithm based on dual-tree complex wavelet transform (DTCWT) and frequency domain U-Net, which improves the accuracy of tumor segmentation. Zhang Tianqi et al. [14] combined the local difference method with U-Net. Yang et al. [15] proposed a method for combining level set and U-Net. Zhang et al. [16] presented a combination of random walk and U-Net. Liu et al. [17] proposed a combination of the graph partition method and U-Net. Man et al. [18] proposed a combination of deep reinforcement learning and U-Net.
Semantic segmentation tasks and medical image segmentation tasks are used to classify each pixel in the image. All segmentation tasks face two problems. The first problem is how to improve the feature extraction ability of the network. If the feature extraction ability is improved by increasing the depth of the network blindly, a lot of detailed information will be lost, and detailed information is extremely important for image segmentation. Second, most methods based on U-Net using skip-connection add the features extracted by the encoder to the output of the corresponding layer of the decoder indiscriminately, and this will inevitably introduce noise information, thus interfering with the subsequent segmentation results. Almost all network improvements are essentially designed to address these two problems. Promoting the feature extraction ability of the network and compensating for lost information to the greatest extent is very important for the segmentation of medical images. The main contributions of this paper are:
- Based on U-Net, we propose an improved network EFEN, which further enhances the feature extraction ability of the network by adding feature extraction processes.
- In this paper, the skip-connection is improved. Each skip-connection uses positional encoding and a cross-attention mechanism. By embedding positional information, absolute information and relative information between organs can be captured. Each skip-connection uses a cross-attention mechanism to select information so that the network can automatically give useful features a larger weight while suppressing useless information. Then, the target boundaries or small targets can be better segmented.
- For medical image segmentation tasks, the proportion of target pixels in the whole image may be much less than that of background pixels, thus causing a class imbalance problem. In this paper, we adopt binary cross-entropy and dice loss to jointly optimize the training learning.
2. Related Works
Ronneberger et al. [7] proposed a network that is called U-Net because its shape is a “U.” It is an encoder–decoder symmetric network that is composed of convolution, down-sampling, up-sampling, and skip-connection. Down-sampling is the process of feature extraction, and it will inevitably lead to the high compression of features and information loss. Up-sampling is the process of restoring a feature map to the same size as the input image for subsequent classification of each pixel. Up-sampling cannot fully restore the original image details. This is because it is performed on the output of down-sampling, and down-sampling is an irreversible process. In order to reduce the training cost, the encoder of VGG-19 is often used as the encoder of U-Net, so that the pre-trained encoder parameters can be used. VGG-19 was originally designed for image classification, so its structure is not symmetrical, which is different from U-Net. However, both image classification and image segmentation require feature extract processing, and the trained encoder parameters of VGG-19 are often used to initialize the encoder parameters of U-Net. Skip-connection is an important application in U-Net that alleviates the problem of gradient disappearance. It can compensate for the information loss in the process of feature extraction to a certain extent. However, this skip-connection will add information from the encoder to the features obtained by the decoder indiscriminately, which will introduce noise. This is very unfavorable for image segmentation.
The end-to-end network with a “U” shape performs best in medical image segmentation tasks, and many scholars have made improvements to it, such as Y-Net [19], Ψ-Net [20], and multi-path dense U-Net [21]. All three of these networks increase the number of encoders for different tasks to promote the feature extraction ability. Y-Net is composed of two encoders and one decoder to extract more features. Ψ-Net uses three encoders in the encoding stage to further improve the feature extraction ability, but the three encoders have to process three slices, respectively. At the same time, the self-attention block and the context attention block are used in the encoding stage and the decoding stage, respectively. However, the relationship between encoders in these networks is not explored, and there is a lack of constraints between encoders, which may increase the number of parameters in the network but improve its limited performance. Multi-path dense U-Net is a multi-modal segmentation model proposed by Dolz et al. Aiming to analyze images of ischemic stroke, its multiple input images include diffusion-weighted imaging (DWI), cerebral blood volume (CBV), CT perfusion imaging (CTP), and mean transit time (MTT). This network mainly alleviates the effect of gradient disappearance and over-fitting. However, the model is limited to specific tasks and needs additional multimodal data, which is unfavorable in the case of scarce medical data.
Xia and Kulis et al. [22] proposed W-Net for image segmentation tasks. Xu et al. [23] and Das et al. [24] proposed DW-Net and WRC-Net, respectively. DW-Net adds dilated convolution on the basis of W-net, which can increase the ability to obtain multiscale context information. The first U-Net of the WRC-Net is employed for boundary prediction, while the second U-Net is utilized to generate the image segmentation result. However, it is important to note that these two U-Nets operate independently of each other. Tang et al. [25] proposed CU-Net (Coupled U-Net), which is the combination of dense U-Net and stacked U-Net, and it improves the efficiency of stacked U-Net. Kang et al. [26] proposed CMU-Net (cascading modular U-Nets). In this method, pre-trained U-Nets are modularized and cascaded to binarize images, which solves the problem of too few samples. In addition, many scholars combine U-Net in parallel, assigning distinct functions to each U-Net. For example, Zhao et al. [27] proposed triple U-Net for nuclear segmentation in pathological cancer. Lee et al. [28] proposed Multi-scale U-Net (Mu-Net), which incorporates multiple U-Nets operating at different scales in parallel and each U-Net handles images at different scales. All of the aforementioned approaches enhance U-Net. However, they all ignore the impact of the relationship between network structures on specific tasks.
In this paper, a cross-attention mechanism is used to select the features of skip-connection. SeNet [29] compresses each feature map in different channels into a single value to induct the network to learn a set of weights that reflect the importance of each feature map in their channel. The authors proposing GSoP-Net [30] and FcaNet [31] argued that using only global average pooling is insufficient, as it limits the modeling ability of the attention mechanism. GSoP-Net improves the squeeze module in SeNet and a global second-order pooling (GSOP) block is proposed to model high-order statistics while collecting global information. FcaNet reconsiders the global information captured from the perspective of compression, and analyzes the global average pooling in the frequency domain. The authors of these studies proved that the global average pooling is a special case of a discrete cosine transform. Building upon this insight, they proposed a novel multi-spectral channel attention. SRM (Style-based Recycling Module) [32] improves the squeeze module and excitement module by incorporating the mean and standard deviation of input features, thereby enhancing the network’s capability to capture global information. It also uses a lightweight channel full connection layer (CFC) instead of the original full connection layer (FC) to reduce the computing requirements. GCT [33] comprises a general transformation unit for visual recognition tasks, which uses interpretable variables to visualize and model channel correlation. These variables determine the competitive or cooperative relationship between neurons and can be jointly trained with network parameters. ECANet [34] replaces the fully connection layer in SENet with one-dimensional convolution, which can replace the SE block well without adding additional parameters, and can also exceed the original performance of SE. Inspired by SENet, EncNet [35] contains a context encoding module, which is combined with the semantic encoding loss to model the relationship between the scene context and the probability of object categories, and then uses the global scene context information for semantic segmentation. There are many ways to combine an attention mechanism with U-Net. For example, Jin et al. [36] proposed RA-UNet for segmenting CT images of liver tumors. This model obtains multi-scale attention information through the network to fuse shallow and deep features. In addition, Ding et al. [37] proposed category attention boosting U-Net (CAB U-Net). Hariyani et al. [38] proposed dual attention CapNet (DA-CapNet). In this paper, we use a cross-attention mechanism to filter out the noise from the encoder information as much as possible. This mechanism aims to enhance the network’s ability to accurately identify the category of each pixel. Compared with the above methods, the cross-attention mechanism has a better ability to consider global information. Therefore, it can reduce the negative impact of complex backgrounds, fuzzy boundaries and small objects in medical image segmentation tasks.
To sum up, deep CNN algorithms are widely used in the field of medical image segmentation tasks and have made great achievements. However, artificial intelligence in medicine image analysis is still a new field. The main challenges in the medical image analysis field are the lack of datasets and the imbalance of datasets. In this field, accurate auxiliary diagnosis is often crucial, as a wrong or inaccurate diagnosis can lead to delays in treatment or even exacerbate the illness. This paper presents an enhanced network derived from U-Net for medical image segmentation tasks. The network takes into account the influence of network structures, resulting in improved feature extraction capabilities compared to traditional U-Net. Furthermore, it fully considers the impact of valuable information within the shallow network on segmentation results.
3. Proposed Method
In this section, we will present the details of the EFEN proposed in our work and explain the role of each part. In this paper, two key issues in the tasks of medical image segmentation are considered when designing the network: first, whether the feature extraction capability of the network can be improved, and second, whether the highly extracted features can be compensated by useful information, and whether the compensation information can avoid noise or useless details as much as possible. Since the U-Net performs best in the medical image segmentation tasks, we design the network on the basis of U-Net. Figure 1 shows an overview of the designed network architecture on the basis of U-Net. In Figure 1, U-Net is used as the basic network for the feature extraction of the first stage, and during up sampling the improved skip-connection with a cross-attention mechanism is used to supervise the information from the encoder. After obtaining the outputs of the base U-Net, we merge them with the inputs of the feature re-extraction network to further extract features. In the up-sampling process of the feature re-extraction network, we also use the cross-attention mechanism to supervise the information of the skip-connection. It is worth noting that the information of the skip-connection only comes from the corresponding layer features which are extracted in the down-sampling process of U-Net. In this way, not only can the ability of the network to extract features be improved, but also more accurate boundary features can be obtained. Finally, because we perform medical image segmentation, in order to further enhance the feature extraction ability, we adopt binary cross-entropy and dice loss to jointly optimize the training learning. Section 3.1 introduces the structure of proposed EFEN. Section 3.2 introduces the improved skip-connection. Section 3.3 introduces the loss function.
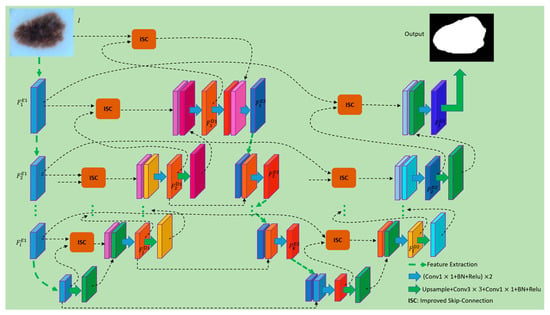
Figure 1.
Structure of the EFEN. The feature maps of different colors represent the results obtained via different methods of processing.
3.1. Structure of EFEN
Compared with traditional medical image analysis methods, using fully convolutional networks can achieve better performance. Fully convolutional networks are more accurate and robust in tasks such as cardiac MR [39], brain tumors [40] and abdominal CT [41,42] image segmentation. Among them, the fully convolutional network U-Net has been proved to be the best performing medical image segmentation architecture. The network proposed in this paper improves the structure on the basis of U-Net. The encoder of VGG-19 is used as a sub-network of our encoder, and a symmetrical decoder is designed. The decoding part of U-Net adopts an improved skip-connection for feature compensation, which can supervise the compensation information and reduce noise or interference effectively. Given an input image , is mapped to a set of feature maps . The process can be formulated as follows:
where is the stage in the processes of feature extraction and refers to the first encoding process. , where is the channel number of the stage during encoding. and are the number of pixels along the height and width of the feature map, respectively. When , is the input image . refers to the weights of the convolutional network during encoding. refers to the operations of mapping feature maps to during the encoding process, which usually includes convolution, pooling, and ReLu, etc. The decoding process begins when we obtain the features from the deepest layer, and it can be formulated as follows:
where is the stage in the process of decoding and refers to the first decoding process. refers to the feature map in the stage during decoding process. refers to the operation of transposed convolution. is the operation of improved skip-connection and is described in detail in Section 3.2. includes two layers of convolution and one layer of convolution.
First, we discard the last convolution layer of the U-Net decoder, and then fuse its outputs with the inputs of the encoder using improved skip-connection. The fused features have both semantic information and shallow information. After that, we continue to build a feature extraction network to enhance the feature extraction capability, and features in each stage in the second feature extraction process are fused with the corresponding layer features which are extracted in the down-sampling process of U-Net. This process can be formulated as follows:
where is the stage in the process of feature extraction, refers to the feature re-extraction process, and . refers to the operations of mapping feature maps using the weights of , which usually include convolution, pooling, and ReLu, etc. The size of is the same as the outputs of . is the fusion method which includes the operations of two layers of convolution and one layer of convolution. The inputs of consist of two parts: one part comes from the features obtained at each encoding stage in the feature re-extraction process, and the other part comes from the features of each corresponding stage in U-Net decoder. It should be noted that the features in each stage of the U-Net decoding part includes the information of the corresponding stage in the encoder because improved skip-connection is used. Therefore, the process of will further improve the feature extraction capability of the entire network.
Like the decoding part of U-Net, we perform up-sampling to complete the final per-pixel classification task. We describe this process as follows:
where is the stage in the process of decoding and refers to the second decoding process. refers to the operation of transposed convolution in the second decoding process. The outputs of can be the inputs of and can also be used to generate the weights for the cross-attention mechanism. is the operation of improved skip-connection and it is detailed in Section 3.2. includes two layers of convolution and one layer of convolution.
3.2. Improved Skip-Connection
In the encoding process of U-Net, higher dimensional features are extracted by processing local information layer by layer. In general, the process of feature extraction requires filters to convolve the inputs of each layer. The size of the convolution kernels can vary, including sizes such as , , , and . Among them, kernels with a size of are used most commonly. Each stage of the encoder in U-Net consists of several layers of convolutional, pooling, and activation. Among them, the pooling layer can keep feature invariance, and to some extent, it can also prevent overfitting, reduce dimensionality, remove redundant information, compress features, simplify network complexity, reduce the amount of computation, reduce memory consumption, etc. Since the pooling layer compresses the features, it inevitably leads to the loss of some information, which is an irreversible process. This operation is beneficial for feature extraction, but is not suitable for pixel classification in the final stage. Because end-to-end image segmentation tasks in the deep learning field are ultimately tasks of pixel classification, they will inevitably lead to inaccurate pixel classification of this part if the lost information cannot be recovered, so an information compensation operation is required.
Skip-connection was originally designed to solve the problem of gradient vanishing. Neural networks use the method of gradient descent to calculate gradient value layer by layer in the direction from the output layer back to the input layer of the network when updating the parameters. However, the gradient is usually a value smaller than 1. The more layers there are in the network, the smaller the gradient value will be. When the gradient value is infinitely close to 0, the network cannot update its parameters. The skip-connection operation involves adding shallow information to the deeper layers of the network, providing a shortcut for gradient backpropagation. This prevents the network from encountering the issue of gradient vanishing, ensuring that parameter updates continue throughout the network. In addition, in the process of continuous feature extraction, details including edge information and small-target information will gradually be lost. However, this detailed information is crucial to the classification of pixels for the segmentation tasks. In each stage of decoding, features from the encoding process are used to fuse with the features from the decoder of the same size. Although the feature maps obtained by such up-sampling cannot completely restore the original features, the features are compensated to a certain extent. However, the information used to compensate for the decoding features also creates noise or interference, and the shallower the layer of the compensation information, the more noise it contains.
Therefore, it is necessary to improve the skip-connection. In this paper, our improved skip-connection includes two aspects, namely positional embedding and cross-attention. Positional embedding is crucial for medical image segmentation tasks because different tissue structures are in different fixed positions in the image. Cross-attention enables the network to obtain a set of weights, allowing it to capture the most important features in skip-connections and effectively incorporate the absolute and relative information between organs. The network strengthens target features by assigning larger weights to the features that are beneficial for segmentation tasks, while reducing the weights of features that are less relevant to segmentation, thus reducing noise and interference. Through continuous forward propagation and back-propagation, the network can improve its ability to identify categories. In this paper, each skip-connection is embedded with positional information and cross-attention mechanism to minimize the impact of information such as noise or interference on medical image segmentation tasks. The improved skip-connection can be formulated as follows:
The skip-connection is related to the encoder and decoder. The final output of improved skip-connection is the result of , which includes the output of cross attention and the result of . is the input of the skip-connection and it comes from the encoder feature . is another input of the skip-connection which comes from the decoder feature . represents the pixel-wise multiplication operation. includes operations such as , batch normalization, Relu and up-sampling. involves up-sampling, one layer of , one layer of , batch normalization and Relu. establishes a correlation between each pixel of the input image. is , which comes from the output features of each stage of the decoder part. The output features from the decoder are more purified, and it is more instructive to use the signal generated by those features to supervise the signal of skip-connection. In fact, is the weights of cross-attention. are obtained according to the following formulas:
where in Equations (6) and (7) is the operation of positional encoding. and in Equation (8) are calculated from , and they are obtained by multiplying with the learnable parameters and in the matrix, respectively. Of course, we also need some necessary dimensional transformation. is the channel number of . in Equation (9) is also a set of learnable parameters, includes max-pooling, , batch normalization and Relu, so the output feature map size of the will be halved. Each variable is shown in Figure 2.
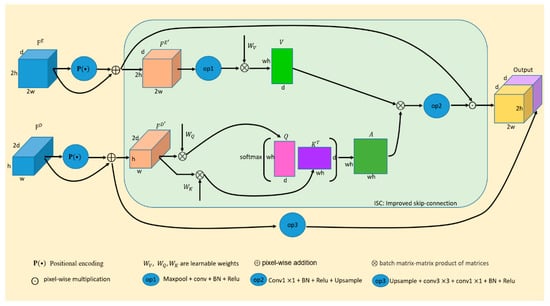
Figure 2.
The improved skip-connection (ISC). The feature maps of different colors represent the results obtained by different ways of processing.
In order to provide a detailed explanation of the contents, Box 1 presents the pseudocode for position encoding, while Box 2 presents the pseudocode for cross-attention. In Box 1, sequence_length represents the length of the input sequence, and d_model represents the hidden dimension of the model. This function returns a position coding matrix with the shape of (sequence_length, d_model). The position coding uses sine and cosine functions to generate coded values for each position in the sequence. The coded value for each dimension is generated using both a sine function and a cosine function, and the frequency and offset are calculated by div_term. Then, according to the parity of position and dimension, the sine coding value and cosine coding value are assigned to the corresponding positions of the position coding matrix, respectively. The position coding matrix can be added to the embedding vector of the input sequence to combine positional and semantic information. In this way, the model can better understand the relevance and importance of different positions in the sequence through the self-attention mechanism. The query and key in Box 2 are derived from the same tensor and are multiplied to calculate the similarity. The model can assign a weight corresponding to the key to each query and use these weights to sum the value to obtain the final representation. By using this method, the network can obtain important information from each stage of the encoder when using skip-connection.
Box 1. Pseudocode of position encoding.
function positional_encoding(sequence_length, d_model):
- position = torch.arange(sequence_length).unsqueeze(1)
- div_term = torch.exp(torch.arange(0, d_model, 2) * − (math.log(10000.0)/d_model))
- positional_encoding = torch.zeros(sequence_length, d_model)
- positional_encoding[:, 0::2] = torch.sin(position * div_term)
- positional_encoding[:, 1::2] = torch.cos(position * div_term)
return positional_encoding
Box 2. Pseudocode of cross-attention.
function cross_attention(query, key, value, mask = None):
- scores = dot_product(query, key)
- if mask is not None:
- scores = apply_mask(scores, mask)
- attention_weights = softmax(scores)
- output = elementwise_multiply(attention_weights, value)
return output
By using this method to solve the medical image segmentation tasks, the essence of the task, pixel-wise classification, is considered. As the cross-attention mechanism selects compensative information, it effectively suppresses the noise or interference in medical images, thereby reducing the risk of misdiagnosis or inaccurate segmentation. This connection method is also used in the second decoding stage, and no noisy information is arbitrarily added to any stage of the entire network, which makes the segmentation results of medical images more accurate.
3.3. Loss Function
Our network is an end-to-end deep learning system. The image segmentation task turns into a classification task for each pixel in the image eventually. Compared with the natural image segmentation, the medical image segmentation task is relatively simple, and it is mostly a two-class problem. It only needs to segment the background and target pixels. However, the proportion of target pixels in the whole image may be much less than that of background pixels, thus causing a class imbalance problem. The most commonly used loss function is cross-entropy, but it may not be the best choice for class imbalance problems. Dice loss can alleviate the class-imbalance problem because it is insensitive to the number of foreground or background pixels. However, dice loss will affect back propagation adversely and can make training unstable. In this paper, binary cross-entropy and dice loss are adopted to jointly optimize the training learning. Under the constraint of improved loss function, our network updates the neuron parameters through gradient descent, and the parameters of the network are optimized through continuous forward propagation and back propagation. Our loss function is formulated as follows:
where denotes the predicted image and denotes the corresponding ground-truth. is the binary cross-entropy loss and is the dice loss. is given as follows:
where is a Laplace smoothing item to avoid the case where the denominator is 0 during division. In this paper, we set . is the position of the pixel.
4. Experiment
We implemented our method on three medical image datasets: the lesion boundary segmentation dataset from ISIC-2018 [43], the CVC-ClinicDB [44], and the 2018 Data Science Bowl challenge [45]. We used these datasets to evaluate the effectiveness of our proposed segmentation network. In this chapter, we first introduce the experimental setup and evaluation metrics. Then, we report our accuracy, comparing its performance to the implementation results of other approaches on the same dataset. Furthermore, we discuss the impact of our method through visual analysis.
4.1. Experimental Setup and Evaluation Metrics
In this section, we mainly introduce the experimental settings in the process of training and testing according to the following aspects. All models were implemented using the Keras framework 2.3.0 [46] with Tensorflow 2.2.0 [47] as the backend. The training and testing were based on the Ubuntu 16.04 system with four NVidia GeForce Titan graphics cards, which have 62 gigabytes of memory. For all models, the SGD optimizer was chosen to train 300 epochs, and the batch size and learning rate were set to 8 and 1 ×10−4, respectively. During training, both Early Stopping and ReduceLROnPlateau were used. In this paper, the evaluation metrics such as Sørensen-Dice Coefficient (DSC), mean Intersection over Union (mIoU), Precision, and Recall were adopted. However, we mainly focused on DSC and mIoU, which are recognized as indicators of the challenge of lesion boundary segmentation. Equations (12)–(15) are our evaluation metrics:
where , , and represent the number of true positives, true negatives, false positives and false negatives, respectively. Additionally, in Equation (13) is the number of target classes to be predicted.
4.2. Datasets and Results
When the training samples are insufficient, the network may overfit, and since medical images are often scarce, it becomes crucial to increase the number and diversity of samples to improve the network’s robustness. To achieve this, we applied 25 types of data augmentation methods, including center crop, random rotation, transpose, elastic transform, etc., to each dataset. Figure 3 shows an original image in CVC-ClinicDB and its corresponding augmented images by 25 kinds of algorithms. For each dataset, the samples were split into three subsets: 80% for training, 10% for validation, and the remaining 10% for testing. We first divided the dataset into training, validation and testing sets and then used 25 kinds of methods to augment them.
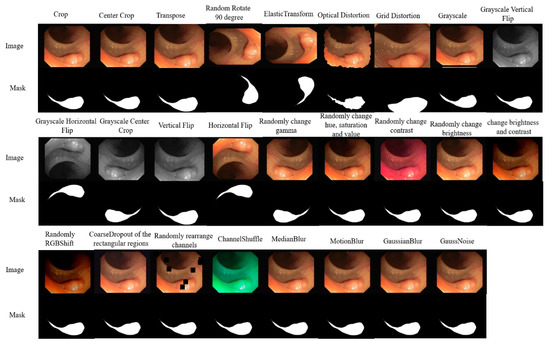
Figure 3.
Images augmented by 25 kinds of methods.
4.2.1. Ablation Studies before and after Network Improvement
In this section, in order to verify the effectiveness of our method, we compare the network before and after improvement. We adopted the U-Net with VGG19 pre-trained model as our framework. The experiments were mainly carried out on the CVC-ClinicDB dataset. The key objectives of the experiments were as follows:
- (1)
- Examining whether feature re-extraction enhances the network’s segmentation performance on medical images.
- (2)
- Evaluating whether the improved skip-connection can effectively filter out interfering information and further improve the performance of the network.
The specific experimental results are shown in Table 1.

Table 1.
Ablation studies before and after network improvement on CVC-ClinicDB dataset.
From Table 1, it is shown that adding the feature re-extraction process and improved skip-connection can improve the performance of the baseline network continuously. Compared with the baseline U-Net, employing feature re-extraction yields a result of 91.52% and 86.9% in DSC and mean IOU, which brings 3.71% and 8.09% improvement, respectively. This is because the feature re-extraction process can further calibrate the fuzzy information on the basis of the initial feature extraction, so the feature extraction ability can be improved. Based on U-Net+Process1, using improved skip-connection can further improve the performance of the network. Because skip-connection in U-Net adds the features extracted by the encoder to the output of the corresponding layer of the decoder indiscriminately, it will inevitably introduce noise information into it, thus interfering with the subsequent segmentation results. The improved skip-connection designed in this paper uses the cross-attention mechanism on the input feature map from the encoder, because the supervision information comes from deep semantic information, so it can greatly reduce the noise. Compared with the baseline U-Net, EFEN has a DSC of 93.04%, which is superior to U-Net (87.81%) by 5.23%. This experiment proves that our method is effective.
4.2.2. Results on CVC-ClinicDB Dataset
The segmentation of polyp images is a challenging task, which is mainly because the demarcation is indistinct between the polyp and its surrounding mucosa and the different sizes, colors and textures of the polyps with the same type. CVC-ClinicDB is an open access colonoscopy image database which is used in our experiments. The CVC-ClinicDB dataset contains 612 images with a resolution of 384 × 288 from 31 colonoscopy sequences. We first split the dataset into training, validation, and testing sets at a ratio of 8:1:1. Then, we used data augmentation methods to obtain 12,714 training images and 1586 validation images. The experimental results of each method in this dataset are shown in Table 2.

Table 2.
Comparisons on CVC-ClinicDB testing set.
Table 2 shows the results on the CVC-ClinicDB testing dataset. Compared with U-Net and the recent works, we observe that the EFEN improves performance remarkably. The comparison methods in this table can be summarized into two categories: one is those based on CNN, and the other is those based on transformer. The methods based on CNN are almost similar to U-Net when using skip-connection; that is, they all add the features extracted by the encoder to the output of the corresponding layer of the decoder indiscriminately. It will inevitably introduce noise information into the decoder, thus interfering with the subsequent segmentation results. However, there are still gaps compared with the state-of-the-art methods. For example, SSFormer-L achieves a DSC of 94.47%. This is because SSFormer-L is a method based on transformer which lacks inductive bias in CNN, and it is based on two datasets. This kind of method needs a lot of pre-training data to obtain better results, or it will be worse. For example, the results of TransUNet and LeViT-UNet are even lower than U-Net. For the smaller dataset, CNN is still the prior choice. However, compared with the baseline U-Net, EFEN outperforms U-Net by a large margin, with 5.23% improvement on DSC and 8.44% improvement on mIoU. In addition, compared with recent works, EFEN has a DSC of 93.04%, which is superior to DoubleU-Net (92.39%) by 0.65%.
A careful visual analysis of the result is shown in Figure 4. In the first two columns of Figure 4, we can see that U-Net misclassified the parts that do not belong to the target. It can be seen from columns 3–6 in Figure 4 that because the boundary between polyps and their surrounding mucosa is unclear, and polyps have different sizes, colors, and textures, it is difficult for U-Net to segment the target accurately. However, our network can segment some smaller targets and targets with unclear edges more accurately. See the red boxes in Figure 4 for more details. Although our improved skip-connection can make up for some missing information, compared with the mask, our method still has some inaccuracies, such as the second, fourth and last columns. There are two main reasons for this issue. First, the quality of samples includes the quality of samples collected and the quality marked by professional doctors should be enhanced. Second, the performance of the model itself also needs to be further improved, which mainly includes the ability of the network to extract features and the further improvement of the compensation method for lost information in the process of feature extraction.
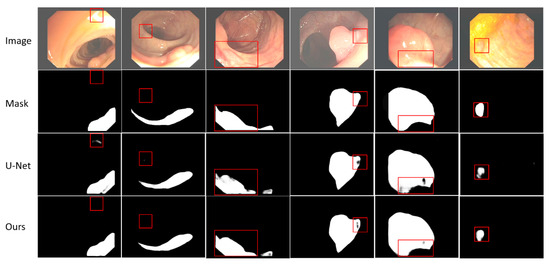
Figure 4.
Visualization results of EFEN on CVC-ClinicDB test set. Each column includes the original input image and its corresponding mask, the result of baseline model U-Net and EFEN. The red box shows the comparisons in the same position.
Furthermore, we introduce the differences between our model and the baseline model U-Net and a recent work, DoubleU-Net, in terms of computational efficiency, utilization of computational resources, and model complexity. The comparison results are presented in Table 3.

Table 3.
More comparisons.
As can be seen from Table 3, compared with U-Net and DoubleU-Net, our model has slightly increased parameters and processing time per image. However, this trade-off results in significantly improved segmentation accuracy.
4.2.3. Results on Lesion Boundary Segmentation Dataset from ISIC-2018
The lesion boundary segmentation dataset is a large-scale dataset which is published by the International Skin Imaging Collaboration (ISIC). It contains 2594 original dermoscopy images and 2594 corresponding binary masks. Lesion boundary segmentation takes its Task 1 from ISIC. It is very useful to analyze dermatoscope images, which can help doctors find potential skin diseases such as skin cancer in advance. Similar to other approaches utilizing this dataset, we randomly partitioned it into three subsets, following the proportions of 80%, 10%, and 10% for training, validation, and testing, respectively. Then, 53,950 training samples and 6734 validation samples are obtained by applying 25 different data augmentation methods. The experimental results of each method in this dataset are shown in Table 4.

Table 4.
Comparisons on ISIC-2018.
Table 4 shows the results on the lesion boundary segmentation dataset from ISIC-2018. EFEN is compared with the baseline network and recent works with the same settings on the same dataset for evaluation. We observe that EFEN achieves a validation DSC of 89.92%, exceeding U-Net (87.46%) by 2.46%. In addition, our method outperforms DoubleU-Net with 0.3% and 0.13% improvements on DSC and mean IOU, respectively. Of course, it should be noted that our method still has room for improvement compared to the state-of-the-art method BAT, which is based on a transformer and requires a larger amount of training data. We acknowledge this gap and remain committed to further exploring and enhancing our method in future research.
The results of visual analysis are shown in Figure 5. We need to segment the target with possible skin diseases from dermoscopy images, even when the skin diseases have different characteristics. Although the same disease appears in different people, the size, boundary and color of the affected area may be different, and there may be some other problems such as hair interference. We select some results with obvious contrast effects to show. Through the comparison of visualization results, it is not difficult to find that our method is better than U-Net in segmenting skin diseases with unclear boundaries and unclear targets. For the segmentation of small targets, such as the first column, our method is better than the baseline method. More details are shown in Figure 5. The red box shows the segmentation results of the same area in the same image using different methods.
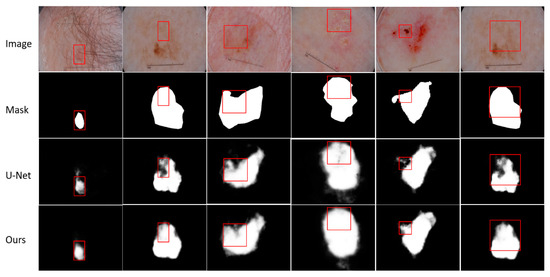
Figure 5.
Visualization results of EFEN on ISIC-2018 test set. Each column includes the original input image and its corresponding mask, the result of baseline model U-Net and EFEN. The red box shows the comparisons in the same position.
4.2.4. Results on 2018 Data Science Bowl Challenge
The main task of this challenge is to detect nuclei in images. Cell nucleus identification helps researchers to identify each cell in an image, thus helping researchers understand potential biological processes. By automatically segmenting the nucleus, doctors can quickly diagnose the disease and treat it. Most genetic disease analysis is based on identifying the nucleus, because most of the 30 trillion cells in the human body contain a nucleus filled with DNA, and DNA is the genetic code for programming each cell. The dataset includes 670 nuclei images. The images are of varied resolutions, and we resize all images to . We first divide the dataset into a training set, verification set and testing set according to the ratio of 8:1:1. By using 25 kinds of data augmentation methods to expand the dataset, we obtain 13,936 training samples and 1742 verification samples. Table 5 shows the experimental comparisons of various methods on this dataset.

Table 5.
Comparisons on 2018 Data Science Bowl challenge.
Table 5 shows the results for the 2018 Data Science Bowl challenge. We observe that EFEN achieves a validation DSC of 91.65%, exceeding baseline U-Net (88.75%) by 2.9%. Compared with recent works, EFEN outperforms DoubleU-Net, with 0.32% and 0.24% improvements on DSC and mean IOU, respectively. SSFormer-L is based on a transformer which needs more data for training, and CNN will be more suitable for the smaller dataset.
In Figure 6, we visualize the segmentation results of our method compared to baseline U-Net. Our method exhibits an advantage in effectively segmenting small objects. As shown in the first, fourth and sixth columns, we observe that the nuclei missed by U-Net can be identified by our method. In the second and third columns, we observe that our method is better than U-Net in identifying targets. Although our method may not be optimal, as shown by previous experiments, our method is better than the baseline method and some recent work.
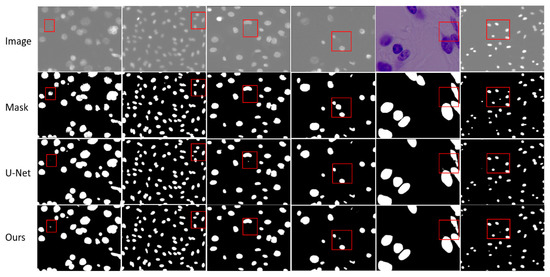
Figure 6.
Visualization results of EFEN on the 2018 Data Science Bowl challenge test set. Each column includes the original input image and its corresponding mask, the result of baseline model U-Net and EFEN. The red box shows the comparisons in the same position.
5. Conclusions
In this paper, we present EFEN for medical image segmentation tasks, which is designed by adding the feature re-extraction process and using improved skip-connection based on U-Net. By adding the feature re-extraction process, the feature extraction ability of the network is enhanced. Improved skip-connection can not only help the network to identify segmentation targets, but also further help the network to reduce the interference information from shallow features. Experiments on the CVC-ClinicDB, the ISIC-2018, and the 2018 Data Science Bowl challenge datasets show that although there are still some gaps between EFEN and the most advanced method, our method is superior to U-Net and other recent excellent networks. Furthermore, from the visual analysis we can see that EFEN can improve the segmentation result of medical images with blurred boundaries or complex backgrounds. The EFEN proposed in this paper can provide doctors with auxiliary diagnosis, help doctors locate lesions and help them to diagnose and evaluate the disease more accurately. In the future, this method or its idea can be applied to cross-modal medical image segmentation, personalized medicine and automated workflows to further explore its advantages.
Author Contributions
Conceptualization, Y.G. and X.C.; methodology, H.X. and Y.G.; investigation, H.X. and M.B.; writing—original draft preparation, Y.G., H.X. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by National Natural Science Foundation of China under Grant 62172184, Science and Technology Development Plan of Jilin Province of China under Grant 20200401077GX, 20200201292JC, Social Science Research of the Education Department of Jilin Province (JJKH20210901SK), and Humanities and Social Science Foundation of Changchun Normal University (2020[011]).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Awan, K.A.; Din, I.U.; Almogren, A.; Almajed, H.; Mohiuddin, l.; Guizani, M. NeuroTrust—Artificial-Neural-Network-Based Intelligent Trust Management Mechanism for Large-Scale Internet of Medical Things. IEEE Internet Things J. 2020, 8, 15672–15682. [Google Scholar] [CrossRef]
- Khan, M.A.; Din, I.U.; Kim, B.S.; Almogren, A. Visualization of Remote Patient Monitoring System Based on Internet of Medical Things. Sustainability 2023, 15, 8120. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; Lange, T.D.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In Proceedings of the International Conference on Multimedia Modeling, Daejeon, Republic of Korea, 5–8 January 2020; Springer: Cham, Switzerland, 2020; pp. 451–462. [Google Scholar]
- Zhao, F.; Xie, X. An overview of interactive medical image segmentation. Ann. BMVA 2013, 2013, 1–22. [Google Scholar]
- Lê, M.; Unkelbach, J.; Ayache, N.; Delingette, H. Gpssi: Gaussian process for sampling segmentations of images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 38–46. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Liu, L.; Cheng, J.; Quan, Q.; Wu, F.X.; Wang, Y.P.; Wang, J.X. A survey on U-shaped networks in medical image segmentations. Neurocomputing 2020, 409, 244–258. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Li, C.; Tan, Y.; Chen, W.; Luo, X.; He, Y.; Gao, Y.; Li, F. ANU-Net: Attention-based Nested U-Net to exploit full resolution features for medical image segmentation. Comput. Graph. 2020, 90, 11–20. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, C.; Coleman, S.; Kerr, D. DENSE-INception U-net for medical image segmentation. Comput. Methods Programs Biomed. 2020, 192, 105395. [Google Scholar] [CrossRef]
- Jha, D.; Riegler, M.A.; Johansen, D.; Halvorsen, P.; Johansen, H.D. Doubleu-net: A deep convolutional neural network for medical image segmentation. In Proceedings of the 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, 28–30 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 558–564. [Google Scholar]
- Li, J.F.; Chen, C.X.; Wang, L. Fusion algorithm of multi-spectral images based on dual-tree complex wavelet transform and frequency-domain U-Net. J. Biomed. Eng. Res. 2020, 39, 145–150. [Google Scholar]
- Zhang, T.Q.; Kang, T.Q.; Meng, X.F.; Liu, Y.; Zhou, Y. U-Net Based Intracranial Hemorrhage Recognition. J. Beijing Univ. Posts Telecommun. 2020, 43, 92. [Google Scholar]
- Yang, Y.; Feng, C.; Wang, R. Automatic segmentation model combining U-Net and level set method for medical images. Expert Syst. Appl. 2020, 153, 113419. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, H.; Ling, X. Polar coordinate sampling-based segmentation of overlapping cervical cells using attention U-Net and random walk. Neurocomputing 2020, 383, 212–223. [Google Scholar] [CrossRef]
- Liu, Z.; Song, Y.Q.; Sheng, V.S.; Wang, L.; Jiang, R.; Zhang, X.; Yuan, D. Liver CT sequence segmentation based with improved U-Net and graph cut. Expert Syst. Appl. 2019, 126, 54–63. [Google Scholar] [CrossRef]
- Man, Y.; Huang, Y.; Feng, J.; Li, X.; Wu, F. Deep Q learning driven CT pancreas segmentation with geometry-aware U-Net. IEEE Trans. Med. Imaging 2019, 38, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Jiang, D.; Yang, C.; Gao, F.; Gao, F. Y-Net: Hybrid deep learning image reconstruction for photoacoustic tomography in vivo. Photoacoustics 2020, 20, 100197. [Google Scholar] [CrossRef]
- Kuang, Z.; Deng, X.; Yu, L.; Wang, H.; Li, T.; Wang, S. Ψ-Net: Focusing on the border areas of intracerebral hemorrhage on CT images. Comput. Methods Programs Biomed. 2020, 194, 105546. [Google Scholar] [CrossRef]
- Dolz, J.; Ben Ayed, I.; Desrosiers, C. Dense multi-path U-Net for ischemic stroke lesion segmentation in multiple image modalities. In Proceedings of the International MICCAI Brainlesion Workshop, Granada, Spain, 16 September 2018; Springer: Cham, Switzerland, 2018; pp. 271–282. [Google Scholar]
- Xia, X.; Kulis, B. W-net: A deep model for fully unsupervised image segmentation. arXiv 2017, arXiv:1711.08506. [Google Scholar]
- Xu, L.; Liu, M.; Shen, Z.; Wang, H.; Liu, X.; Wang, X.; Wang, S.; Li, T.; Yu, S.; Hou, M.; et al. DW-Net: A cascaded convolutional neural network for apical four-chamber view segmentation in fetal echocardiography. Comput. Med. Imaging Graph. 2020, 80, 101690. [Google Scholar] [CrossRef]
- Das, S.; Deka, A.; Iwahori, Y.; Bhuyan, M.K.; Iwamoto, T.; Ueda, J. Contour-aware residual W-Net for nuclei segmentation. Procedia Comput. Sci. 2019, 159, 1479–1488. [Google Scholar] [CrossRef]
- Tang, Z.; Peng, X.; Li, K.; Metaxas, D.N. Towards efficient u-nets: A coupled and quantized approach. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 42, 2038–2050. [Google Scholar] [CrossRef]
- Kang, S.; Iwana, B.K.; Uchida, S. Complex image processing with less data—Document image binarization by integrating multiple pre-trained U-Net modules. Pattern Recognit. 2021, 109, 107577. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, X.; Li, Z.; Yu, Z.; Yao, S.; Yan, L.; Wang, Y.; Liu, Z.; Liang, C.; Han, C. Triple U-net: Hematoxylin-aware nuclei segmentation with progressive dense feature aggregation. Med. Image Anal. 2020, 65, 101786. [Google Scholar] [CrossRef]
- Lee, S.; Negishi, M.; Urakubo, H.; Kasai, H.; Ishii, S. Mu-net: Multi-scale U-net for two-photon microscopy image denoising and restoration. Neural Netw. 2020, 125, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Gao, Z.; Xie, J.; Wang, Q.; Li, P. Global second-order pooling convolutional networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 3024–3033. [Google Scholar]
- Qin, Z.; Zhang, P.; Wu, F.; Li, X. Fcanet: Frequency channel attention networks. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 783–792. [Google Scholar]
- Lee, H.J.; Kim, H.E.; Nam, H. Srm: A style-based recalibration module for convolutional neural networks. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1854–1862. [Google Scholar]
- Yang, Z.; Zhu, L.; Wu, Y.; Yang, Y. Gated channel transformation for visual recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 11794–11803. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. Supplementary material for ‘ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 13–19. [Google Scholar]
- Zhang, H.; Dana, K.; Shi, J.; Zhang, Z.; Wang, X.; Tyagy, A.; Agrawal, A. Context encoding for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7151–7160. [Google Scholar]
- Jin, Q.; Meng, Z.; Sun, C.; Cui, H.; Su, R. RA-UNet: A hybrid deep attention-aware network to extract liver and tumor in CT scans. Front. Bioeng. Biotechnol. 2020, 8, 1471. [Google Scholar] [CrossRef]
- Ding, X.; Peng, Y.; Shen, C.; Zeng, T. CAB U-Net: An end-to-end category attention boosting algorithm for segmentation. Comput. Med. Imaging Graph. 2020, 84, 101764. [Google Scholar] [CrossRef]
- Hariyani, Y.S.; Eom, H.; Park, C. DA-CapNet: Dual attention deep learning based on U-Net for nailfold capillary segmentation. IEEE Access 2020, 8, 10543–10553. [Google Scholar] [CrossRef]
- Bai, W.; Sinclair, M.; Tarroni, G.; Oktay, O.; Rajchl, M.; Vaillant, G.; Lee, A.M.; Aung, N.; Lukaschuk, E.; Sanghvi, M.M.; et al. Human-level CMR image analysis with deep fully convolutional networks. arXiv 2017, arXiv:1710.09289v1. [Google Scholar]
- Kamnitsas, K.; Bai, W.; Ferrante, E.; McDonagh, S.; Sinclair, M.; Pawlowski, N.; Rajchl, M.; Lee, M.; Kainz, B.; Rueckert, D.; et al. Ensembles of multiple models and architectures for robust brain tumour segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Quebec City, QC, Canada, 14 September 2017; Springer: Cham, Switzerland, 2017; pp. 450–462. [Google Scholar]
- Roth, H.R.; Lu, L.; Lay, N.; Harrison, A.P.; Farag, A.; Sohn, A.; Summers, R.M. Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med. Image Anal. 2018, 45, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.R.; Oda, H.; Hayashi, Y.; Oda, M.; Shimizu, N.; Fujiwara, M.; Misawa, K.; Mori, K. Hierarchical 3D fully convolutional networks for multi-organ segmentation. arXiv 2017, arXiv:1704.06382. [Google Scholar]
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 168–172. [Google Scholar]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Goodman, A.; Karhohs, K.W.; Cimini, B.A.; Ackerman, J.; Haghighi, M.; Heng, C.K.; Becker, T.; Doan, M.; McQuin, C.; et al. Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Ketkar, N. Introduction to keras. In Deep Learning with Python; Apress: Berkeley, CA, USA, 2017; pp. 97–111. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. {TensorFlow}: A system for {Large-Scale} machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Xu, G.; Wu, X.; Zhang, X.; He, X. Levit-unet: Make faster encoders with transformer for medical image segmentation. arXiv 2021, arXiv:2107.08623. [Google Scholar] [CrossRef]
- Gao, G.; Yu, Y.; Yang, M.; Huang, P.; Ge, Q.; Yue, D. Multi-scale patch based representation feature learning for low-resolution face recognition. Appl. Soft Comput. 2020, 90, 106183. [Google Scholar] [CrossRef]
- Jha, D.; Smedsrud, P.H.; Johansen, D.; Lange, T.D.; Johansen, H.D.; Halvorsen, P.; Riegler, M.A. A comprehensive study on colorectal polyp segmentation with ResUNet++, conditional random field and test-time augmentation. IEEE J. Biomed. Health Inform. 2021, 25, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, B.; Wang, L.; Zu, C.; Lalush, D.S.; Lin, W.; Wu, X.; Zhou, J.; Shen, D.; Zhou, L. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage 2018, 174, 550–562. [Google Scholar] [CrossRef]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Pranet: Parallel reverse attention network for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Cham, Switzerland, 2020; pp. 263–273. [Google Scholar]
- Jha, D.; Ali, S.; Tomar, N.K.; Johansen, H.D.; Johansen, D.; Rittscher, J.; Riegler, M.A.; Halvorsen, P. Real-time polyp detection, localization and segmentation in colonoscopy using deep learning. IEEE Access 2021, 9, 40496–40510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, H.; Hu, Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Cham, Switzerland, 2021; pp. 14–24. [Google Scholar]
- Viet Sang, D.; Quang Chung, T.; Lan, P.N.; Hang, D.V.; Long, D.V.; Thuy, N.Y. AG-CUResNeSt: A Novel Method for Colon Polyp Segmentation. arXiv 2021, arXiv:2105.00402. [Google Scholar]
- Kim, T.; Lee, H.; Kim, D. Uacanet: Uncertainty augmented context attention for polyp segmentation. In Proceedings of the 29th ACM International Conference on Multimedia, Virtual, China, 20–24 October 2021; pp. 2167–2175. [Google Scholar]
- Wang, J.; Huang, Q.; Tang, F.; Meng, J.; Su, J.; Song, S. Stepwise Feature Fusion: Local Guides Global. arXiv 2022, arXiv:2203.03635. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Wang, J.; Wei, L.; Wang, L.; Zhou, Q.; Zhu, L.; Qin, J. Boundary-aware transformers for skin lesion segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Cham, Switzerland, 2021; pp. 206–216. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).