1. Introduction
Livestock farming has become an important industrial sector as well as a side occupation for people engaged in agriculture in rural areas. Thanks to practices such as cooperatives, producer unions, registered breeding, artificial insemination practices, and livestock supports, the place of the livestock sector in the country’s economy has started to gain more importance. It is necessary to determine the weight of the animals raised in cattle breeding farms and to follow them regularly. Increasing the profitability of the business depends on the regular follow-up of live weight [
1].
The most common method of measuring the live weight of farm animals is traditional measurement using a scale. Although this direct approach is very accurate, it comes with various difficulties and limitations. Firstly, animals are required to be moved to the site of measurement scale, which can be time-consuming and laborious, especially in farms with a large number of animals. Secondly, this whole operation with the separation of animals from their natural environment causes stress, and therefore negatively affects their health and milk yield. Due to those drawbacks of direct measurement approaches, a variety of indirect measurement approaches have been proposed in the literature [
2]. In indirect measurement, the true value of animal live weight is estimated by a regression model trained on various features extracted from measurements obtained from several sensors such as 2D [
3] and 3D cameras [
4], thermal cameras [
5], and ultrasonic sensors [
6].
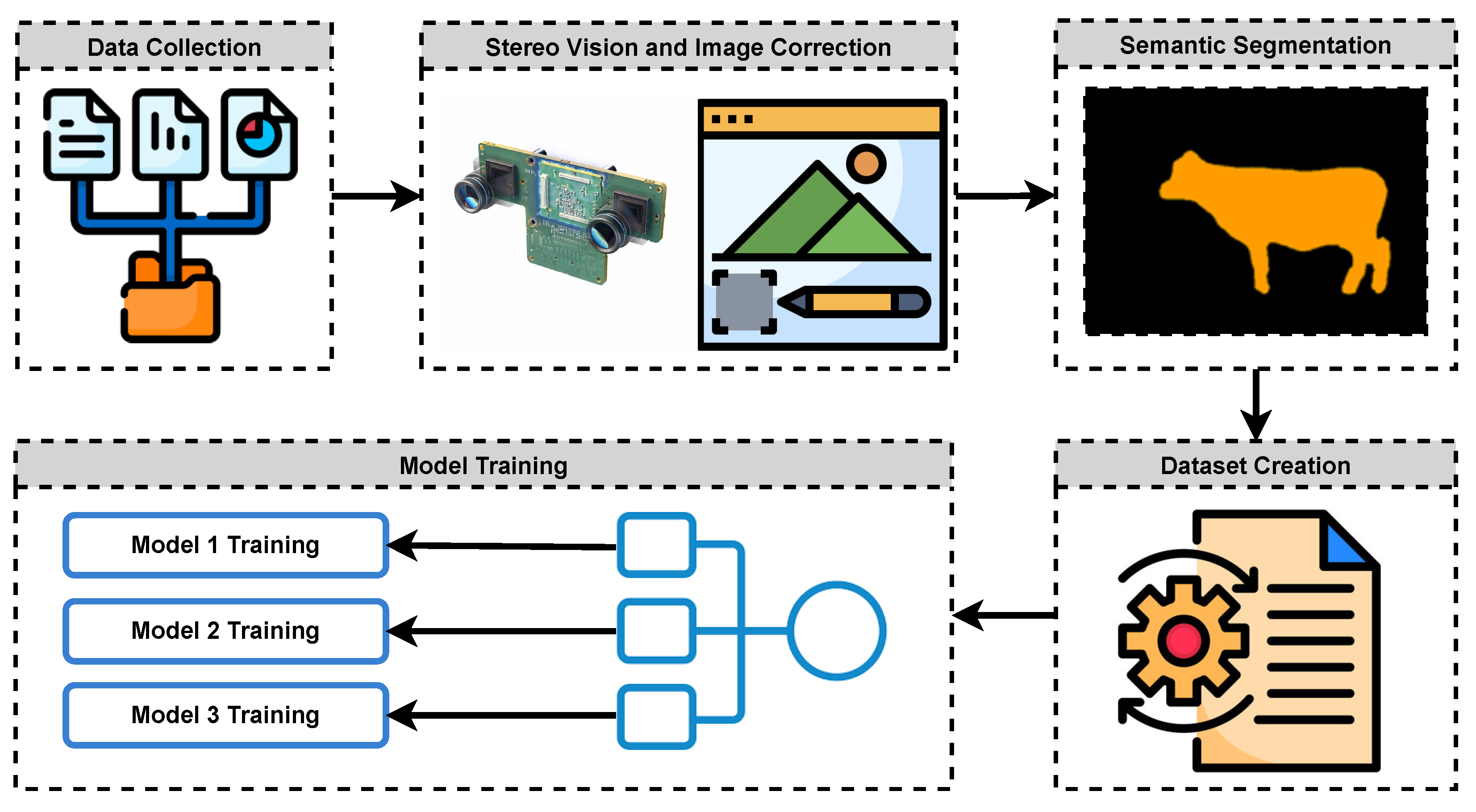
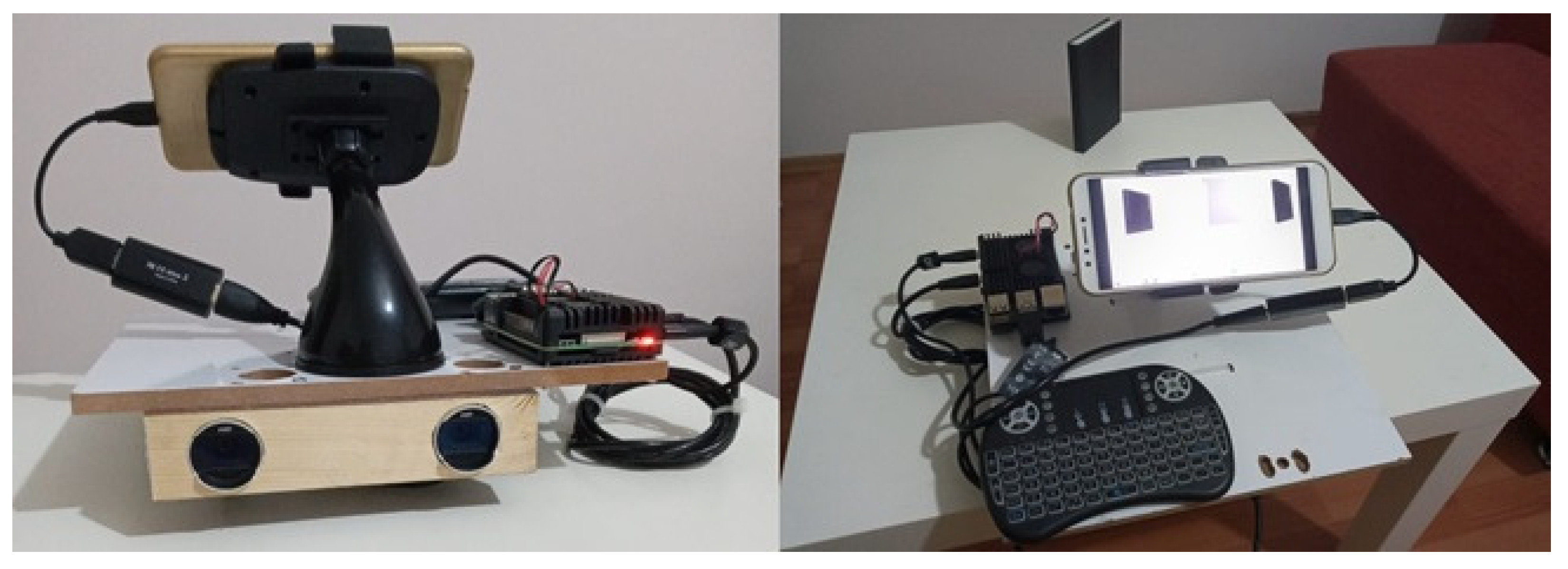
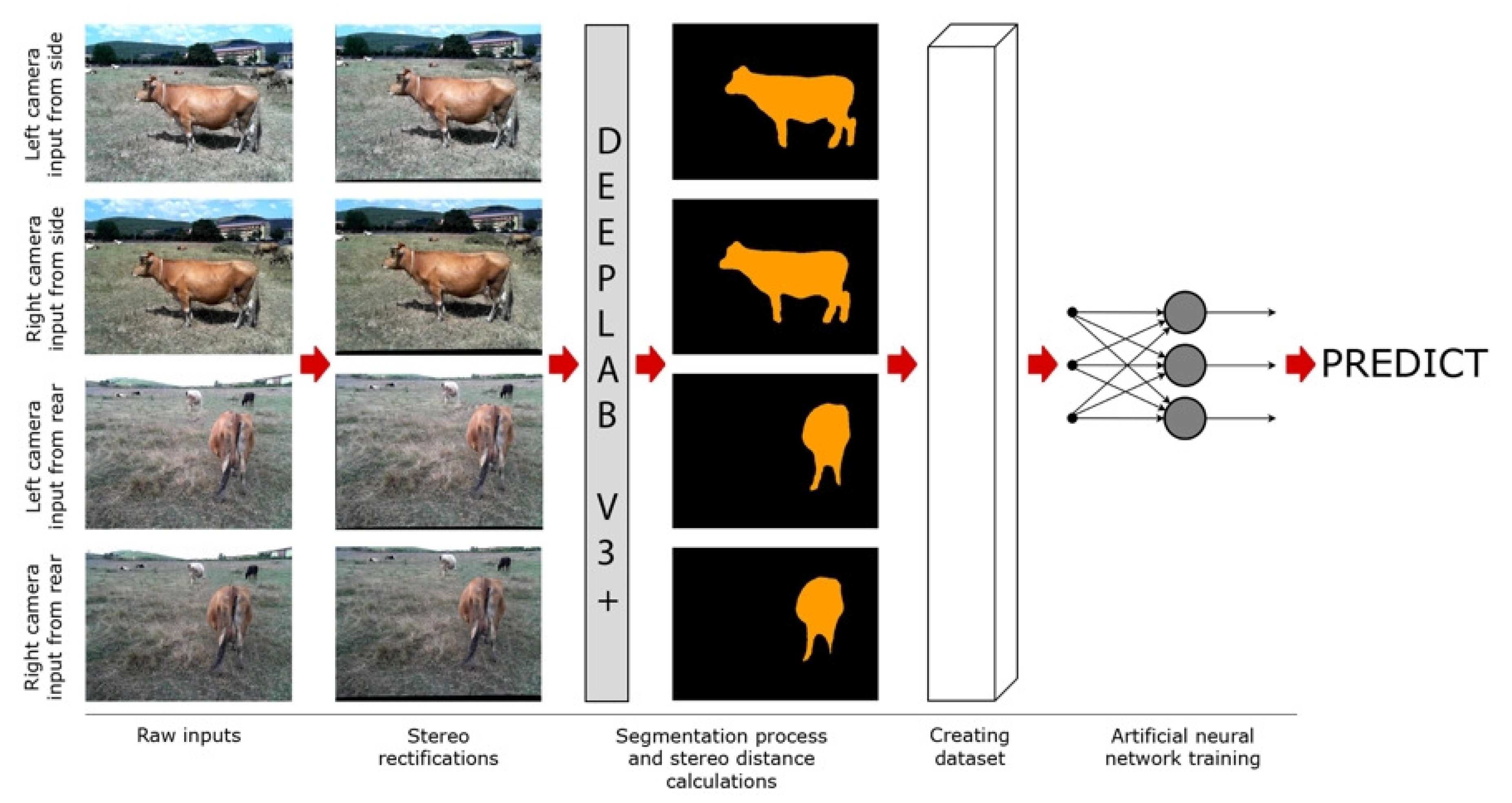
In this study, we consider the determination of the live weight of farm animals as a computer vision and a regression problem. First, we obtain the images of farm animals using a stereo setup. Then, applying deep learning-based semantic segmentation techniques, we extract distance and size data from images to feed into a regression model. Finally, we obtain the weight estimates from the regression model as a proxy for the actual weights of the animals. The main motivation for our study was to apply state-of-the-art image processing techniques using modern deep learning approaches to propose an effective solution to the problem considered. The main contributions and novelty of our study can be summarized as follows:
We propose an effective indirect measurement method for determining the live weight of farm animals based on stereo vision and state-of-the-art semantic segmentation techniques using deep learning.
Our method is particularly important in that animals’ body measurements are taken without the need for separating them from their natural environments and thus not adversely affecting their health and milk yield.
We propose a very simple yet effective system and setup composed of relatively cheaper hardware that is accessible and affordable for many farms of small to large scale.
We investigate and compare the performances of three different Artificial Neural Network (ANN) architectures in estimating live animal weight.
The rest of this paper is organized as follows. The related work is reviewed in
Section 2. In
Section 3, we present the materials and methods used in the study. We present our experimental results and discussion in
Section 4 and
Section 5, respectively. Finally, in
Section 6, we conclude the paper.
2. Related Work
In this section, we provide essential background on livestock weight estimation with a review of significant past research. Our focus in this review is on the work with indirect measurement approaches based on image processing techniques. We also summarize them in
Table 1.
There are several studies in the literature that are based on image processing techniques on 2D images. In a study by Weber et al., the live body weight of cattle was estimated using dorsal area images taken from above using a kind of fence system [
7]. Their system first performs segmentation and then generates a convex hull around the segmented area to obtain features to feed a Random Forest-based regression model. Tasdemir and Ozkan performed a study where they predicted the live weight of cows using an ANN-based regression model [
8]. They determined various body dimensions such as wither height, hip height, body length, and hip width applying photogrammetric techniques on images of cows captured from various angles. Wang et al. developed an image processing-based system to estimate the body weight of pigs [
9]. Their main approach was to process images captured from above to extract features such as area, convex area, perimeter, and so on. Then, using these features, they trained an ANN-based regression model for weight prediction. A Fuzzy Rule-Based System was also utilized in cattle weight estimation by Anifah and Haryanto [
10]. They obtained 2D side images of cattle from a very close distance of 1.5 m. After applying the Gabor filter to the images, they obtained body length and circumference as features. Finally, they designed a fuzzy logic system to estimate body weight.
Three-dimensional imaging techniques also found application in body weight estimation systems. Hansen et al. used a 3D Kinect-like depth camera to obtain the views of cows from above as they passed along a fence [
11]. Applying thresholding, they obtained the segmented area of cows to reach a body weight estimate. In another study where a 3D Kinect camera was used, Fernandes et al. processed images taken from above of pigs by applying two segmentation steps [
12]. Then, they extracted features from segmented images such as body area, volume, width, and height to feed a linear regression model to obtain the weight estimate. In a similar study, Cominotte et al. developed a system to capture images of cattle using a 3D Kinect camera [
13]. They trained and compared a number of linear and non-linear regression models by feeding them with features extracted from segmented images. In a study by Martins et al., a 3D Kinect camera was used to capture images of cows from lateral and dorsal perspectives [
14]. They used several measurements obtained from these images to run a Lasso regression model to estimate body weight. Nir et al. used a 3D Kinect camera as well to take images of dairy heifers to estimate height and body mass [
15]. Their approach was to fit an ellipse to the body image to calculate some features. Then, they used these features to train various linear regression models. Song et al. created a system to estimate the body weight of cows using a 3D camera system [
16]. Similar to previous studies, they extracted morphological features from 3D images such as hip height, hip width, and rump length. Combining these features with some other cow data such as days in milk, age, and parity, they trained multiple linear regression models. Another study that employed a 3D Kinect camera is the one conducted by Pezzuolo et al. [
17]. They captured body images of pigs using two cameras from top and side, and then extracted body dimensions from images such as heart girth, length, and height using image processing techniques. They developed linear and non-linear regression models based on these dimensions to predict weight.
Advanced scanning devices were also introduced in body weight estimation studies. Le Cozler et al. used a 3D full-body scanning device to obtain very detailed body images of cows [
18]. Then, they computed body measures from these 3D images such as volume, area, and other morphological traits. Using these measures, they trained and compared several regression models. Stajnko et al. developed a system to make use of thermal camera images of cows to extract body features and then used them in several linear regression models to estimate body weight [
19].
Stereo vision techniques are also used in the determination of live animal weight. Shi et al. developed a regression model to analyze and estimate the body size and live weight of farm pigs under indoor conditions in a farm [
20]. Their system was based on a binocular stereo vision system and a special fence system through which animals passed for taking the measurements. They segmented the images obtained from the stereo system using a depth threshold and predicted the body length and withers height, then the body weight. Some other notable studies using stereo vision are by Nishide et al. and Yamashita et al. [
21,
22].
Deep learning-based approaches are very popular today due to their success in image-processing applications. Deep learning is a special form of neural network algorithm. Although it has achieved the most advanced results in many fields, its use in determining the weight of livestock is limited [
23]. There are studies that apply deep learning algorithms and determine the weight of pigs [
24,
25].
When we examine the prior research on the estimation of live body weight of farm animals such as pigs, cattle, cows, and heifers, there is a common approach to capturing images of animals that the animals are forced to move into special types of boxes or fences, or they are forced to pass through a special passage. This operation is very similar to traditional weight measurement with scales, and therefore, it also requires the separation of animals from their natural environment, and it causes stress-related problems in their health and milk yield [
3]. Our proposed approach is superior to this in that animals’ pictures are taken in their natural environments without the need for a special measurement station. Additionally, our approach is totally contactless and pictures do not need to be taken from very close proximity, unlike previous studies. One other advantage of our proposed approach provides a simpler structure and setup composed of relatively cheaper hardware that can be accessible and affordable for many farms of small to large scale. Last but not least, we employ modern and state-of-the-art deep learning-based image processing techniques in our system, which is one of the few such studies.
4. Results
In this section, we present the prediction performances of the neural networks trained in a comparative manner. The performance levels of the networks are shown in
Figure 11.
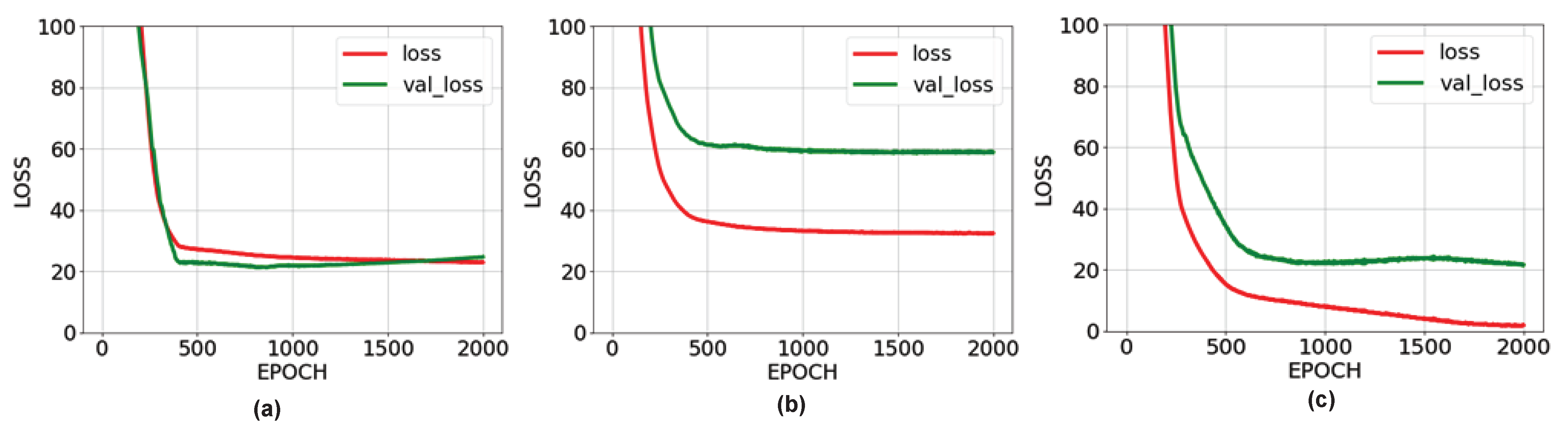
The success rate of the ANN-1 network is higher than the ANN-2 network. The reason for this is the inability of the images taken from the back to reveal the general body dimensions of the animal. On the other hand, the performance rate of the ANN-3 network is higher than the other two networks. This is because the network was trained with data from images taken from both angles of animals. Randomly, 10% of the taken images were not used in the training but in the testing of the estimated animal weights. Weight estimation was made separately for the three proposed networks and the results are shown in
Table 5,
Table 6 and
Table 7.
As seen in
Table 5, the estimations for the test data made by the ANN-1 network vary between approximately ±50 kg. Note that ANN-1 is only trained with data obtained from the side. In
Table 6, the error amounts in the estimations made by the ANN-2 network, which was trained only with photographs taken from the back, vary between approximately ±50 kg. However, the error rates increased dramatically in animals with id numbers 36, 70, and 81. This significantly reduces the accuracy of the network trained with images taken from behind. The reason for this is the inability of the images taken from the back to reveal the general body dimensions of the animal.
Table 7 shows the results obtained from the ANN-3 network trained with the entire dataset. In most cases, the predictions were made with a margin of error of approximately ±20 kg, and much more successful results were obtained than the first two networks. The animal image taken in prediction number 36 with a high amount of error is very close to the camera plane. The image of the animal taken very close to the camera plane causes serious errors as it cannot be adequately represented in the dataset. For this reason, it would be more appropriate to take the images to be obtained at reasonable distances not very close to the camera plane.
In this study, the K-fold cross-validation technique was used to test the validity of the proposed method and the accuracy of the results obtained. K-fold cross-validation is one of the methods of splitting the dataset for evaluation of classification models and training of the model [
34,
35]. This method is used to generate random layers. Each layer represents a combination of training data subset and test data subset sections for training and validating machine learning models. For each layer, a certain accuracy value is obtained for the model. For example, in the case of 10-fold cross-validation, the overall accuracy is estimated by averaging the accuracy values produced by all 10 folds. For any dataset with a given number of samples, there are many possible combinations of training and test datasets that can be generated. Some of these datasets are used to train the model and some are used to test the success of the model. Therefore, it allows each divided part to be used separately for both training and testing. The representation of the K-fold cross-validation method for K = 10 is given in
Figure 12.
Training and testing the model up to K can take a long time and can be costly in terms of computation and time for large datasets. On the other hand, it provides a reliable result. In this study, the K value was accepted as 10, and validity tests were carried out. Here, the test and training images at each step are meaningfully segmented. A similar situation was repeated at each K step and validity tests were performed on different images. In this study, the validity of the ANN-3 architecture, which was trained using both side and rear images, was tested with K-fold. The results obtained are given in
Figure 12. When the predicted values obtained in each K step are compared with the actual values in
Table 8, it can be concluded that the proposed model is quite successful. It is thought that 85 animals are not enough to successfully train a neural network. In addition, the weight distribution of the animals, whose images were taken with the stereo device, is generally around 400 kg. Therefore, the estimates made by nets are generally more successful for animals weighing 400 kg. Another weakness of the dataset we created is that animal images are generally taken from 6 to 8 m away. During the image acquisition phase, it was mostly not possible to take images from closer distances, such as 2–3 m, due to frightening the animals. At these distances, stereo vision works more successfully than at distances of 6–8 m. Utilizing all this information, more successful results can be obtained from a trained network with more animal images whose weights are normally distributed. In order to train a neural network successfully, the dataset on which the neural network is trained must be large enough, that is, it must consist of a sufficient number of observations [
36]. All known possible variations of the problem area should be added to the dataset. Adequate data delivery to a system is necessary to obtain a robust and reliable network [
37,
38]. For example, the generated third neural network is trained with data created with images taken from both the side and the back. The amount of error in the weight estimations made by this neural network decreased to the range of ±20 kg.
5. Discussion
In this study, an attempt was made to estimate live animal weight by using stereo vision and semantic segmentation methods in the literature. Within the scope of the study, a stereo vision device was prepared, and stereo images of 85 cattle whose weights were known beforehand were obtained with this setup. Segmentation maps of the animals in these images were created with the Deeplab v3+ deep learning model, which is one of the semantic segmentation models.
Using the segmentation maps, the number of pixels covered by each animal in the image and their distance to the camera plane were calculated using the stereo distance calculation technique. A dataset was created by combining these obtained data. The dataset was created from the data obtained from photographs of animals taken from two different angles, from the side, and from the back.
Using this dataset, three different artificial neural networks, which are architecturally similar to each other, are trained. When the trained neural networks were compared, it was seen that the third neural network trained with the whole dataset was significantly more successful than the first two neural networks. At this point, it is clear that neural networks to be trained with datasets created with images taken from more angles will be more successful. For example, top images of cattle contain important information about the animal’s body structure. It can be said that networks trained with a dataset that includes top-shot data, if possible, will be more successful.
In addition, it is possible to say that neural networks will make more successful predictions if the quality and quantity of the dataset are increased. In the resulting estimations, although rare, dramatically incorrect estimations were observed. Weight estimations of animals that were limited in number in the dataset, that were light in weight, and whose stereo distance was very different from the rest of the dataset were found to be relatively unsuccessful. Therefore, it is clear that creating a more comprehensive and homogeneously distributed dataset will significantly increase the performance of the models.
Moreover, characteristics such as race and gender of animals directly affect their weight. For example, if the body sizes of two animals of different breeds are assumed to be exactly the same, it will be seen that the weights of these two animals are different from each other. At this point, in the study, a deep learning method that recognizes the breed and gender of the animal can be developed and the performance in weight estimation can be increased with a separate training model for each breed.