NMR-Based Metabolomics for a More Holistic and Sustainable Research in Food Quality Assessment: A Narrative Review
Abstract
1. Introduction
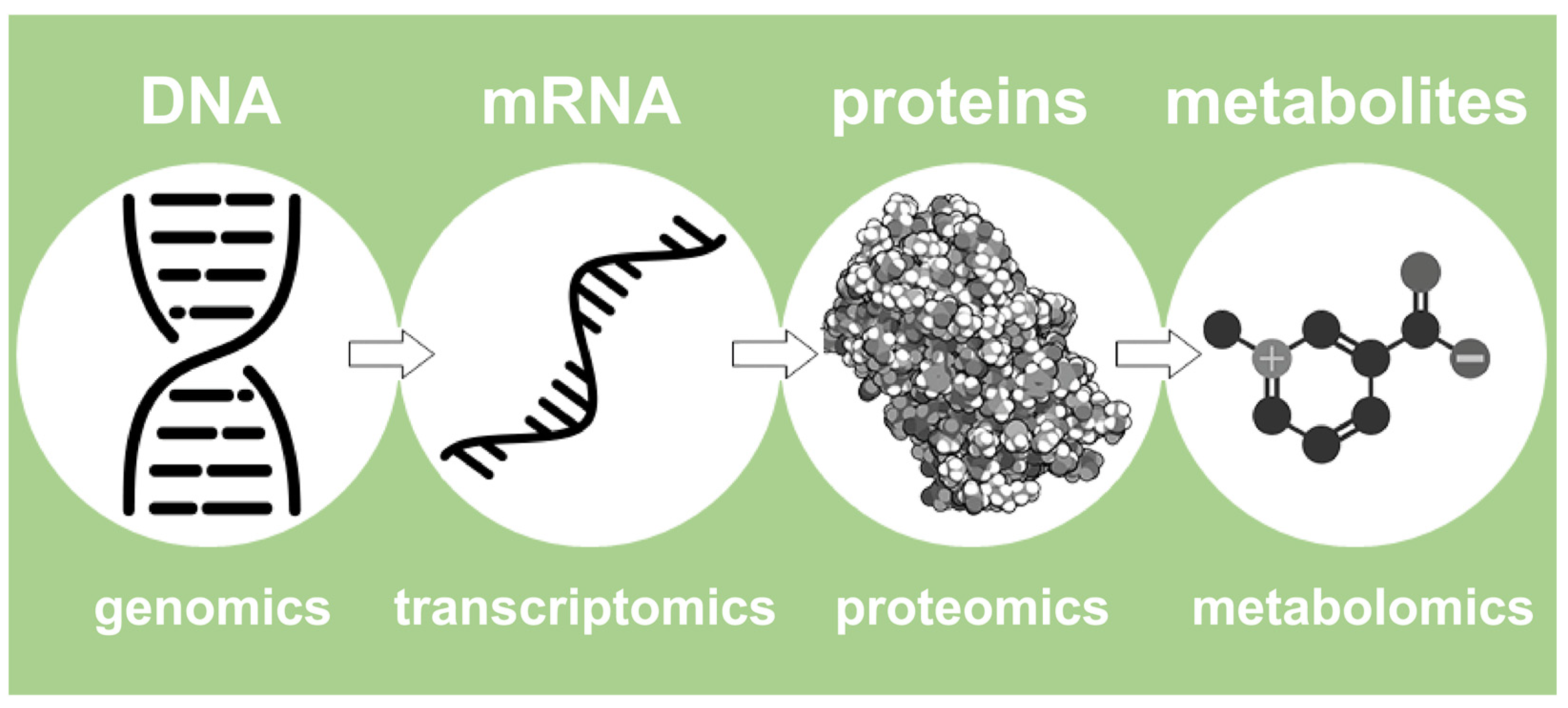
2. The NMR-Based Metabolomics in Food Science and the Foodomics Approach
2.1. The NMR-Based Foodomics Approach for Food Bio-Waste or By-Products Analysis
2.2. The NMR-Based Foodomics Approach for the Simulated Digestion and Absorption of Food
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, S.; Tian, Y.; Jiang, P.; Lin, Y.; Liu, X.; Yang, H. Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. Nutr. 2021, 61, 1448–1469. [Google Scholar] [CrossRef] [PubMed]
- Fernández-García, E.; Carvajal-Lérida, I.; Pérez-Gálvez, A. In vitro bioaccessibility assessment as a prediction tool of nutritional efficiency. Nutr. Res. 2009, 29, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, K.C.; Agache, I.; Jutel, M.; Annesi Maesano, I.; Akdis, M.; Sampath, V.; d’Amato, G.; Cecchi, L.; Traidl-Hoffmann, C.; Akdis, C.A. Climate change: A call to action for the united nations. Allergy 2022, 77, 1087–1090. [Google Scholar] [CrossRef] [PubMed]
- Tobiszewski, M.; Mechlińska, A.; Namieśnik, J. Green analytical chemistry—Theory and practice. Chem. Soc. Rev. 2010, 39, 2869–2878. [Google Scholar] [CrossRef]
- Płotka-Wasylka, J.; Mohamed, H.M.; Kurowska-Susdorf, A.; Dewani, R.; Fares, M.Y.; Andruch, V. Green analytical chemistry as an integral part of sustainable education development. Curr. Opin. Green Sustain. Chem. 2021, 31, 100508. [Google Scholar] [CrossRef]
- Poliakoff, M.; Licence, P.; George, M.W. UN sustainable development goals: How can sustainable/green chemistry contribute? By doing things differently. Curr. Opin. Green Sustain. Chem. 2018, 13, 146–149. [Google Scholar] [CrossRef]
- Poliakoff, M.; Licence, P.; George, M.W. A new approach to sustainability: A Moor’s law for chemistry. Angew. Chem. Int. Ed. 2018, 57, 12590–12591. [Google Scholar] [CrossRef]
- Gottlieb, H.E.; Graczyk-Millbrandt, G.; Inglis, G.G.; Nudelman, A.; Perez, D.; Qian, Y.; Shuster, L.E.; Sneddon, H.F.; Upton, R.J. Development of GSK’s NMR guides—A tool to encourage the use of more sustainable solvents. Green Chem. 2016, 18, 3867–3878. [Google Scholar] [CrossRef]
- Mielko, K.A.; Pudełko-Malik, N.; Tarczewska, A.; Młynarz, P. NMR spectroscopy as a “green analytical method” in metabolomics and proteomics studies. Sustain. Chem. Pharm. 2021, 22, 100474. [Google Scholar] [CrossRef]
- Trimigno, A.; Münger, L.; Picone, G.; Freiburghaus, C.; Pimentel, G.; Vionnet, N.; Pralong, F.; Capozzi, F.; Badertscher, R.; Vergères, G. GC-MS based metabolomics and NMR spectroscopy investigation of food intake biomarkers for milk and cheese in serum of healthy humans. Metabolites 2018, 8, 26. [Google Scholar] [CrossRef]
- Münger, L.H.; Trimigno, A.; Picone, G.; Freiburghaus, C.; Pimentel, G.g.; Burton, K.J.; Pralong, F.o.P.; Vionnet, N.; Capozzi, F.; Badertscher, R. Identification of urinary food intake biomarkers for milk, cheese, and soy-based drink by untargeted GC-MS and NMR in healthy humans. J. Proteome Res. 2017, 16, 3321–3335. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Laghi, L.; Picone, G.; Capozzi, F. Nuclear magnetic resonance for foodomics beyond food analysis. TrAC Trends Anal. Chem. 2014, 59, 93–102. [Google Scholar] [CrossRef]
- Ciampa, A.; Laghi, L.; Picone, G. Validation of a 1H-NMR Spectroscopy Quantitative Method to Quantify Trimethylamine Content and K-Index Value in Different Species of Fish. J. Food Qual. 2022, 2022, 3612095. [Google Scholar] [CrossRef]
- Bisht, B.; Kumar, V.; Gururani, P.; Tomar, M.S.; Nanda, M.; Vlaskin, M.S.; Kumar, S.; Kurbatova, A. The potential of nuclear magnetic resonance (NMR) in metabolomics and lipidomics of microalgae—A review. Arch. Biochem. Biophys. 2021, 710, 108987. [Google Scholar] [CrossRef] [PubMed]
- Picone, G.; Mengucci, C.; Capozzi, F. The NMR added value to the green foodomics perspective: Advances by machine learning to the holistic view on food and nutrition. Magn. Reson. Chem. 2022, 60, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. In Metabonomics; Springer: Cham, Switzerland, 2015; pp. 161–193. [Google Scholar]
- Letertre, M.P.; Giraudeau, P.; De Tullio, P. Nuclear magnetic resonance spectroscopy in clinical metabolomics and personalized medicine: Current challenges and perspectives. Front. Mol. Biosci. 2021, 8, 698337. [Google Scholar] [CrossRef]
- Robosky, L.C.; Reily, M.D.; Avizonis, D. Improving NMR sensitivity by use of salt-tolerant cryogenically cooled probes. Anal. Bioanal. Chem. 2007, 387, 529–532. [Google Scholar] [CrossRef]
- Lee, J.H.; Okuno, Y.; Cavagnero, S. Sensitivity enhancement in solution NMR: Emerging ideas and new frontiers. J. Magn. Reson. 2014, 241, 18–31. [Google Scholar] [CrossRef]
- Ghorbani, B.; Amidpour, M. Energy, exergy, and sensitivity analyses of a new integrated system for generation of liquid methanol, liquefied natural gas, and crude helium using organic Rankine cycle, and solar collectors. J. Therm. Anal. Calorim. 2021, 145, 1485–1508. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. In Functional Genomic; Springer: Cham, Switzerland, 2002; pp. 155–171. [Google Scholar]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.D.; Theodoridis, G.; Virgiliou, C. A perspective on the standards describing mass spectrometry-based metabolic phenotyping (metabolomics/metabonomics) studies in publications. J. Chromatogr. B 2021, 1164, 122515. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Camargo, C.A., Jr.; Hasegawa, K. Metabolomics in the prevention and management of asthma. Expert Rev. Respir. Med. 2019, 13, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Sturm, S. Analytical Aspects of Plant Metabolite Profiling Platforms: Current Standings and Future Aims. J. Proteome Res. 2007, 6, 480–497. [Google Scholar] [CrossRef]
- Mannina, L.; Sobolev, A.P.; Capitani, D.; Iaffaldano, N.; Rosato, M.P.; Ragni, P.; Reale, A.; Sorrentino, E.; D’Amico, I.; Coppola, R. NMR metabolic profiling of organic and aqueous sea bass extracts: Implications in the discrimination of wild and cultured sea bass. Talanta 2008, 77, 433–444. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, E.T.; Eom, J.S.; Choi, Y.Y.; Lee, S.J.; Lee, S.S.; Chung, C.D.; Lee, S.S. Exploration of metabolite profiles in the biofluids of dairy cows by proton nuclear magnetic resonance analysis. PLoS ONE 2021, 16, e0246290. [Google Scholar] [CrossRef]
- Antonelo, D.S.; Cônsolo, N.R.; Gómez, J.F.; Beline, M.; Pavan, B.; Souza, C.; Goulart, R.S.; Colnago, L.A.; Silva, S.L. Meat metabolomic pathway of Nellore and crossbred Angus x Nellore cattle. In Proceedings of the 65th International Congress of Meat Scienece and Technology, Berlin, Germany, 4–9 August 2019. [Google Scholar]
- Picone, G.; Mezzetti, B.; Babini, E.; Capocasa, F.; Placucci, G.; Capozzi, F. Unsupervised Principal Component Analysis of NMR Metabolic Profiles for the Assessment of Substantial Equivalence of Transgenic Grapes (Vitis vinifera). J. Agric. Food Chem. 2011, 59, 9271–9279. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.R. The potentiality of NMR-based metabolomics in food science and food authentication assessment. Magn. Reson. Chem. MRC 2019, 57, 558–578. [Google Scholar] [CrossRef]
- Horlings, L.G.; Marsden, T.K. Towards the real green revolution? Exploring the conceptual dimensions of a new ecological modernisation of agriculture that could ‘feed the world’. Glob. Environ. Change 2011, 21, 441–452. [Google Scholar] [CrossRef]
- Collins, C.M.; Vaskou, P.; Kountouris, Y. Insect food products in the western world: Assessing the potential of a new ‘green’ market. Ann. Entomol. Soc. Am. 2019, 112, 518–528. [Google Scholar] [CrossRef]
- Lamsal, B.; Wang, H.; Pinsirodom, P.; Dossey, A.T. Applications of insect-derived protein ingredients in food and feed industry. J. Am. Oil Chem. Soc. 2019, 96, 105–123. [Google Scholar] [CrossRef]
- Varelas, V. Food wastes as a potential new source for edible insect mass production for food and feed: A review. Fermentation 2019, 5, 81. [Google Scholar] [CrossRef]
- Di Nunzio, M.; Picone, G.; Pasini, F.; Caboni, M.F.; Gianotti, A.; Bordoni, A.; Capozzi, F. Olive oil industry by-products. Effects of a polyphenol-rich extract on the metabolome and response to inflammation in cultured intestinal cell. Food Res. Int. 2018, 113, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Di Nunzio, M.; Picone, G.; Pasini, F.; Chiarello, E.; Caboni, M.F.; Capozzi, F.; Gianotti, A.; Bordoni, A. Olive oil by-product as functional ingredient in bakery products. Influence of processing and evaluation of biological effects. Food Res. Int. 2020, 131, 108940. [Google Scholar] [CrossRef]
- Pampuri, A.; Casson, A.; Alamprese, C.; Di Mattia, C.D.; Piscopo, A.; Difonzo, G.; Conte, P.; Paciulli, M.; Tugnolo, A.; Beghi, R.; et al. Environmental Impact of Food Preparations Enriched with Phenolic Extracts from Olive Oil Mill Waste. Foods 2021, 10, 980. [Google Scholar] [CrossRef]
- Masetti, O.; Sorbo, A.; Nisini, L. NMR Tracing of Food Geographical Origin: The Impact of Seasonality, Cultivar and Production Year on Data Analysis. Separations 2021, 8, 230. [Google Scholar] [CrossRef]
- Longobardi, F.; Ventrella, A.; Napoli, C.; Humpfer, E.; Schütz, B.; Schäfer, H.; Kontominas, M.G.; Sacco, A. Classification of olive oils according to geographical origin by using 1H NMR fingerprinting combined with multivariate analysis. Food Chem. 2012, 130, 177–183. [Google Scholar] [CrossRef]
- Mannina, L.; Marini, F.; Gobbino, M.; Sobolev, A.; Capitani, D. NMR and chemometrics in tracing European olive oils: The case study of Ligurian samples. Talanta 2010, 80, 2141–2148. [Google Scholar] [CrossRef]
- Mannina, L.; Patumi, M.; Proietti, N.; Bassi, D.; Segre, A.L. Geographical characterization of Italian extra virgin olive oils using high-field 1H NMR spectroscopy. J. Agric. Food Chem. 2001, 49, 2687–2696. [Google Scholar] [CrossRef]
- Ingallina, C.; Cerreto, A.; Mannina, L.; Circi, S.; Vista, S.; Capitani, D.; Spano, M.; Sobolev, A.P.; Marini, F. Extra-virgin olive oils from nine Italian regions: An 1H NMR-chemometric characterization. Metabolites 2019, 9, 65. [Google Scholar] [CrossRef]
- Shintu, L.; Caldarelli, S. Toward the determination of the geographical origin of emmental (er) cheese via high resolution MAS NMR: A preliminary investigation. J. Agric. Food Chem. 2006, 54, 4148–4154. [Google Scholar] [CrossRef] [PubMed]
- Brescia, M.; Monfreda, M.; Buccolieri, A.; Carrino, C. Characterisation of the geographical origin of buffalo milk and mozzarella cheese by means of analytical and spectroscopic determinations. Food Chem. 2005, 89, 139–147. [Google Scholar] [CrossRef]
- Piras, C.; Marincola, F.C.; Savorani, F.; Engelsen, S.B.; Cosentino, S.; Viale, S.; Pisano, M.B. A NMR metabolomics study of the ripening process of the Fiore Sardo cheese produced with autochthonous adjunct cultures. Food Chem. 2013, 141, 2137–2147. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.C.; Fernández, I. NMR metabolomics applied on the discrimination of variables influencing tomato (Solanum lycopersicum). Molecules 2020, 25, 3738. [Google Scholar] [CrossRef]
- Masetti, O.; Nisini, L.; Ciampa, A.; Dell’Abate, M.T. 1H NMR spectroscopy coupled with multivariate analysis was applied to investigate Italian cherry tomatoes metabolic profile. J. Chemom. 2020, 34, e3191. [Google Scholar] [CrossRef]
- Capozzi, F.; Savorani, F.; Engelsen, S.; Dell’Abate, M.; Sequi, P. Pomodoro di Pachino: An authentication study using 1H-NMR and chemometrics-protecting its PGI European certification. Magn. Reson. Food Sci. 2009, 106–166. [Google Scholar] [CrossRef]
- Sobolev, A.P.; Carradori, S.; Capitani, D.; Vista, S.; Trella, A.; Marini, F.; Mannina, L. Saffron samples of different origin: An NMR study of microwave-assisted extracts. Foods 2014, 3, 403–419. [Google Scholar] [CrossRef]
- Consonni, R.; Ordoudi, S.A.; Cagliani, L.R.; Tsiangali, M.; Tsimidou, M.Z. On the traceability of commercial saffron samples using 1H-NMR and FT-IR metabolomics. Molecules 2016, 21, 286. [Google Scholar] [CrossRef]
- Di Matteo, G.; Spano, M.; Esposito, C.; Santarcangelo, C.; Baldi, A.; Daglia, M.; Mannina, L.; Ingallina, C.; Sobolev, A.P. Nmr characterization of ten apple cultivars from the piedmont region. Foods 2021, 10, 289. [Google Scholar] [CrossRef]
- Hamid, N.A.A.; Mediani, A.; Maulidiani, M.; Abas, F.; Park, Y.S.; Leontowicz, H.; Leontowicz, M.; Namiesnik, J.; Gorinstein, S. Characterization of metabolites in different kiwifruit varieties by NMR and fluorescence spectroscopy. J. Pharm. Biomed. Anal. 2017, 138, 80–91. [Google Scholar] [CrossRef]
- Do Prado Apparecido, R.; Lopes, T.I.B.; Alcantara, G.B. NMR-based foodomics of common tubers and roots. J. Pharm. Biomed. Anal. 2022, 209, 114527. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Kim, M.; Kim, B.H.; Ahn, S. Identification of the geographical origin of Asian red pepper (Capsicum annuum L.) powders using 1H NMR spectroscopy. Bull. Korean Chem. Soc. 2020, 41, 317–322. [Google Scholar] [CrossRef]
- Lau, H.; Laserna, A.K.C.; Li, S.F.Y. 1H NMR-based metabolomics for the discrimination of celery (Apium graveolens L. var. dulce) from different geographical origins. Food Chem. 2020, 332, 127424. [Google Scholar] [CrossRef] [PubMed]
- Spiteri, C.; Lia, F.; Farrugia, C. Determination of the geographical origin of Maltese honey using 1H NMR fingerprinting. Foods 2020, 9, 1455. [Google Scholar] [CrossRef] [PubMed]
- Schievano, E.; Stocchero, M.; Morelato, E.; Facchin, C.; Mammi, S. An NMR-based metabolomic approach to identify the botanical origin of honey. Metabolomics 2012, 8, 679–690. [Google Scholar] [CrossRef]
- Gerginova, D.; Simova, S.; Popova, M.; Stefova, M.; Stanoeva, J.P.; Bankova, V. NMR profiling of North Macedonian and Bulgarian honeys for detection of botanical and geographical origin. Molecules 2020, 25, 4687. [Google Scholar] [CrossRef]
- Karabagias, I.K.; Vlasiou, M.; Kontakos, S.; Drouza, C.; Kontominas, M.G.; Keramidas, A.D. Geographical discrimination of pine and fir honeys using multivariate analyses of major and minor honey components identified by 1H NMR and HPLC along with physicochemical data. Eur. Food Res. Technol. 2018, 244, 1249–1259. [Google Scholar] [CrossRef]
- Brescia, M.; Caldarola, V.; de Giglio, A.; Benedetti, D.; Fanizzi, F.; Sacco, A. Characterization of the geographical origin of Italian red wines based on traditional and nuclear magnetic resonance spectrometric determinations. Anal. Chim. Acta 2002, 458, 177–186. [Google Scholar] [CrossRef]
- Ogrinc, N.; Košir, I.J.; Kocjančič, M.; Kidrič, J. Determination of authenticy, regional origin, and vintage of Slovenian wines using a combination of IRMS and SNIF-NMR analyses. J. Agric. Food Chem. 2001, 49, 1432–1440. [Google Scholar] [CrossRef]
- Mascellani, A.; Hoca, G.; Babisz, M.; Krska, P.; Kloucek, P.; Havlik, J. 1H NMR chemometric models for classification of Czech wine type and variety. Food Chem. 2021, 339, 127852. [Google Scholar] [CrossRef]
- Gougeon, L.; da Costa, G.; Guyon, F.; Richard, T. 1H NMR metabolomics applied to Bordeaux red wines. Food Chem. 2019, 301, 125257. [Google Scholar] [CrossRef] [PubMed]
- Magdas, D.A.; Pirnau, A.; Feher, I.; Guyon, F.; Cozar, B.I. Alternative approach of applying 1H NMR in conjunction with chemometrics for wine classification. LWT 2019, 109, 422–428. [Google Scholar] [CrossRef]
- Girelli, C.R.; Del Coco, L.; Fanizzi, F.P. Tunisian Extra Virgin Olive Oil Traceability in the EEC Market: Tunisian/Italian (Coratina) EVOOs Blend as a Case Study. Sustainability 2017, 9, 1471. [Google Scholar] [CrossRef]
- Šmejkalová, D.; Piccolo, A. High-power gradient diffusion NMR spectroscopy for the rapid assessment of extra-virgin olive oil adulteration. Food Chem. 2010, 118, 153–158. [Google Scholar] [CrossRef]
- Petrakis, E.A.; Cagliani, L.R.; Tarantilis, P.A.; Polissiou, M.G.; Consonni, R. Sudan dyes in adulterated saffron (Crocus sativus L.): Identification and quantification by 1H NMR. Food Chem. 2017, 217, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, A.P.; Thomas, F.; Donarski, J.; Ingallina, C.; Circi, S.; Marincola, F.C.; Capitani, D.; Mannina, L. Use of NMR applications to tackle future food fraud issues. Trends Food Sci. Technol. 2019, 91, 347–353. [Google Scholar] [CrossRef]
- Milani, M.I.; Rossini, E.L.; Catelani, T.A.; Pezza, L.; Toci, A.T.; Pezza, H.R. Authentication of roasted and ground coffee samples containing multiple adulterants using NMR and a chemometric approach. Food Control 2020, 112, 107104. [Google Scholar] [CrossRef]
- Yong, C.-H.; Muhammad, S.A.; Nasir, F.I.; Mustafa, M.Z.; Ibrahim, B.; Kelly, S.D.; Cannavan, A.; Seow, E.-K. Detecting adulteration of stingless bee honey using untargeted 1H NMR metabolomics with chemometrics. Food Chem. 2022, 368, 130808. [Google Scholar] [CrossRef]
- Schmitt, C.; Bastek, T.; Stelzer, A.; Schneider, T.; Fischer, M.; Hackl, T. Detection of peanut adulteration in food samples by nuclear magnetic resonance spectroscopy. J. Agric. Food Chem. 2020, 68, 14364–14373. [Google Scholar] [CrossRef]
- Rysova, L.; Legarova, V.; Pacakova, Z.; Hanus, O.; Nemeckova, I.; Klimesova, M.; Havlik, J. Detection of bovine milk adulteration in caprine milk with N-acetyl carbohydrate biomarkers by using 1H nuclear magnetic resonance spectroscopy. J. Dairy Sci. 2021, 104, 9583–9595. [Google Scholar] [CrossRef]
- Horn, B.; Esslinger, S.; Fauhl-Hassek, C.; Riedl, J. 1H NMR spectroscopy, one-class classification and outlier diagnosis: A powerful combination for adulteration detection in paprika powder. Food Control 2021, 128, 108205. [Google Scholar] [CrossRef]
- Kuballa, T.; Brunner, T.S.; Thongpanchang, T.; Walch, S.G.; Lachenmeier, D.W. Application of NMR for authentication of honey, beer and spices. Curr. Opin. Food Sci. 2018, 19, 57–62. [Google Scholar] [CrossRef]
- Cifuentes, A. Food analysis and foodomics. J. Chromatogr. A 2009, 1216, 7109. [Google Scholar] [CrossRef] [PubMed]
- Balkir, P.; Kemahlioglu, K.; Yucel, U. Foodomics: A new approach in food quality and safety. Trends Food Sci. Technol. 2021, 108, 49–57. [Google Scholar] [CrossRef]
- Cifuentes, A. Advanced separation methods in food analysis. J. Chromatogr. A 2009, 1216, 7109–7358. [Google Scholar]
- Capozzi, F.; Bordoni, A. Foodomics: A new comprehensive approach to food and nutrition. Genes Nutr. 2013, 8, 1–4. [Google Scholar] [CrossRef]
- Valdés, A.; Álvarez-Rivera, G.; Socas-Rodríguez, B.; Herrero, M.; Ibáñez, E.; Cifuentes, A. Foodomics: Analytical opportunities and challenges. Anal. Chem. 2021, 94, 366–381. [Google Scholar] [CrossRef]
- Cifuentes, A. Food analysis: Present, future, and foodomics. Int. Sch. Res. Not. 2012, 2012, 801607. [Google Scholar] [CrossRef]
- García-Cañas, V.; Simó, C.; Herrero, M.; Ibáñez, E.; Cifuentes, A. Present and future challenges in food analysis: Foodomics. Anal. Chem. 2012, 84, 10150–10159. [Google Scholar] [CrossRef]
- Cifuentes, A. Comprehensive Foodomics; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Gallo, M.; Ferranti, P. The evolution of analytical chemistry methods in foodomics. J. Chromatogr. A 2016, 1428, 3–15. [Google Scholar] [CrossRef]
- Castro-Puyana, M.; Mendiola, J.A.; Ibañez, E. Strategies for a cleaner new scientific discipline of green foodomics. TrAC Trends Anal. Chem. 2013, 52, 23–35. [Google Scholar] [CrossRef]
- Díaz-de-Cerio, E.; Aguilera-Saez, L.M.; Gómez-Caravaca, A.M.; Verardo, V.; Fernández-Gutiérrez, A.; Fernández, I.; Arráez-Román, D. Characterization of bioactive compounds of Annona cherimola L. leaves using a combined approach based on HPLC-ESI-TOF-MS and NMR. Anal. Bioanal. Chem. 2018, 410, 3607–3619. [Google Scholar] [CrossRef] [PubMed]
- Helkar, P.B.; Sahoo, A.K.; Patil, N. Review: Food industry by-products used as a functional food ingredients. Int. J. Waste Resour. 2016, 6, 1–6. [Google Scholar]
- Azizan, A.; Lee, A.X.; Abdul Hamid, N.A.; Maulidiani, M.; Mediani, A.; Abdul Ghafar, S.Z.; Zolkeflee, N.K.Z.; Abas, F. Potentially bioactive metabolites from pineapple waste extracts and their antioxidant and α-glucosidase inhibitory activities by 1H NMR. Foods 2020, 9, 173. [Google Scholar] [CrossRef]
- Cardullo, N.; Leanza, M.; Muccilli, V.; Tringali, C. Valorization of Agri-Food Waste from Pistachio Hard Shells: Extraction of Polyphenols as Natural Antioxidants. Resources 2021, 10, 45. [Google Scholar] [CrossRef]
- Farooque, S.; Rose, P.M.; Benohoud, M.; Blackburn, R.S.; Rayner, C.M. Enhancing the Potential Exploitation of Food Waste: Extraction, Purification, and Characterization of Renewable Specialty Chemicals from Blackcurrants (Ribes nigrum L.). J. Agric. Food Chem. 2018, 66, 12265–12273. [Google Scholar] [CrossRef]
- Hutkins, R.W.; Krumbeck, J.A.; Bindels, L.B.; Cani, P.D.; Fahey, G.; Goh, Y.J.; Hamaker, B.; Martens, E.C.; Mills, D.A.; Rastal, R.A.; et al. Prebiotics: Why definitions matter. Curr. Opin. Biotechnol. 2016, 37, 1–7. [Google Scholar] [CrossRef]
- Kadum, H.; Hamid, A.A.; Abas, F.; Ramli, N.S.; Mohammed, A.K.S.; Muhialdin, B.J.; Jaafar, A.H. Bioactive compounds responsible for antioxidant activity of different varieties of date (Phoenix dactylifera L.) elucidated by 1H-NMR based metabolomics. Int. J. Food Prop. 2019, 22, 462–476. [Google Scholar] [CrossRef]
- Gibson, G.R.; Probert, H.M.; van Loo, J.; Rastall, R.A.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr. Res. Rev. 2004, 17, 259–275. [Google Scholar] [CrossRef]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Haltrich, D. 18—Microbial production of prebiotic oligosaccharides. In Microbial Production of Food Ingredients, Enzymes and Nutraceuticals; McNeil, B., Archer, D., Giavasis, I., Harvey, L., Eds.; Woodhead Publishing: Sawston, UK, 2013; pp. 494–530. [Google Scholar]
- Al-Sheraji, S.H.; Ismail, A.; Manap, M.Y.; Mustafa, S.; Yusof, R.M.; Hassan, F.A. Prebiotics as functional foods: A review. J. Funct. Foods 2013, 5, 1542–1553. [Google Scholar] [CrossRef]
- Liburdi, K.; Esti, M. Galacto-Oligosaccharide (GOS) Synthesis during Enzymatic Lactose-Free Milk Production: State of the Art and Emerging Opportunities. Beverages 2022, 8, 21. [Google Scholar] [CrossRef]
- Awasthi, M.K.; Tarafdar, A.; Gaur, V.K.; Amulya, K.; Narisetty, V.; Yadav, D.K.; Sindhu, R.; Binod, P.; Negi, T.; Pandey, A. Emerging trends of microbial technology for the production of oligosaccharides from biowaste and their potential application as prebiotic. Int. J. Food Microbiol. 2022, 368, 109610. [Google Scholar] [CrossRef]
- Palai, T.; Dubey, K.K. Valorization of dairy industry waste into functional foods using lactase. In Thermochemical and Catalytic Conversion Technologies for Future Biorefineries; Springer: Cham, Switzerland, 2022; pp. 161–183. [Google Scholar]
- Jana, U.K.; Kango, N. Characteristics and bioactive properties of mannooligosaccharides derived from agro-waste mannans. Int. J. Biol. Macromol. 2020, 149, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Narisetty, V.; Parhi, P.; Mohan, B.; Hakkim Hazeena, S.; Naresh Kumar, A.; Gullón, B.; Srivastava, A.; Nair, L.M.; Paul Alphy, M.; Sindhu, R.; et al. Valorization of renewable resources to functional oligosaccharides: Recent trends and future prospective. Bioresour. Technol. 2022, 346, 126590. [Google Scholar] [CrossRef]
- Hussin, F.S.; Chay, S.Y.; Hussin, A.S.M.; Wan Ibadullah, W.Z.; Muhialdin, B.J.; Abd Ghani, M.S.; Saari, N. GABA enhancement by simple carbohydrates in yoghurt fermented using novel, self-cloned Lactobacillus plantarum Taj-Apis362 and metabolomics profiling. Sci. Rep. 2021, 11, 9417. [Google Scholar] [CrossRef]
- Cerulli, A.; Masullo, M.; Piacente, S. Metabolite Profiling of Helichrysum italicum Derived Food Supplements by 1H-NMR-Based Metabolomics. Molecules 2021, 26, 6619. [Google Scholar] [CrossRef]
- Cerulli, A.; Masullo, M.; Montoro, P.; Hošek, J.; Pizza, C.; Piacente, S. Metabolite profiling of “green” extracts of Corylus avellana leaves by 1H NMR spectroscopy and multivariate statistical analysis. J. Pharm. Biomed. Anal. 2018, 160, 168–178. [Google Scholar] [CrossRef]
- Price, N.; Fei, T.; Clark, S.; Wang, T. Extraction of phospholipids from a dairy by-product (whey protein phospholipid concentrate) using ethanol. J. Dairy Sci. 2018, 101, 8778–8787. [Google Scholar] [CrossRef]
- Subratti, A.; Lalgee, L.J.; Jalsa, N.K. Liquified dimethyl ether (DME): A green solvent for the extraction of hemp (Cannabis sativa L.) seed oil. Sustain. Chem. Pharm. 2019, 12, 100144. [Google Scholar] [CrossRef]
- Cerulli, A.; Masullo, M.; Pizza, C.; Piacente, S. Metabolite Profiling of Green Extracts of Cynara cardunculus subsp. scolymus, Cultivar “Carciofo di Paestum” PGI by 1H NMR and HRMS-Based Metabolomics. Molecules 2022, 27, 3328. [Google Scholar] [CrossRef] [PubMed]
- Fratianni, F.; Tucci, M.; de Palma, M.; Pepe, R.; Nazzaro, F. Polyphenolic composition in different parts of some cultivars of globe artichoke (Cynara cardunculus L. var. scolymus (L.) Fiori). Food Chem. 2007, 104, 1282–1286. [Google Scholar] [CrossRef]
- Bordoni, A.; Picone, G.; Babini, E.; Vignali, M.; Danesi, F.; Valli, V.; Di Nunzio, M.; Laghi, L.; Capozzi, F. NMR comparison of in vitro digestion of Parmigiano Reggiano cheese aged 15 and 30 months. Magn. Reson. Chem. 2011, 49, S61–S70. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Vadillo, C.; Nau, F.; Dubiard, C.G.; Cheynier, V.; Meudec, E.; Sanz-Buenhombre, M.; Guadarrama, A.; Tóth, T.; Csavajda, É.; Hingyi, H. In vitro digestion of dairy and egg products enriched with grape extracts: Effect of the food matrix on polyphenol bioaccessibility and antioxidant activity. Food Res. Int. 2016, 88, 284–292. [Google Scholar] [CrossRef]
- Sęczyk, Ł.; Sugier, D.; Świeca, M.; Gawlik-Dziki, U. The effect of in vitro digestion, food matrix, and hydrothermal treatment on the potential bioaccessibility of selected phenolic compounds. Food Chem. 2021, 344, 128581. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Lerma, J.; Fornés-Ferrer, V.; Heredia, A.; Andrés, A. In vitro digestion of lipids in real foods: Influence of lipid organization within the food matrix and interactions with nonlipid components. J. Food Sci. 2018, 83, 2629–2637. [Google Scholar] [CrossRef] [PubMed]
- Čakarević, J.; Torbica, A.; Belović, M.; Tomić, J.; Sedlar, T.; Popović, L. Pumpkin oil cake protein as a new carrier for encapsulation incorporated in food matrix: Effect of processing, storage and in vitro digestion on bioactivity. Int. J. Food Sci. Technol. 2021, 56, 3400–3408. [Google Scholar] [CrossRef]
- Marcolini, E.; Babini, E.; Bordoni, A.; di Nunzio, M.; Laghi, L.; Maczó, A.; Picone, G.; Szerdahelyi, E.; Valli, V.; Capozzi, F. Bioaccessibility of the Bioactive Peptide Carnosine during in Vitro Digestion of Cured Beef Meat. J. Agric. Food Chem. 2015, 63, 4973–4978. [Google Scholar] [CrossRef]
- Bordoni, A.; Laghi, L.; Babini, E.; di Nunzio, M.; Picone, G.; Ciampa, A.; Valli, V.; Danesi, F.; Capozzi, F. The foodomics approach for the evaluation of protein bioaccessibility in processed meat upon in vitro digestion. Electrophoresis 2014, 35, 1607–1614. [Google Scholar] [CrossRef]
- Mahomoodally, M.F.; Dilmar, A.S.; Khadaroo, S.K. Chapter 4.2—Carnosine. In Antioxidants Effects in Health; Nabavi, S.M., Silva, A.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 251–268. [Google Scholar]
- Fonteles, T.V.; Alves Filho, E.d.G.; Karolina de Araújo Barroso, M.; de Fátima Dantas Linhares, M.; Rabelo, M.C.; Silva, L.M.A.e.; Sousa de Brito, E.; Wurlitzer, N.J.; Rodrigues Pereira, E.P.; Ferreira, B.M.; et al. Protective effect of inulin on thermally treated acerola juice: In vitro bioaccessibility of bioactive compounds. Food Biosci. 2021, 41, 101018. [Google Scholar] [CrossRef]
- Vidal, N.P.; Picone, G.; Goicoechea, E.; Laghi, L.; Manzanos, M.J.; Danesi, F.; Bordoni, A.; Capozzi, F.; Guillén, M.D. Metabolite release and protein hydrolysis during the in vitro digestion of cooked sea bass fillets. A study by 1H NMR. Food Res. Int. 2016, 88, 293–301. [Google Scholar] [CrossRef]
- Nieva-Echevarría, B.; Goicoechea, E.; Manzanos, M.J.; Guillén, M.D. A method based on 1H NMR spectral data useful to evaluate the hydrolysis level in complex lipid mixtures. Food Res. Int. 2014, 66, 379–387. [Google Scholar] [CrossRef]
- Nieva-Echevarria, B.; Goicoechea, E.; Manzanos, M.; Guillen, M. Usefulness of 1H NMR to study the food lipolysis during in vitro digestion. In Magnetic Resonance in Food Science: Defining Food by Magnetic Resonance; Royal Society of Chemistry: London, UK, 2015; pp. 31–38. [Google Scholar]
- Bordoni, A.; Capozzi, F. Foodomics for healthy nutrition. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Smith, F.; Cliff, M.T.; Capozzi, F.; Mills, E.C. The application of nutrimetabolomics to investigating the bioaccessibility of nutrients in ham using a batch in vitro digestion model. Food Nutr. Sci. 2014, 2014, 41706. [Google Scholar]
| NMR | MS | Is NMR ‘Greener’ than MS? | |
|---|---|---|---|
| Sensitivity and Selectivity | Low sensitivity (can be improved using microfluidics, dynamic nuclear polarization, …); Generally used for nonselective analysis | High sensitivity (nanomolar); Can be used for both selective (targeted) and nonselective (nontargeted) analyses | |
| Sample measurement | All metabolites that have NMR concentration level can be detected in one measurement | Usually needs different chromatography techniques for different classes of metabolites |  |
| Number of detectable metabolites | 40–200 depending on spectral resolution | ≥300 (depending on MS techniques, whether GC-MS or LC-MS is used) | |
| Reproducibility | Very high | Moderate | |
| Sample preparation | Minimal | Complex (needs different columns and ionization methods) |  |
| Tissue extraction | Not required (tissues can be analyzed directly using HRMAS NMR) | Yes, requires tissue extraction |  |
| Sample recovery | Nondestructive; sample can be recovered and stored for a long time; several analyses can be carried out on the same sample | Destructive technique but need a small amount of sample |  |
| Sample analysis time | Fast (the entire sample can be analyzed in one measurement) | Longer (requires different chromatography techniques depending on the metabolites analyzed) |  |
| Sample cost | Low cost per sample | High cost per sample, more expensive than NMR |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciampa, A.; Danesi, F.; Picone, G. NMR-Based Metabolomics for a More Holistic and Sustainable Research in Food Quality Assessment: A Narrative Review. Appl. Sci. 2023, 13, 372. https://doi.org/10.3390/app13010372
Ciampa A, Danesi F, Picone G. NMR-Based Metabolomics for a More Holistic and Sustainable Research in Food Quality Assessment: A Narrative Review. Applied Sciences. 2023; 13(1):372. https://doi.org/10.3390/app13010372
Chicago/Turabian StyleCiampa, Alessandra, Francesca Danesi, and Gianfranco Picone. 2023. "NMR-Based Metabolomics for a More Holistic and Sustainable Research in Food Quality Assessment: A Narrative Review" Applied Sciences 13, no. 1: 372. https://doi.org/10.3390/app13010372
APA StyleCiampa, A., Danesi, F., & Picone, G. (2023). NMR-Based Metabolomics for a More Holistic and Sustainable Research in Food Quality Assessment: A Narrative Review. Applied Sciences, 13(1), 372. https://doi.org/10.3390/app13010372









