Abstract
The ability of nuclear magnetic resonance spectroscopy (NMR) to extract chemical information from a complex mixture is invaluable and widely described in literature. Many applications of this technique in the foodomics field have highlighted how NMR could characterize food matrices, and it can be used all along its “life chain”: from farm to fork and from fork to the digestion process. The aim of this review is an attempt to show, firstly, the potential of NMR as a method based on green chemistry in sample preparation, and then in characterizing the nutritional qualities of agri-food products (with particular attention to their by-products) from a sustainable point of view. For instance, the NMR-based metabolomics approach has been used to enhance the nutritional properties of bio-products waste naturally rich in antioxidants and prebiotics. The reintroduction of these products in the food supply chain as functional foods or ingredients answers and satisfies the consumer demand for more food with high nutritional quality and more respect for the environment.
1. Introduction
Food quality, food authenticity, and food safety are the most valuable targets to ensure and guarantee the needs of a world population that is increasing higher and higher. Consumers ask for much more security and, at the same time, for better information on food chemical and nutritional composition, origin, authenticity, and the effect of technological transformation on both the food’s molecular profile and human health [1]. Furthermore, the bioavailability and bioaccessibility of the nutrients and/or bioactive compounds (BC), and the prediction of nutrition efficiency are additional key concepts in which consumers begin to be interested in [2]. They also become more sensitive to the environmental aspect, considering the deep global warming alarm in 2022, one of the warmest years since 2005 [3]. Nowadays, the attention on “what we eat” reflects not only the food’s nutritional-health aspects but also “avoiding derivatization and favoring substances based on renewable sources” [4] by respecting the environment. In light of these global warming events, academics together with industry must work to ensure consumers’ requests and to improve their human life. These pathways go hand-hand with sustainable development inevitably.
Thus, how can academics answer to the consumers’ needs while being at the same time environmentally friendly? Secondly, how can NMR-based metabolomics play a key role in this scenario?
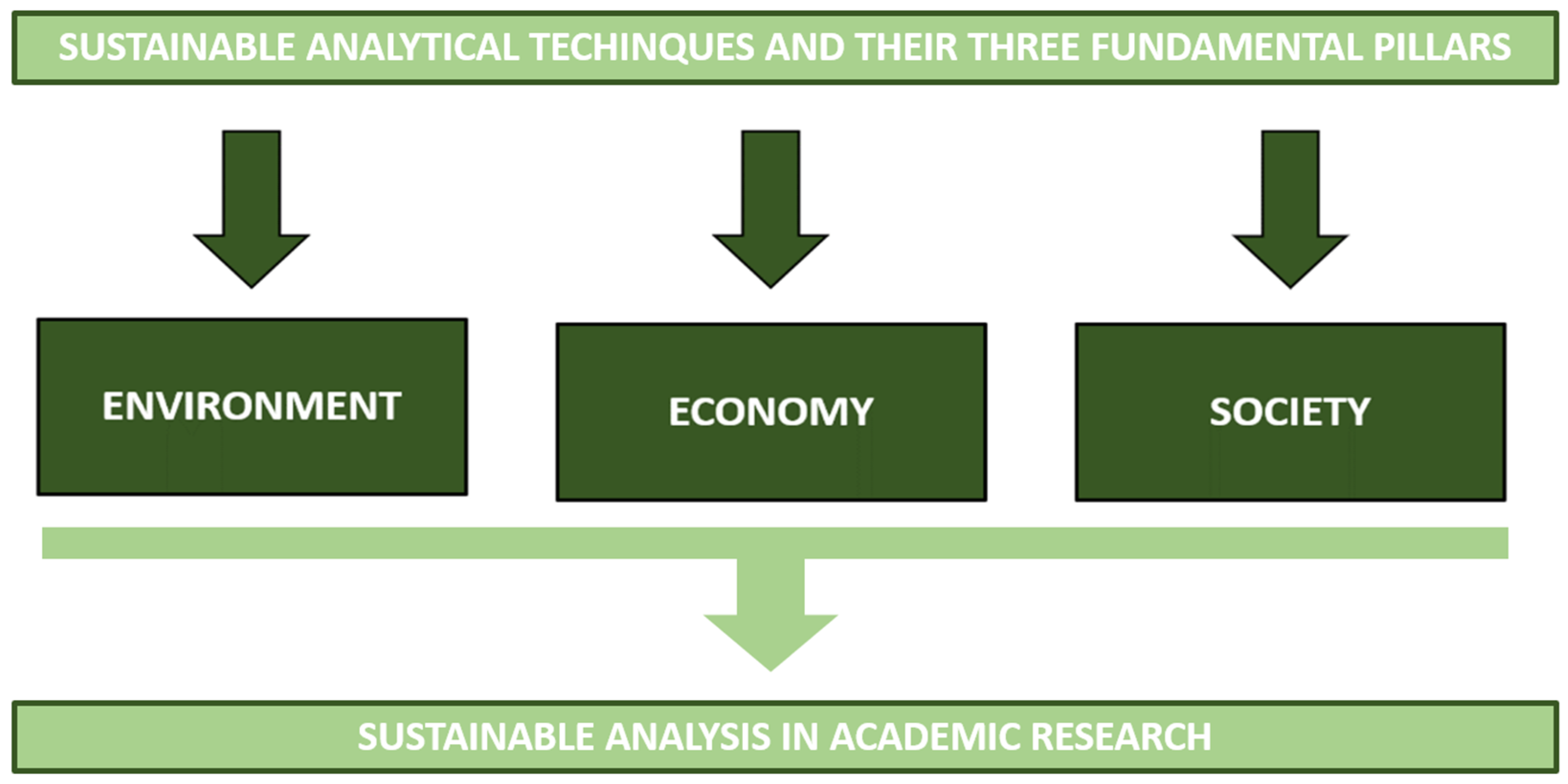
Before answering these two questions it is important to keep in mind the meaning of the word sustainable when we take into account academic research and, thus, analytical techniques. Płotka-Wasylka, et al. [5] defined sustainable as those analytical techniques that take into consideration three fundamental pillars: (1) the environment, (2) the economy, and (3) the society (Figure 1).
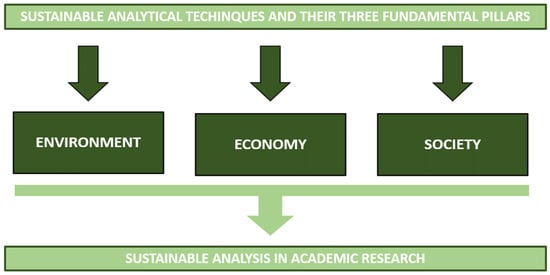
Figure 1.
Sustainable analysis in academic research starts from sustainable analytical techniques that have to take into consideration three fundamental pillars: environment, economy, and society.
Pillar 1: Sustainability can be achieved by waste minimization, adopting green solvents, or focusing on solventless methods, energy savings, and trying to avoid the use of auxiliary reagents and chemicals [5]. Sustainable analytical techniques should also be considered to limit pollution.
Pillar 2: Academic research is generally recognized as curiosity-driven. However, there has recently been a shift in this, which was also influenced by the types of funding available. Research has become more industry-oriented and practical than conceptual. It has also started to suggest green solutions to problems and have an economic impact. Therefore, in the current context, food analysis and quality evaluation need to be supported by procedures and analytical techniques whose challenge is to reduce the environmental footprint [5].
Pillar 3: The main educational task is to transmit a clear message to society that would shed light on chemistry as a fundamental part of the solution to pollution problems and not just part of the problem. Information on the possibility of “making science” with low environmental impact is a fundamental leitmotif that society should have clearly in mind.
In the context of analytical chemistry, as well as in food analytical chemistry, some well-established methods require hazardous chemicals and/or a high energy demand for sample preservation, pretreatment, calibration, and analyte determination. Frequently, the analytical methods yield large amounts of waste with even higher toxicity than the target analytes. Thus, analytical chemistry plays an important role in the sustainable development of the planet [5]. For this reason, green chemistry (GC) is considered an important tool for achieving sustainability since it aims at the development of chemicals and chemical processes to reduce the impact on human health and the environment. But how can it be achieved? Poliakoff, et al. [6] suggest a deep change in attitudes and behavior in the chemical industry all along the chemical supply chain. For example, at the start of the chain, laboratories, where research is carried out, should be rethought and built to minimize the use of energy. At the other end of the chain, wherever possible, the amount of chemical(s) used to achieve a given effect should be decreased by a factor of two every five years [6], according to Moore’s Law for Chemistry (MLFC) [7]. In this scenario, scientists are thus encouraged to develop or adopt new green analytical methods that (1) contribute to the reduction of pollution, including real-time analysis of pollutants; (2) avoid sample pretreatment or involve greener approaches for sample pretreatment, including safe solvents and auxiliaries; (3) use miniaturization and automation, including flow analysis and microfluidics and (4) use green separation techniques.
Based on these aspects, it is now possible to answer the second question: how can NMR-based metabolomics play a key role in this scenario? The High Resolution (HR)-NMR is a successful and functional research technology because of both its peculiarities which are listed in pillar 1 above. HR-NMR spectroscopy is one of the main analytical technologies that operates following the green chemistry guidelines in sample preparation [8]. Mielko, et al. [9] recognized the NMR as a green method as it requires for metabolomics studies only 10% D2O (deuterated water) in 90% of distilled water (H2Odd). The solution has the characteristic of being versatile as it can be used in different studies. For example, this aqueous solution has been adopted for urine and serum fluid analysis, as shown by Trimigno, et al. [10] and Münger, et al. [11]. The authors used simply a phosphate-buffered saline (PBS) which also includes sodium azide (NaN3) as an antibacterial agent and a 20 mM 2-chloropyrimidine-5-carboxylic acid (2CLPYR5CA) as the reference standard. Also, in the case of food metabolomics analysis, the NMR spectroscopic method requires simplicity in the analysis of polar metabolites. Their extraction can be performed by using a solution of trichloroacetic acid (TCA) 7% including 10% D2O [12,13]. This application has been described in the work of Ciampa, et al. [14], where the authors show how the NMR satisfies, for the quantification of trimethylamine (TMA) content in fish, all the validation requirements at the same level as the most frequently used methods (as HPLC). Furthermore, the technique has the advantage of being faster and more repeatable, avoiding the use of solvents, such as toluene and formaldehyde, or dangerous reagents, such as picric acid.
In addition to the lower environmental impacts in sample preparations, other peculiar characteristics make NMR spectroscopic methods very attractive for metabolomics analysis. Several papers highlight its capability to offer qualitative, as well as quantitative, knowledge about complex biological samples [15] and their high reproducibility. At the same level the NMR weaknesses, such as limited sensitivity and resolution of the spectra, are largely documented [9,16,17,18], and summarized in Table 1 in which we compared the two most important platforms used for metabolomics analysis, such as Mass Spectroscopy (MS) and NMR as well.

Table 1.
Key differences between NMR and MS as green tools for metabolomics research (adapted from Emwas [17]).
Anyhow, significant developments have been made to enhance the NMR sensitivity, including microprobes, cryogenically cooled probes [19], and the dynamic nuclear polarization (DNP) approach [20]. Another critical point about this technique regards the huge amounts of cryogens (helium and nitrogen) required that are impacting the environment quite deeply. The increasing growth of cryogen consumption and their limited resources are the challenges that industries will face in the future [21]. Anyhow, the present review is focused on the green aspect concerning NMR-based metabolomics samples’ preparation so the cryogens matter has not been considered, even if their reduction is still a big deal.
2. The NMR-Based Metabolomics in Food Science and the Foodomics Approach
It is important, before exploring the potentiality of the NMR in metabolomics studies, to clarify what metabolomics studies are. The term “metabolomics” was introduced by Oliver Fiehn and defined as a comprehensive and quantitative analysis of all metabolites in a system [22]. On the other hand, the term “metabonomics” was coined by J. K. Nicholson in 1999 [23], and it represents “the quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification.” Nowadays, modern metabolic phenotyping (metabotyping) is known as metabolomics, encompassing both the comprehensive analysis of the small molecule content of the tested samples, and the changes that occur in response to a stimulus of one sort or another (physiological, pharmacological, or toxicological) [24].
The given definition points out the importance of the HR-NMR in “the augmentation and complementation of the information provided by measuring the genetic and proteomic responses to xenobiotic exposure” as it is appropriate “for investigating abnormal body fluid compositions”, as a wide range of metabolites can be quantified simultaneously with no sample preparation and “without prejudice” [23].
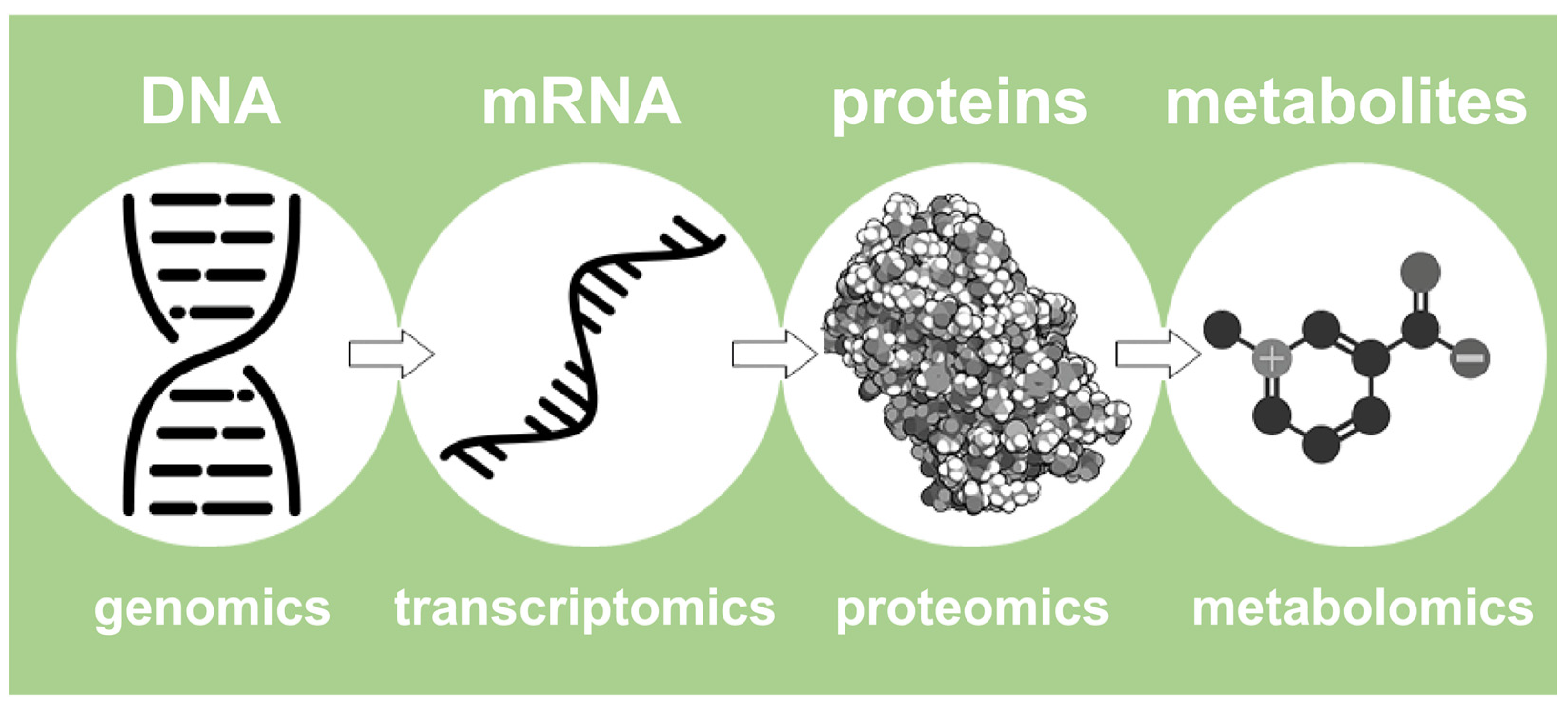
In this light, Nicholson describes de facto the NMR-based metabolomics approach. In summary, it represents a high-performance fingerprinting process that examines the entire collection of small molecules (typically <1800 Dalton) that are present in a concentration above 10 µM, including sugars, amino acids, organic acids, and lipids, and is called ‘metabolome’ [25]. In summary, metabolomics is the final step of a more structured pipeline that involves more omics platforms, such as proteomics, transcriptomics, and genomics [22], as represented in Figure 2.
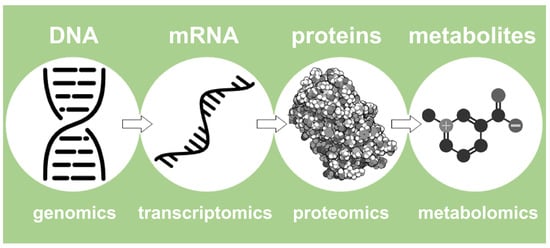
Figure 2.
An outline of the four most important omics fields, ranging from genomics to metabolomics.
This integrated “snapshot” may change throughout the exposure to external factors or during organism development. When dealing with this technique, almost two kinds of approaches can be used for the analysis of the metabolome: (i) the target analysis, which is metabolic profiling, and (ii) pattern analysis, which is metabolic and metabonomic fingerprinting. Target analysis, or metabolic profiling, is the identification and quantification of given and predefined metabolites [26]. It can be a specific metabolite or a selected number of metabolites. In the first case, a selective extraction is necessary before the NMR analysis to concentrate the selected metabolite. The latter case is used when the attention is focused on the specific role of a selected metabolic pathway; this approach is called metabolic profiling [27]. The pattern analysis is metabolic profiling obtained by identifying and quantifying all metabolites. This operation is led by using specific software, such as Chenomx [28,29], and it is called ‘metabonomics.’ On the other hand, metabolic profiling is used when sample classification without quantification of individual-specific metabolites is required [27]. In this case, the NMR spectrum becomes a fingerprint of the product, and all the NMR resonances/signals are measured without any identification [30], and it is called ‘fingerprinting’. Due to the above-all-mentioned characteristics, it appears clear why in the last 15 years academic researchers have employed NMR-based metabolomics in food science more and more. With this technology, academics meet the consumers’ requests in one shot: a green approach that both pays attention to the environment in sample preparation and can assess and guarantee the quality of foodstuff. It is not a coincidence that with the opening of the food markets at a worldwide level, the number of research papers on the evaluation of food quality by using an NMR-metabolomics approach increased. Consonni and Cagliani [31] explain that the globalization phenomenon changes the definition of food quality that have to include also other aspects like geographical origin, sophisticated frauds, and adulteration practices, etc. The expansion of the food quality concept is also determined nowadays by the introduction on the market of new foods formulated to be much more sustainable and green [32]. These innovative foods are designed considering unconventional sources of nutrients, such as insects [33,34,35], for example, but also converting waste products into second-life products [36,37,38]. For this reason, consumers have to be reassured about food safety and from a nutritional value point of view. As these new aspects overcome the official analytical determination focused on specific compounds investigation, the role of metabolomics in this new quality assessment has become very important [31]. The importance is proved by the tremendous number of papers dealing with metabolomics and NMR that appear in the bibliography. Among them, geographical origin plays a key role as food quality is strongly affected by the particular conditions of production areas, which give unrepeatable organoleptic and nutritional properties to agricultural food products [39]. Extensive studies have been performed on several different foods to find out biomarkers able to classify them concerning their geographic origin: extra virgin olive oil (EVOO) [40,41,42,43], cheese [44,45,46], tomato [47,48,49], saffron [50,51], fruits and vegetables [52,53,54,55,56], honey [57,58,59,60] and wine [61,62,63,64,65], etc. Food frauds and adulteration have been largely taken in consideration in NMR-based metabolomics studies as well [66,67,68,69,70,71,72,73,74,75]. The application of NMR-based metabolomics in a food quality context can be included in the so-called foodomics, and we can talk about NMR-based foodomics. The first definition of this omics approach appeared in 2009 by Cifuentes [76], and it put attention on the investigation of all the possible connections among food, diet, and the individual, including health and ill impact [16]. These connections are included in the suffix “omics”, which comes from the Latin word “omne” and it means everything, totality, wholeness, and entirety [77]. A more holistic definition by Cifuentes [78] described foodomics as “the discipline that studies the food and nutrition domains through the application and integration of advanced omics technologies to improve consumer’s well-being, health, and confidence”. This new approach then had much more resonance during the first International Conference on Foodomics, held in Cesena in 2009, where scientists were invited to contribute to the holistic definition of food in a multidisciplinary environment [79]. The academics’ contribution to this new discipline is largely demonstrated by several papers describing in proper matter the aim and the applications of foodomics [77,80,81,82,83,84].
2.1. The NMR-Based Foodomics Approach for Food Bio-Waste or By-Products Analysis
The valorization of the green aspects in the NMR-based foodomics leads to a green NMR-based foodomics approach [13], and it answers much more to the consumers’ “2.0” needs in terms of both food safety and quality, and sustainability. The green attribution has a twofold meaning: (1) green because, as it has been previously underlined, it involves “environment-friendly” chemicals (green chemistry). Castro-Puyana, et al. [85] demonstrate how foodomics can emerge as a green discipline by applying the basic concepts of Green Chemistry; (2) it is green because its challenge is to preserve sustainability, understood as a way of using and preserving what nature gives us in the most productive, environmentally safe way [85]. In the last years, foodomics started dealing with food ingredients able to improve our health, which means nutrients and BC. Thus, the extraction of BC from natural sources (such as plants, algae, food bio-waste, and food by-products, among others) and the quality analysis of functional foods (FF) is a key step in the foodomics workflow [16,85].
Food bio-waste or by-products from various sources can be fundamental for BC production and FF formulation. The high added value of these wastes is due to the content of BC, which, most of the time, is greater than in the edible part of the fruit [86]. For this reason, their proper waste management plays a vital role in the growth of food industries as they can contain valuable components such as polysaccharides, proteins, fats, fibers, and flavor compounds [87]. Azizan, et al. [88], by using 1H NMR, described bioactive metabolites from pineapple waste. These compounds have antioxidant capacity and they can inhibit the activity of a carbohydrate-active enzyme (α-glucosidase). In conclusion, it emerges that 3-methylglutaric acid, threonine, valine, and α-linolenic acid were the main contributors to the antioxidant activities, whereas epicatechin was responsible for the α-glucosidase inhibitory activity. The metabolic composition of cherimoya leaves (Annona cherimola Mill.) is also characterized by the presence of BC, such as phenolic compounds. It is a deciduous tree from the Annonaceae family, typical of Peru and Ecuador, and its fruits are largely produced in Spain [86]. In this case, the metabolic composition has been profiled by using the 1H NMR coupled with high-performance liquid chromatography coupled with time-of-flight mass spectrometry (HPLC-TOF-MS). The first (1H NMR) achieved 23 primary compounds, classified as amino acids, organic acids, carbohydrates, choline, phenolic acid derivatives, and flavonoids. The second (HPLC-TOF-MS) profiled 66 secondary metabolites among carbohydrates, amino acids, phenolic acids and derivatives, flavonoids, phenylpropanoids, and other polar compounds [86]. Similar antioxidant proprieties have also been identified in pistachio’s hard shells [89], blackcurrant skin [90], olive oil by-products [36,37], and date palm by-products [91]. In this last case, a strong antioxidant activity has also been attributed to date fruits, whose bioactive metabolites have been profiled by 1H NMR by Kadum, et al. [92]. In this work, five different cultivars have been compared to find the one with the most antioxidant activity. The obtained results demonstrated that the extract from the Piyarom variety had both the highest total phenolic and flavonoid content and exhibited good antioxidant activity. The metabolites responsible for the antioxidant activity, glucose, ascorbic acid, epicatechin, gallic acid, and citric acid, were successfully identified using 1H NMR-based metabolomics [92]. Among the BC, a particular interest is covered by prebiotics which are defined as “a nondigestible food ingredient that beneficially affects the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria in the colon, and thus improves host health” [93,94]. To guarantee their bioactivity, prebiotics have to (i) be resistant to gastric acidity; (ii) avoid absorption in the gastrointestinal tract; (iii) not be subjected to enzymatic activity; (iv) be fermented by intestinal microflora to produce short-chain fatty acids, and (v) be a selective substrate for the growth of intestinal bacteria associated to human health and well-being [95,96]. The main prebiotic agents consist of nondigestible carbohydrates, such as polysaccharides (POS), oligosaccharides (OS), and soluble dietary fibers (mainly inulin) [96]. Prebiotic foods are largely requested on the market; thus, food industries are working on the development of sustainable bioprocesses that can guarantee OS from a waste of different sources [97,98,99]. An important resource of OS comes from lignocellulosic agricultural waste disposals. Apart from bioethanol generation, several studies demonstrated the conversion of lignocellulosic biomass into oligosaccharides, through both microbial fermentation and enzymatic hydrolysis, by microorganisms or enzymes [100,101]. Jana and Kango [100], through enzymatic hydrolysis, produced Mannooligosaccharides (MOS) from agricultural waste and their structural aspects were assessed using both 1H and 13C NMR spectroscopy. MOS respect all the important criteria of being a prebiotic as they are (i) gastrointestinal resistant, (ii) an active substrate promoting the growth of healthy bacteria, and (iii) able to confer antineoplastic properties without any cytotoxicity to normal cells [100]. The capability of prebiotics to enhance the growth of healthy bacteria can be exploited for the production of important physiological metabolites. An example is given by recent research from Hussin, et al. [102]. The study investigated the effect of different commercial prebiotics on enhancing natural gamma-aminobutyric acid (GABA) production in cultured yogurt. In addition, the metabolomics profile of the fermentation-derived biomolecules in yogurt has been obtained via NMR-based metabolomics. This approach was useful for comparing the major metabolite profile of freeze-dried GABA-rich yogurt (GY) and standard freeze-dried yogurt (SY). A total of 16 and 13 compounds were detected in GY and SY, respectively, and this difference may be due to the strain-specific metabolic activities of GABA [102]. According to pillar 1, described in the introduction, the choice of extraction methods to obtain any kind of BC is fundamental. Ethanol is an example of a green and sustainable bio-solvent as it is completely biodegradable and obtained by the fermentation of sugar-rich materials [103,104,105]. Also, dimethyl ether (DME), having a high cetane number and favorable carbon/oxygen ratio, emerged in the spotlight as a cleaner, environmentally friendly, and high-efficiency ignition fuel [106]. Thus, green solvents are among the preferred ones for extraction processes. For example, Cerulli, et al. [107], compared several ethanol-based solutions to find out the best one for the extraction of primary metabolites and inulin from “Carciofo di Paestum” (C. cardunculus subsp. scolymus) PGI heads. For the first time, a comprehensive investigation of green extracts was performed by LC-MS and NMR analysis to highlight the occurrence of both specialized and primary metabolites, respectively. Among all the extracts, hydroalcoholic, infusion, and decoction extracts were the most interesting, showing higher peaks for flavonoids, quinic acid derivatives, and inulin than MeOH extract [107]. Inulin is a polymer found widely distributed in nature as a plant storage carbohydrate with a high value in human nutrition. It has prebiotic proprieties for its resistance to digestion in the small intestine, it can be fermented by colonic microflora, and it can stimulate the proliferation of commensal bacteria [107]. Hence, artichokes are a source of prebiotic dietary fibers but also a source of metabolites with antioxidant activity associated with their high phenolic content [108].
2.2. The NMR-Based Foodomics Approach for the Simulated Digestion and Absorption of Food
Food is a biological matrix where different food components are organized in a complex structure and interact with each other. The digestion process is needed to break the structure to release nutrients and other BC. Thus, the overall nutritional value of food and bioactivity of its components relies on the bioavailability, not only on the amount of both nutrients and BC [109]. Their bioavailability depends on several factors, including their bioaccessibility, which means the amount that can be released from the matrix during digestion and pass into the soluble fraction, becoming available for absorption. The composition of the food matrix and the processing which foods are subjected can influence digestion and, in turn, the bioaccessibility of its components. From this point of view, the evaluation of the in vitro digestibility of different food matrices subjected to different types of processing or transformation, for example, enriched with BC from by-products, represents a fundamental parameter for the determination of the nutritional value of foods, and the possible effect of nutrients and BC [110,111,112,113]. NMR-based metabolomics showed a fundamental role in the determination of both the bioavailability and bioaccessibility of BC in natural or enriched foods during simulated digestion and absorption.
The application of NMR methodology in this field respects the aim described in pillar 1: both the in vitro digestion and NMR analysis follow the green chemistry guidelines in sample preparation [109]. For the NMR, samples taken during digestion are prepared by adding to 1 mL of each sample, 160 µL of 1 m phosphate buffer in deuterium oxide (D2O), containing 10 mm 3-TSP as internal standard. According to the evaluation of the bioaccessibility of BC, a work by Marcolini, et al. [114] demonstrated how it depends on several conditions, such as (1) digestion conditions (pH, enzyme concentration, and activity), (2) compound stability, and (3) interactions with other food components, as well as the supramolecular organization of the food molecules [115]. In particular, the work focused on the effect of pH on the solubility and bioaccessibility of carnosine obtained by the digestion of bresaola, a typical beef-based product of northern Italy (Valtellina) [114]. Carnosine is a water-soluble dipeptide with functional and biological properties, especially as an antioxidant [116].
The dependence of BC bioaccessibility and bioavailability from the supramolecular organization of molecules in food, in summary, the food matrix, is also well described by Fonteles, et al. [117]. The research paper described the effect of inulin on the bioaccessibility and bioavailability of BC naturally present in acerola juice by applying both in vitro digestion and NMR protocols. After inulin addition, the prebiotic juice and a non-prebiotic one were first treated by to two different thermal processes and then subjected to in vitro digestion to evaluate the bioactive juice compounds’ bioaccessibility and the juice composition. Inulin, beyond its prebiotic proprieties, also has a protective effect in preserving the organic compounds after industrial processes such as thermal processing, and also improving the bioaccessibility of BC in the treated juice [117].
The combination of in vitro digestion and 1H NMR has also been developed and employed in a work by Vidal, et al. [118], where the first aim of the paper was the study of the hydrophilic metabolites of raw and cooked sea bass (Dicentrarchus labrax) through NMR. First of all, the spectrometry technique has been used for both metabolites’ identification and quantification, as well as to evaluate the effect of microwave cooking on them. The interest in this study was great because the results provided information concerning the presence and concentration of BCs among the hydrophilic metabolites of raw and cooked sea bass, and because it enabled monitoring of the advance of the proteolysis and of the release of hydrophilic metabolites at different points of the in vitro digestion of cooked sea bass fillets. Other interesting works based on 1H NMR spectral data to study qualitatively and quantitatively the hydrolysis level in complex lipid mixtures, such as fish samples, have been published by Nieva-Echevarría, et al. [119,120].
3. Conclusions
Different applications in food assessment, from quality to digestibility, can be successfully addressed by the application of the 1H NMR-based metabolomics together with approaches such as in vitro digestion. Along with the power of the NMR technique, its attention to sustainability and low environmental impact in sample preparation were also emphasized. The green (NMR-based metabolomics) foodomics makes a valuable contribution to metabolites’ profiling of complex samples, such as foods and food by-products. Bioactive compounds like prebiotics or antioxidant metabolites can easily extract from these products by following and respecting the green chemistry protocols, for example, by using water/ethanol solutions. The applications of this approach are not limited to what has been described as food metabolomics, but several works in the nutritional field have been reported in the literature, and it goes under the name of “nutri-metabonomics”. This new “omics” approach can be considered the foodomics for healthy nutrition, and it offers valid tools to investigate the effect of nutrients or specific dietary components (BC, for example) on the human metabolome and metabolic regulation [121,122]. Therefore, it is clear that the NMR approach can meet the consumers’ needs in an environmentally friendly way while also being a good method to detect food quality parameters. The next step to make NMR much greener towards the environment’s needs is also to limit the use of cryogens liquids which are a serious problem considering their scarcity.
Author Contributions
Conceptualization, G.P.; writing—original draft preparation, G.P.; writing—review and editing, F.D. and A.C.; visualization, G.P., F.D. and A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially supported by the Italian Ministry of University and Research MUR—NRRP funding (project number SOE_0000116, funded to G. Picone).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, S.; Tian, Y.; Jiang, P.; Lin, Y.; Liu, X.; Yang, H. Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. Nutr. 2021, 61, 1448–1469. [Google Scholar] [CrossRef] [PubMed]
- Fernández-García, E.; Carvajal-Lérida, I.; Pérez-Gálvez, A. In vitro bioaccessibility assessment as a prediction tool of nutritional efficiency. Nutr. Res. 2009, 29, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, K.C.; Agache, I.; Jutel, M.; Annesi Maesano, I.; Akdis, M.; Sampath, V.; d’Amato, G.; Cecchi, L.; Traidl-Hoffmann, C.; Akdis, C.A. Climate change: A call to action for the united nations. Allergy 2022, 77, 1087–1090. [Google Scholar] [CrossRef] [PubMed]
- Tobiszewski, M.; Mechlińska, A.; Namieśnik, J. Green analytical chemistry—Theory and practice. Chem. Soc. Rev. 2010, 39, 2869–2878. [Google Scholar] [CrossRef]
- Płotka-Wasylka, J.; Mohamed, H.M.; Kurowska-Susdorf, A.; Dewani, R.; Fares, M.Y.; Andruch, V. Green analytical chemistry as an integral part of sustainable education development. Curr. Opin. Green Sustain. Chem. 2021, 31, 100508. [Google Scholar] [CrossRef]
- Poliakoff, M.; Licence, P.; George, M.W. UN sustainable development goals: How can sustainable/green chemistry contribute? By doing things differently. Curr. Opin. Green Sustain. Chem. 2018, 13, 146–149. [Google Scholar] [CrossRef]
- Poliakoff, M.; Licence, P.; George, M.W. A new approach to sustainability: A Moor’s law for chemistry. Angew. Chem. Int. Ed. 2018, 57, 12590–12591. [Google Scholar] [CrossRef]
- Gottlieb, H.E.; Graczyk-Millbrandt, G.; Inglis, G.G.; Nudelman, A.; Perez, D.; Qian, Y.; Shuster, L.E.; Sneddon, H.F.; Upton, R.J. Development of GSK’s NMR guides—A tool to encourage the use of more sustainable solvents. Green Chem. 2016, 18, 3867–3878. [Google Scholar] [CrossRef]
- Mielko, K.A.; Pudełko-Malik, N.; Tarczewska, A.; Młynarz, P. NMR spectroscopy as a “green analytical method” in metabolomics and proteomics studies. Sustain. Chem. Pharm. 2021, 22, 100474. [Google Scholar] [CrossRef]
- Trimigno, A.; Münger, L.; Picone, G.; Freiburghaus, C.; Pimentel, G.; Vionnet, N.; Pralong, F.; Capozzi, F.; Badertscher, R.; Vergères, G. GC-MS based metabolomics and NMR spectroscopy investigation of food intake biomarkers for milk and cheese in serum of healthy humans. Metabolites 2018, 8, 26. [Google Scholar] [CrossRef]
- Münger, L.H.; Trimigno, A.; Picone, G.; Freiburghaus, C.; Pimentel, G.g.; Burton, K.J.; Pralong, F.o.P.; Vionnet, N.; Capozzi, F.; Badertscher, R. Identification of urinary food intake biomarkers for milk, cheese, and soy-based drink by untargeted GC-MS and NMR in healthy humans. J. Proteome Res. 2017, 16, 3321–3335. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Laghi, L.; Picone, G.; Capozzi, F. Nuclear magnetic resonance for foodomics beyond food analysis. TrAC Trends Anal. Chem. 2014, 59, 93–102. [Google Scholar] [CrossRef]
- Ciampa, A.; Laghi, L.; Picone, G. Validation of a 1H-NMR Spectroscopy Quantitative Method to Quantify Trimethylamine Content and K-Index Value in Different Species of Fish. J. Food Qual. 2022, 2022, 3612095. [Google Scholar] [CrossRef]
- Bisht, B.; Kumar, V.; Gururani, P.; Tomar, M.S.; Nanda, M.; Vlaskin, M.S.; Kumar, S.; Kurbatova, A. The potential of nuclear magnetic resonance (NMR) in metabolomics and lipidomics of microalgae—A review. Arch. Biochem. Biophys. 2021, 710, 108987. [Google Scholar] [CrossRef] [PubMed]
- Picone, G.; Mengucci, C.; Capozzi, F. The NMR added value to the green foodomics perspective: Advances by machine learning to the holistic view on food and nutrition. Magn. Reson. Chem. 2022, 60, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. In Metabonomics; Springer: Cham, Switzerland, 2015; pp. 161–193. [Google Scholar]
- Letertre, M.P.; Giraudeau, P.; De Tullio, P. Nuclear magnetic resonance spectroscopy in clinical metabolomics and personalized medicine: Current challenges and perspectives. Front. Mol. Biosci. 2021, 8, 698337. [Google Scholar] [CrossRef]
- Robosky, L.C.; Reily, M.D.; Avizonis, D. Improving NMR sensitivity by use of salt-tolerant cryogenically cooled probes. Anal. Bioanal. Chem. 2007, 387, 529–532. [Google Scholar] [CrossRef]
- Lee, J.H.; Okuno, Y.; Cavagnero, S. Sensitivity enhancement in solution NMR: Emerging ideas and new frontiers. J. Magn. Reson. 2014, 241, 18–31. [Google Scholar] [CrossRef]
- Ghorbani, B.; Amidpour, M. Energy, exergy, and sensitivity analyses of a new integrated system for generation of liquid methanol, liquefied natural gas, and crude helium using organic Rankine cycle, and solar collectors. J. Therm. Anal. Calorim. 2021, 145, 1485–1508. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. In Functional Genomic; Springer: Cham, Switzerland, 2002; pp. 155–171. [Google Scholar]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.D.; Theodoridis, G.; Virgiliou, C. A perspective on the standards describing mass spectrometry-based metabolic phenotyping (metabolomics/metabonomics) studies in publications. J. Chromatogr. B 2021, 1164, 122515. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Camargo, C.A., Jr.; Hasegawa, K. Metabolomics in the prevention and management of asthma. Expert Rev. Respir. Med. 2019, 13, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Sturm, S. Analytical Aspects of Plant Metabolite Profiling Platforms: Current Standings and Future Aims. J. Proteome Res. 2007, 6, 480–497. [Google Scholar] [CrossRef]
- Mannina, L.; Sobolev, A.P.; Capitani, D.; Iaffaldano, N.; Rosato, M.P.; Ragni, P.; Reale, A.; Sorrentino, E.; D’Amico, I.; Coppola, R. NMR metabolic profiling of organic and aqueous sea bass extracts: Implications in the discrimination of wild and cultured sea bass. Talanta 2008, 77, 433–444. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, E.T.; Eom, J.S.; Choi, Y.Y.; Lee, S.J.; Lee, S.S.; Chung, C.D.; Lee, S.S. Exploration of metabolite profiles in the biofluids of dairy cows by proton nuclear magnetic resonance analysis. PLoS ONE 2021, 16, e0246290. [Google Scholar] [CrossRef]
- Antonelo, D.S.; Cônsolo, N.R.; Gómez, J.F.; Beline, M.; Pavan, B.; Souza, C.; Goulart, R.S.; Colnago, L.A.; Silva, S.L. Meat metabolomic pathway of Nellore and crossbred Angus x Nellore cattle. In Proceedings of the 65th International Congress of Meat Scienece and Technology, Berlin, Germany, 4–9 August 2019. [Google Scholar]
- Picone, G.; Mezzetti, B.; Babini, E.; Capocasa, F.; Placucci, G.; Capozzi, F. Unsupervised Principal Component Analysis of NMR Metabolic Profiles for the Assessment of Substantial Equivalence of Transgenic Grapes (Vitis vinifera). J. Agric. Food Chem. 2011, 59, 9271–9279. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.R. The potentiality of NMR-based metabolomics in food science and food authentication assessment. Magn. Reson. Chem. MRC 2019, 57, 558–578. [Google Scholar] [CrossRef]
- Horlings, L.G.; Marsden, T.K. Towards the real green revolution? Exploring the conceptual dimensions of a new ecological modernisation of agriculture that could ‘feed the world’. Glob. Environ. Change 2011, 21, 441–452. [Google Scholar] [CrossRef]
- Collins, C.M.; Vaskou, P.; Kountouris, Y. Insect food products in the western world: Assessing the potential of a new ‘green’ market. Ann. Entomol. Soc. Am. 2019, 112, 518–528. [Google Scholar] [CrossRef]
- Lamsal, B.; Wang, H.; Pinsirodom, P.; Dossey, A.T. Applications of insect-derived protein ingredients in food and feed industry. J. Am. Oil Chem. Soc. 2019, 96, 105–123. [Google Scholar] [CrossRef]
- Varelas, V. Food wastes as a potential new source for edible insect mass production for food and feed: A review. Fermentation 2019, 5, 81. [Google Scholar] [CrossRef]
- Di Nunzio, M.; Picone, G.; Pasini, F.; Caboni, M.F.; Gianotti, A.; Bordoni, A.; Capozzi, F. Olive oil industry by-products. Effects of a polyphenol-rich extract on the metabolome and response to inflammation in cultured intestinal cell. Food Res. Int. 2018, 113, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Di Nunzio, M.; Picone, G.; Pasini, F.; Chiarello, E.; Caboni, M.F.; Capozzi, F.; Gianotti, A.; Bordoni, A. Olive oil by-product as functional ingredient in bakery products. Influence of processing and evaluation of biological effects. Food Res. Int. 2020, 131, 108940. [Google Scholar] [CrossRef]
- Pampuri, A.; Casson, A.; Alamprese, C.; Di Mattia, C.D.; Piscopo, A.; Difonzo, G.; Conte, P.; Paciulli, M.; Tugnolo, A.; Beghi, R.; et al. Environmental Impact of Food Preparations Enriched with Phenolic Extracts from Olive Oil Mill Waste. Foods 2021, 10, 980. [Google Scholar] [CrossRef]
- Masetti, O.; Sorbo, A.; Nisini, L. NMR Tracing of Food Geographical Origin: The Impact of Seasonality, Cultivar and Production Year on Data Analysis. Separations 2021, 8, 230. [Google Scholar] [CrossRef]
- Longobardi, F.; Ventrella, A.; Napoli, C.; Humpfer, E.; Schütz, B.; Schäfer, H.; Kontominas, M.G.; Sacco, A. Classification of olive oils according to geographical origin by using 1H NMR fingerprinting combined with multivariate analysis. Food Chem. 2012, 130, 177–183. [Google Scholar] [CrossRef]
- Mannina, L.; Marini, F.; Gobbino, M.; Sobolev, A.; Capitani, D. NMR and chemometrics in tracing European olive oils: The case study of Ligurian samples. Talanta 2010, 80, 2141–2148. [Google Scholar] [CrossRef]
- Mannina, L.; Patumi, M.; Proietti, N.; Bassi, D.; Segre, A.L. Geographical characterization of Italian extra virgin olive oils using high-field 1H NMR spectroscopy. J. Agric. Food Chem. 2001, 49, 2687–2696. [Google Scholar] [CrossRef]
- Ingallina, C.; Cerreto, A.; Mannina, L.; Circi, S.; Vista, S.; Capitani, D.; Spano, M.; Sobolev, A.P.; Marini, F. Extra-virgin olive oils from nine Italian regions: An 1H NMR-chemometric characterization. Metabolites 2019, 9, 65. [Google Scholar] [CrossRef]
- Shintu, L.; Caldarelli, S. Toward the determination of the geographical origin of emmental (er) cheese via high resolution MAS NMR: A preliminary investigation. J. Agric. Food Chem. 2006, 54, 4148–4154. [Google Scholar] [CrossRef] [PubMed]
- Brescia, M.; Monfreda, M.; Buccolieri, A.; Carrino, C. Characterisation of the geographical origin of buffalo milk and mozzarella cheese by means of analytical and spectroscopic determinations. Food Chem. 2005, 89, 139–147. [Google Scholar] [CrossRef]
- Piras, C.; Marincola, F.C.; Savorani, F.; Engelsen, S.B.; Cosentino, S.; Viale, S.; Pisano, M.B. A NMR metabolomics study of the ripening process of the Fiore Sardo cheese produced with autochthonous adjunct cultures. Food Chem. 2013, 141, 2137–2147. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.C.; Fernández, I. NMR metabolomics applied on the discrimination of variables influencing tomato (Solanum lycopersicum). Molecules 2020, 25, 3738. [Google Scholar] [CrossRef]
- Masetti, O.; Nisini, L.; Ciampa, A.; Dell’Abate, M.T. 1H NMR spectroscopy coupled with multivariate analysis was applied to investigate Italian cherry tomatoes metabolic profile. J. Chemom. 2020, 34, e3191. [Google Scholar] [CrossRef]
- Capozzi, F.; Savorani, F.; Engelsen, S.; Dell’Abate, M.; Sequi, P. Pomodoro di Pachino: An authentication study using 1H-NMR and chemometrics-protecting its PGI European certification. Magn. Reson. Food Sci. 2009, 106–166. [Google Scholar] [CrossRef]
- Sobolev, A.P.; Carradori, S.; Capitani, D.; Vista, S.; Trella, A.; Marini, F.; Mannina, L. Saffron samples of different origin: An NMR study of microwave-assisted extracts. Foods 2014, 3, 403–419. [Google Scholar] [CrossRef]
- Consonni, R.; Ordoudi, S.A.; Cagliani, L.R.; Tsiangali, M.; Tsimidou, M.Z. On the traceability of commercial saffron samples using 1H-NMR and FT-IR metabolomics. Molecules 2016, 21, 286. [Google Scholar] [CrossRef]
- Di Matteo, G.; Spano, M.; Esposito, C.; Santarcangelo, C.; Baldi, A.; Daglia, M.; Mannina, L.; Ingallina, C.; Sobolev, A.P. Nmr characterization of ten apple cultivars from the piedmont region. Foods 2021, 10, 289. [Google Scholar] [CrossRef]
- Hamid, N.A.A.; Mediani, A.; Maulidiani, M.; Abas, F.; Park, Y.S.; Leontowicz, H.; Leontowicz, M.; Namiesnik, J.; Gorinstein, S. Characterization of metabolites in different kiwifruit varieties by NMR and fluorescence spectroscopy. J. Pharm. Biomed. Anal. 2017, 138, 80–91. [Google Scholar] [CrossRef]
- Do Prado Apparecido, R.; Lopes, T.I.B.; Alcantara, G.B. NMR-based foodomics of common tubers and roots. J. Pharm. Biomed. Anal. 2022, 209, 114527. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Kim, M.; Kim, B.H.; Ahn, S. Identification of the geographical origin of Asian red pepper (Capsicum annuum L.) powders using 1H NMR spectroscopy. Bull. Korean Chem. Soc. 2020, 41, 317–322. [Google Scholar] [CrossRef]
- Lau, H.; Laserna, A.K.C.; Li, S.F.Y. 1H NMR-based metabolomics for the discrimination of celery (Apium graveolens L. var. dulce) from different geographical origins. Food Chem. 2020, 332, 127424. [Google Scholar] [CrossRef] [PubMed]
- Spiteri, C.; Lia, F.; Farrugia, C. Determination of the geographical origin of Maltese honey using 1H NMR fingerprinting. Foods 2020, 9, 1455. [Google Scholar] [CrossRef] [PubMed]
- Schievano, E.; Stocchero, M.; Morelato, E.; Facchin, C.; Mammi, S. An NMR-based metabolomic approach to identify the botanical origin of honey. Metabolomics 2012, 8, 679–690. [Google Scholar] [CrossRef]
- Gerginova, D.; Simova, S.; Popova, M.; Stefova, M.; Stanoeva, J.P.; Bankova, V. NMR profiling of North Macedonian and Bulgarian honeys for detection of botanical and geographical origin. Molecules 2020, 25, 4687. [Google Scholar] [CrossRef]
- Karabagias, I.K.; Vlasiou, M.; Kontakos, S.; Drouza, C.; Kontominas, M.G.; Keramidas, A.D. Geographical discrimination of pine and fir honeys using multivariate analyses of major and minor honey components identified by 1H NMR and HPLC along with physicochemical data. Eur. Food Res. Technol. 2018, 244, 1249–1259. [Google Scholar] [CrossRef]
- Brescia, M.; Caldarola, V.; de Giglio, A.; Benedetti, D.; Fanizzi, F.; Sacco, A. Characterization of the geographical origin of Italian red wines based on traditional and nuclear magnetic resonance spectrometric determinations. Anal. Chim. Acta 2002, 458, 177–186. [Google Scholar] [CrossRef]
- Ogrinc, N.; Košir, I.J.; Kocjančič, M.; Kidrič, J. Determination of authenticy, regional origin, and vintage of Slovenian wines using a combination of IRMS and SNIF-NMR analyses. J. Agric. Food Chem. 2001, 49, 1432–1440. [Google Scholar] [CrossRef]
- Mascellani, A.; Hoca, G.; Babisz, M.; Krska, P.; Kloucek, P.; Havlik, J. 1H NMR chemometric models for classification of Czech wine type and variety. Food Chem. 2021, 339, 127852. [Google Scholar] [CrossRef]
- Gougeon, L.; da Costa, G.; Guyon, F.; Richard, T. 1H NMR metabolomics applied to Bordeaux red wines. Food Chem. 2019, 301, 125257. [Google Scholar] [CrossRef] [PubMed]
- Magdas, D.A.; Pirnau, A.; Feher, I.; Guyon, F.; Cozar, B.I. Alternative approach of applying 1H NMR in conjunction with chemometrics for wine classification. LWT 2019, 109, 422–428. [Google Scholar] [CrossRef]
- Girelli, C.R.; Del Coco, L.; Fanizzi, F.P. Tunisian Extra Virgin Olive Oil Traceability in the EEC Market: Tunisian/Italian (Coratina) EVOOs Blend as a Case Study. Sustainability 2017, 9, 1471. [Google Scholar] [CrossRef]
- Šmejkalová, D.; Piccolo, A. High-power gradient diffusion NMR spectroscopy for the rapid assessment of extra-virgin olive oil adulteration. Food Chem. 2010, 118, 153–158. [Google Scholar] [CrossRef]
- Petrakis, E.A.; Cagliani, L.R.; Tarantilis, P.A.; Polissiou, M.G.; Consonni, R. Sudan dyes in adulterated saffron (Crocus sativus L.): Identification and quantification by 1H NMR. Food Chem. 2017, 217, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, A.P.; Thomas, F.; Donarski, J.; Ingallina, C.; Circi, S.; Marincola, F.C.; Capitani, D.; Mannina, L. Use of NMR applications to tackle future food fraud issues. Trends Food Sci. Technol. 2019, 91, 347–353. [Google Scholar] [CrossRef]
- Milani, M.I.; Rossini, E.L.; Catelani, T.A.; Pezza, L.; Toci, A.T.; Pezza, H.R. Authentication of roasted and ground coffee samples containing multiple adulterants using NMR and a chemometric approach. Food Control 2020, 112, 107104. [Google Scholar] [CrossRef]
- Yong, C.-H.; Muhammad, S.A.; Nasir, F.I.; Mustafa, M.Z.; Ibrahim, B.; Kelly, S.D.; Cannavan, A.; Seow, E.-K. Detecting adulteration of stingless bee honey using untargeted 1H NMR metabolomics with chemometrics. Food Chem. 2022, 368, 130808. [Google Scholar] [CrossRef]
- Schmitt, C.; Bastek, T.; Stelzer, A.; Schneider, T.; Fischer, M.; Hackl, T. Detection of peanut adulteration in food samples by nuclear magnetic resonance spectroscopy. J. Agric. Food Chem. 2020, 68, 14364–14373. [Google Scholar] [CrossRef]
- Rysova, L.; Legarova, V.; Pacakova, Z.; Hanus, O.; Nemeckova, I.; Klimesova, M.; Havlik, J. Detection of bovine milk adulteration in caprine milk with N-acetyl carbohydrate biomarkers by using 1H nuclear magnetic resonance spectroscopy. J. Dairy Sci. 2021, 104, 9583–9595. [Google Scholar] [CrossRef]
- Horn, B.; Esslinger, S.; Fauhl-Hassek, C.; Riedl, J. 1H NMR spectroscopy, one-class classification and outlier diagnosis: A powerful combination for adulteration detection in paprika powder. Food Control 2021, 128, 108205. [Google Scholar] [CrossRef]
- Kuballa, T.; Brunner, T.S.; Thongpanchang, T.; Walch, S.G.; Lachenmeier, D.W. Application of NMR for authentication of honey, beer and spices. Curr. Opin. Food Sci. 2018, 19, 57–62. [Google Scholar] [CrossRef]
- Cifuentes, A. Food analysis and foodomics. J. Chromatogr. A 2009, 1216, 7109. [Google Scholar] [CrossRef] [PubMed]
- Balkir, P.; Kemahlioglu, K.; Yucel, U. Foodomics: A new approach in food quality and safety. Trends Food Sci. Technol. 2021, 108, 49–57. [Google Scholar] [CrossRef]
- Cifuentes, A. Advanced separation methods in food analysis. J. Chromatogr. A 2009, 1216, 7109–7358. [Google Scholar]
- Capozzi, F.; Bordoni, A. Foodomics: A new comprehensive approach to food and nutrition. Genes Nutr. 2013, 8, 1–4. [Google Scholar] [CrossRef]
- Valdés, A.; Álvarez-Rivera, G.; Socas-Rodríguez, B.; Herrero, M.; Ibáñez, E.; Cifuentes, A. Foodomics: Analytical opportunities and challenges. Anal. Chem. 2021, 94, 366–381. [Google Scholar] [CrossRef]
- Cifuentes, A. Food analysis: Present, future, and foodomics. Int. Sch. Res. Not. 2012, 2012, 801607. [Google Scholar] [CrossRef]
- García-Cañas, V.; Simó, C.; Herrero, M.; Ibáñez, E.; Cifuentes, A. Present and future challenges in food analysis: Foodomics. Anal. Chem. 2012, 84, 10150–10159. [Google Scholar] [CrossRef]
- Cifuentes, A. Comprehensive Foodomics; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Gallo, M.; Ferranti, P. The evolution of analytical chemistry methods in foodomics. J. Chromatogr. A 2016, 1428, 3–15. [Google Scholar] [CrossRef]
- Castro-Puyana, M.; Mendiola, J.A.; Ibañez, E. Strategies for a cleaner new scientific discipline of green foodomics. TrAC Trends Anal. Chem. 2013, 52, 23–35. [Google Scholar] [CrossRef]
- Díaz-de-Cerio, E.; Aguilera-Saez, L.M.; Gómez-Caravaca, A.M.; Verardo, V.; Fernández-Gutiérrez, A.; Fernández, I.; Arráez-Román, D. Characterization of bioactive compounds of Annona cherimola L. leaves using a combined approach based on HPLC-ESI-TOF-MS and NMR. Anal. Bioanal. Chem. 2018, 410, 3607–3619. [Google Scholar] [CrossRef] [PubMed]
- Helkar, P.B.; Sahoo, A.K.; Patil, N. Review: Food industry by-products used as a functional food ingredients. Int. J. Waste Resour. 2016, 6, 1–6. [Google Scholar]
- Azizan, A.; Lee, A.X.; Abdul Hamid, N.A.; Maulidiani, M.; Mediani, A.; Abdul Ghafar, S.Z.; Zolkeflee, N.K.Z.; Abas, F. Potentially bioactive metabolites from pineapple waste extracts and their antioxidant and α-glucosidase inhibitory activities by 1H NMR. Foods 2020, 9, 173. [Google Scholar] [CrossRef]
- Cardullo, N.; Leanza, M.; Muccilli, V.; Tringali, C. Valorization of Agri-Food Waste from Pistachio Hard Shells: Extraction of Polyphenols as Natural Antioxidants. Resources 2021, 10, 45. [Google Scholar] [CrossRef]
- Farooque, S.; Rose, P.M.; Benohoud, M.; Blackburn, R.S.; Rayner, C.M. Enhancing the Potential Exploitation of Food Waste: Extraction, Purification, and Characterization of Renewable Specialty Chemicals from Blackcurrants (Ribes nigrum L.). J. Agric. Food Chem. 2018, 66, 12265–12273. [Google Scholar] [CrossRef]
- Hutkins, R.W.; Krumbeck, J.A.; Bindels, L.B.; Cani, P.D.; Fahey, G.; Goh, Y.J.; Hamaker, B.; Martens, E.C.; Mills, D.A.; Rastal, R.A.; et al. Prebiotics: Why definitions matter. Curr. Opin. Biotechnol. 2016, 37, 1–7. [Google Scholar] [CrossRef]
- Kadum, H.; Hamid, A.A.; Abas, F.; Ramli, N.S.; Mohammed, A.K.S.; Muhialdin, B.J.; Jaafar, A.H. Bioactive compounds responsible for antioxidant activity of different varieties of date (Phoenix dactylifera L.) elucidated by 1H-NMR based metabolomics. Int. J. Food Prop. 2019, 22, 462–476. [Google Scholar] [CrossRef]
- Gibson, G.R.; Probert, H.M.; van Loo, J.; Rastall, R.A.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr. Res. Rev. 2004, 17, 259–275. [Google Scholar] [CrossRef]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Haltrich, D. 18—Microbial production of prebiotic oligosaccharides. In Microbial Production of Food Ingredients, Enzymes and Nutraceuticals; McNeil, B., Archer, D., Giavasis, I., Harvey, L., Eds.; Woodhead Publishing: Sawston, UK, 2013; pp. 494–530. [Google Scholar]
- Al-Sheraji, S.H.; Ismail, A.; Manap, M.Y.; Mustafa, S.; Yusof, R.M.; Hassan, F.A. Prebiotics as functional foods: A review. J. Funct. Foods 2013, 5, 1542–1553. [Google Scholar] [CrossRef]
- Liburdi, K.; Esti, M. Galacto-Oligosaccharide (GOS) Synthesis during Enzymatic Lactose-Free Milk Production: State of the Art and Emerging Opportunities. Beverages 2022, 8, 21. [Google Scholar] [CrossRef]
- Awasthi, M.K.; Tarafdar, A.; Gaur, V.K.; Amulya, K.; Narisetty, V.; Yadav, D.K.; Sindhu, R.; Binod, P.; Negi, T.; Pandey, A. Emerging trends of microbial technology for the production of oligosaccharides from biowaste and their potential application as prebiotic. Int. J. Food Microbiol. 2022, 368, 109610. [Google Scholar] [CrossRef]
- Palai, T.; Dubey, K.K. Valorization of dairy industry waste into functional foods using lactase. In Thermochemical and Catalytic Conversion Technologies for Future Biorefineries; Springer: Cham, Switzerland, 2022; pp. 161–183. [Google Scholar]
- Jana, U.K.; Kango, N. Characteristics and bioactive properties of mannooligosaccharides derived from agro-waste mannans. Int. J. Biol. Macromol. 2020, 149, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Narisetty, V.; Parhi, P.; Mohan, B.; Hakkim Hazeena, S.; Naresh Kumar, A.; Gullón, B.; Srivastava, A.; Nair, L.M.; Paul Alphy, M.; Sindhu, R.; et al. Valorization of renewable resources to functional oligosaccharides: Recent trends and future prospective. Bioresour. Technol. 2022, 346, 126590. [Google Scholar] [CrossRef]
- Hussin, F.S.; Chay, S.Y.; Hussin, A.S.M.; Wan Ibadullah, W.Z.; Muhialdin, B.J.; Abd Ghani, M.S.; Saari, N. GABA enhancement by simple carbohydrates in yoghurt fermented using novel, self-cloned Lactobacillus plantarum Taj-Apis362 and metabolomics profiling. Sci. Rep. 2021, 11, 9417. [Google Scholar] [CrossRef]
- Cerulli, A.; Masullo, M.; Piacente, S. Metabolite Profiling of Helichrysum italicum Derived Food Supplements by 1H-NMR-Based Metabolomics. Molecules 2021, 26, 6619. [Google Scholar] [CrossRef]
- Cerulli, A.; Masullo, M.; Montoro, P.; Hošek, J.; Pizza, C.; Piacente, S. Metabolite profiling of “green” extracts of Corylus avellana leaves by 1H NMR spectroscopy and multivariate statistical analysis. J. Pharm. Biomed. Anal. 2018, 160, 168–178. [Google Scholar] [CrossRef]
- Price, N.; Fei, T.; Clark, S.; Wang, T. Extraction of phospholipids from a dairy by-product (whey protein phospholipid concentrate) using ethanol. J. Dairy Sci. 2018, 101, 8778–8787. [Google Scholar] [CrossRef]
- Subratti, A.; Lalgee, L.J.; Jalsa, N.K. Liquified dimethyl ether (DME): A green solvent for the extraction of hemp (Cannabis sativa L.) seed oil. Sustain. Chem. Pharm. 2019, 12, 100144. [Google Scholar] [CrossRef]
- Cerulli, A.; Masullo, M.; Pizza, C.; Piacente, S. Metabolite Profiling of Green Extracts of Cynara cardunculus subsp. scolymus, Cultivar “Carciofo di Paestum” PGI by 1H NMR and HRMS-Based Metabolomics. Molecules 2022, 27, 3328. [Google Scholar] [CrossRef] [PubMed]
- Fratianni, F.; Tucci, M.; de Palma, M.; Pepe, R.; Nazzaro, F. Polyphenolic composition in different parts of some cultivars of globe artichoke (Cynara cardunculus L. var. scolymus (L.) Fiori). Food Chem. 2007, 104, 1282–1286. [Google Scholar] [CrossRef]
- Bordoni, A.; Picone, G.; Babini, E.; Vignali, M.; Danesi, F.; Valli, V.; Di Nunzio, M.; Laghi, L.; Capozzi, F. NMR comparison of in vitro digestion of Parmigiano Reggiano cheese aged 15 and 30 months. Magn. Reson. Chem. 2011, 49, S61–S70. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Vadillo, C.; Nau, F.; Dubiard, C.G.; Cheynier, V.; Meudec, E.; Sanz-Buenhombre, M.; Guadarrama, A.; Tóth, T.; Csavajda, É.; Hingyi, H. In vitro digestion of dairy and egg products enriched with grape extracts: Effect of the food matrix on polyphenol bioaccessibility and antioxidant activity. Food Res. Int. 2016, 88, 284–292. [Google Scholar] [CrossRef]
- Sęczyk, Ł.; Sugier, D.; Świeca, M.; Gawlik-Dziki, U. The effect of in vitro digestion, food matrix, and hydrothermal treatment on the potential bioaccessibility of selected phenolic compounds. Food Chem. 2021, 344, 128581. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Lerma, J.; Fornés-Ferrer, V.; Heredia, A.; Andrés, A. In vitro digestion of lipids in real foods: Influence of lipid organization within the food matrix and interactions with nonlipid components. J. Food Sci. 2018, 83, 2629–2637. [Google Scholar] [CrossRef] [PubMed]
- Čakarević, J.; Torbica, A.; Belović, M.; Tomić, J.; Sedlar, T.; Popović, L. Pumpkin oil cake protein as a new carrier for encapsulation incorporated in food matrix: Effect of processing, storage and in vitro digestion on bioactivity. Int. J. Food Sci. Technol. 2021, 56, 3400–3408. [Google Scholar] [CrossRef]
- Marcolini, E.; Babini, E.; Bordoni, A.; di Nunzio, M.; Laghi, L.; Maczó, A.; Picone, G.; Szerdahelyi, E.; Valli, V.; Capozzi, F. Bioaccessibility of the Bioactive Peptide Carnosine during in Vitro Digestion of Cured Beef Meat. J. Agric. Food Chem. 2015, 63, 4973–4978. [Google Scholar] [CrossRef]
- Bordoni, A.; Laghi, L.; Babini, E.; di Nunzio, M.; Picone, G.; Ciampa, A.; Valli, V.; Danesi, F.; Capozzi, F. The foodomics approach for the evaluation of protein bioaccessibility in processed meat upon in vitro digestion. Electrophoresis 2014, 35, 1607–1614. [Google Scholar] [CrossRef]
- Mahomoodally, M.F.; Dilmar, A.S.; Khadaroo, S.K. Chapter 4.2—Carnosine. In Antioxidants Effects in Health; Nabavi, S.M., Silva, A.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 251–268. [Google Scholar]
- Fonteles, T.V.; Alves Filho, E.d.G.; Karolina de Araújo Barroso, M.; de Fátima Dantas Linhares, M.; Rabelo, M.C.; Silva, L.M.A.e.; Sousa de Brito, E.; Wurlitzer, N.J.; Rodrigues Pereira, E.P.; Ferreira, B.M.; et al. Protective effect of inulin on thermally treated acerola juice: In vitro bioaccessibility of bioactive compounds. Food Biosci. 2021, 41, 101018. [Google Scholar] [CrossRef]
- Vidal, N.P.; Picone, G.; Goicoechea, E.; Laghi, L.; Manzanos, M.J.; Danesi, F.; Bordoni, A.; Capozzi, F.; Guillén, M.D. Metabolite release and protein hydrolysis during the in vitro digestion of cooked sea bass fillets. A study by 1H NMR. Food Res. Int. 2016, 88, 293–301. [Google Scholar] [CrossRef]
- Nieva-Echevarría, B.; Goicoechea, E.; Manzanos, M.J.; Guillén, M.D. A method based on 1H NMR spectral data useful to evaluate the hydrolysis level in complex lipid mixtures. Food Res. Int. 2014, 66, 379–387. [Google Scholar] [CrossRef]
- Nieva-Echevarria, B.; Goicoechea, E.; Manzanos, M.; Guillen, M. Usefulness of 1H NMR to study the food lipolysis during in vitro digestion. In Magnetic Resonance in Food Science: Defining Food by Magnetic Resonance; Royal Society of Chemistry: London, UK, 2015; pp. 31–38. [Google Scholar]
- Bordoni, A.; Capozzi, F. Foodomics for healthy nutrition. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Smith, F.; Cliff, M.T.; Capozzi, F.; Mills, E.C. The application of nutrimetabolomics to investigating the bioaccessibility of nutrients in ham using a batch in vitro digestion model. Food Nutr. Sci. 2014, 2014, 41706. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).