Presence of Known and Emerging Honey Bee Pathogens in Apiaries of Veneto Region (Northeast of Italy) during Spring 2020 and 2021
Abstract
1. Introduction
2. Materials and Methods
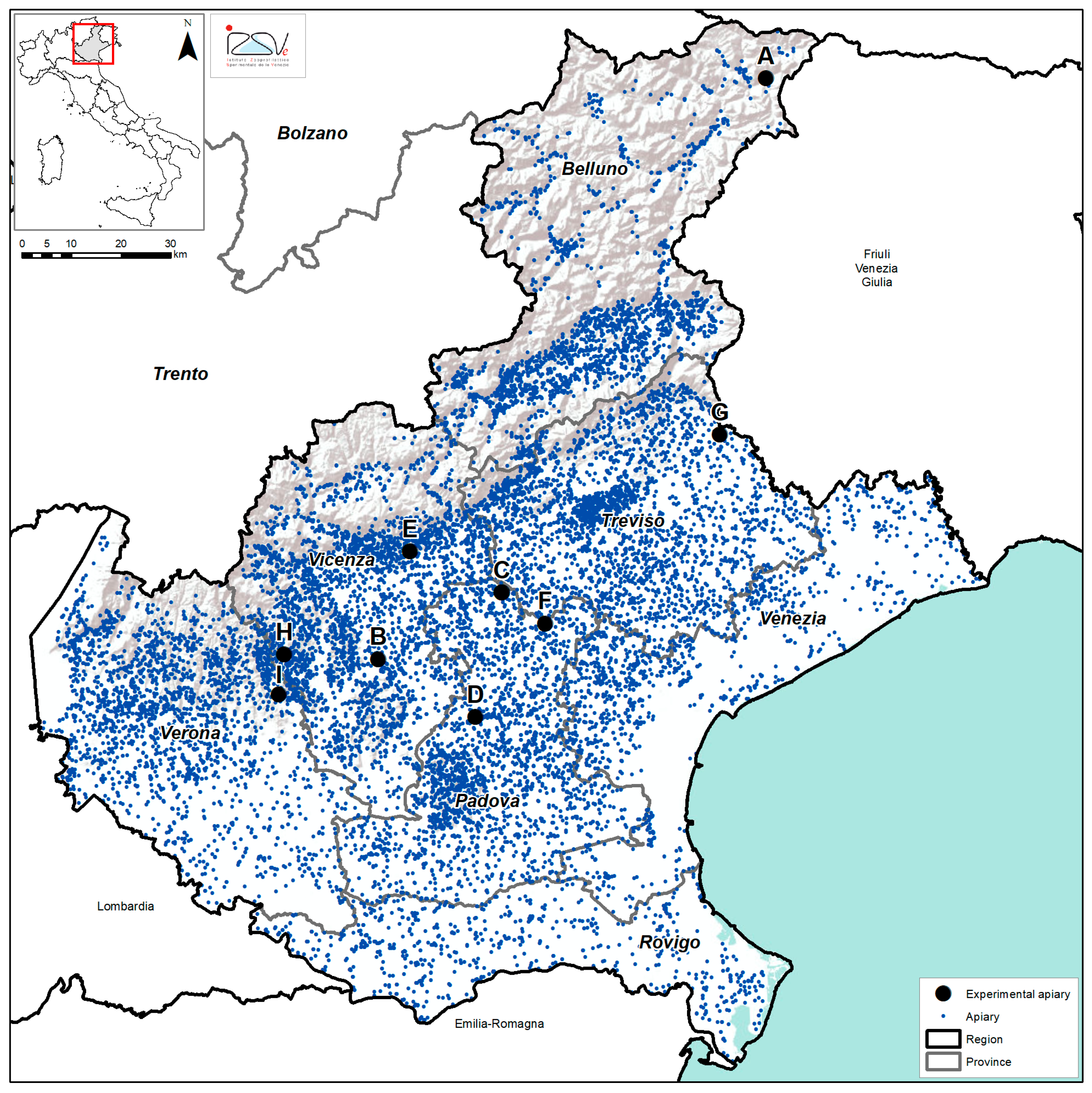
2.1. Sample Collection
2.2. Nosema spp.
2.3. Lotmaria passim and Crithidia mellificae
2.4. Honey Bee Viruses
2.5. Varroa Mite
2.6. Statistical Analysis
3. Results
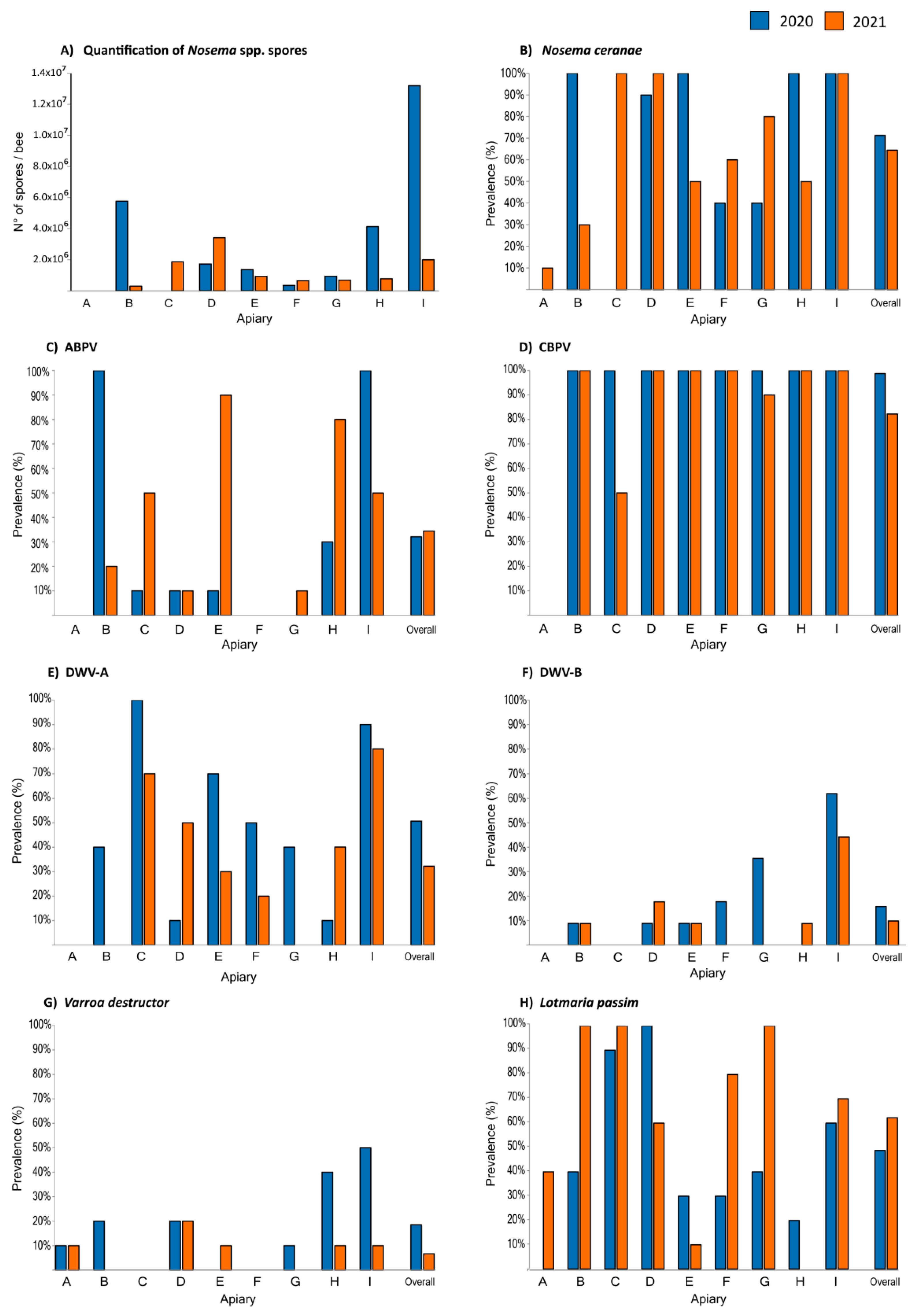
3.1. Nosema spp.
3.2. Lotmaria passim and Crithidia mellificae
3.3. Viruses
3.4. Varroa Destructor Mite
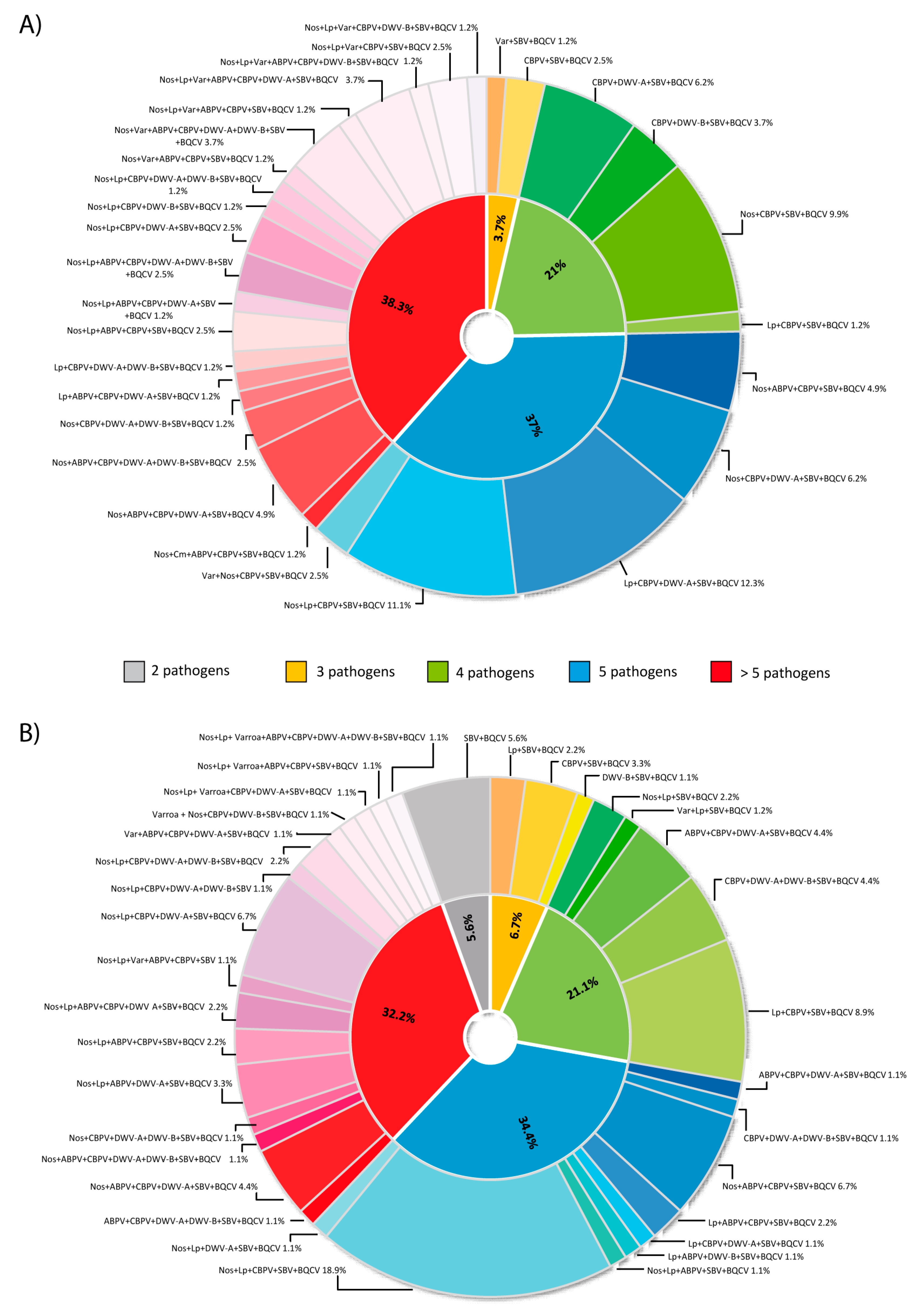
3.5. Associations between Pathogen Occurrences
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Genersch, E. Honey bee pathology: Current threats to honey bees and beekeeping. Appl. Microbiol. Biotechnol. 2010, 87, 87–97. [Google Scholar] [CrossRef]
- Genersch, E.; Evans, J.D.; Fries, I. Honey bee disease overview. J. Invertebr. Pathol. 2010, 103, S2–S4. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, A.J.; Brutscher, L.M.; Glenny, W.; Flenniken, M.L. Abiotic and biotic factors affecting the replication and pathogenicity of bee viruses. Curr. Opin. Insect Sci. 2016, 16, 14–21. [Google Scholar] [CrossRef]
- Regulation (EU) No 1308/2013 of the European Parliament and of the Council of 17 December 2013 Establishing a Common Organisation of the Markets in Agricultural Products and Repealing Council Regulations (EEC) No 922/72, (EEC) No 234/79, (EC) No 1037/2001 and (EC) No 1234/2007. 2013. Available online: https://eur-lex.europa.eu/eli/reg/2013/1308/oj (accessed on 20 January 2022).
- Ellis, J.D.; Munn, P.A. The worldwide health status of honey bees. Bee World 2005, 86, 88–101. [Google Scholar] [CrossRef]
- Beaurepaire, A.; Piot, N.; Doublet, V.; Antunez, K.; Campbell, E.; Chantawannakul, P.; Chejanovsky, N.; Gajda, A.; Heerman, M.; Panziera, D.; et al. Diversity and Global Distribution of Viruses of the Western Honey Bee, Apis mellifera. Insects 2020, 11, 239. [Google Scholar] [CrossRef]
- Steinhauer, N.; Kulhanek, K.; Antúnez, K.; Human, H.; Chantawannakul, P.; Chauzat, M.P.; van Engelsdorp, D. Drivers of colony losses. Curr. Opin. Insect Sci. 2018, 26, 142–148. [Google Scholar] [CrossRef]
- Nanetti, A.; Bortolotti, L.; Cilia, G. Pathogens Spillover from Honey Bees to Other Arthropods. Pathogens 2021, 10, 1044. [Google Scholar] [CrossRef]
- Alaux, C.; Folschweiller, M.; McDonnell, C.; Beslay, D.; Cousin, M.; Dussaubat, C.; Brunet, J.L.; Le Conte, Y. Pathological effects of the microsporidium Nosema ceranae on honey bee queen physiology (Apis mellifera). J. Invertebr. Pathol. 2011, 106, 380–385. [Google Scholar] [CrossRef]
- Fries, I. Nosema ceranae in European honey bees (Apis mellifera). J. Invertebr. Pathol. 2010, 103 (Suppl. 1), 73. [Google Scholar] [CrossRef]
- Bromenshenk, J.J.; Henderson, C.B.; Wick, C.H.; Stanford, M.F.; Zulich, A.W.; Jabbour, R.E.; Deshpande, S.V.; McCubbin, P.E.; Seccomb, R.A.; Welch, P.M.; et al. Iridovirus and microsporidian linked to honey bee colony decline. PLoS ONE 2010, 5, e13181. [Google Scholar] [CrossRef]
- Anderson, D.L. Pests and Pathogens of the Honeybee (Apis mellifera L.) in Fiji. J. Apic. Res. 1990, 29, 53–59. [Google Scholar] [CrossRef]
- Benjeddou, M.; Leat, N.; Allsopp, M.; Davison, S. Detection of acute bee paralysis virus and black queen cell virus from honeybees by reverse transcriptase pcr. Appl. Environ. Microbiol. 2001, 67, 2384–2387. [Google Scholar] [CrossRef]
- Evans, J.D. Genetic evidence for coinfection of honey bees by acute bee paralysis and Kashmir bee viruses. J. Invertebr. Pathol. 2001, 78, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Hammond, J.; Hsu, H.T.; Evans, J.; Feldlaufer, M. Multiple virus infections in the honey bee and genome divergence of honey bee viruses. J. Invertebr. Pathol. 2004, 87, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pettis, J.S.; Feldlaufer, M.F. Detection of multiple viruses in queens of the honey bee Apis mellifera L. J. Invertebr. Pathol. 2005, 90, 118–121. [Google Scholar] [CrossRef]
- Carrillo-Tripp, J.; Dolezal, A.G.; Goblirsch, M.J.; Miller, W.A.; Toth, A.L.; Bonning, B.C. In Vivo and in vitro infection dynamics of honey bee viruses. Sci. Rep. 2016, 6, 22265. [Google Scholar] [CrossRef]
- Christian, P.D.; Scotti, P.D. Picornalike Viruses of Insects. In The Insect Viruses; Miller, L.K., Ball, L.A., Eds.; Springer: Boston, MA, USA, 1998; pp. 301–336. [Google Scholar]
- Valles, S.M.; Strong, C.A.; Hashimoto, Y. A new positive-strand RNA virus with unique genome characteristics from the red imported fire ant, Solenopsis invicta. Virology 2007, 365, 457–463. [Google Scholar] [CrossRef]
- Ribière, M.; Ball, B.; Aubert, M. Natural history and geographical distribution of honey bee viruses. In Virology and the Honey Bee; Aubert, M., Ball, B., Fries, I., Moritz, R., Milani, N., Bernardinelli, I., Eds.; European Communities: Luxembourg, 2008; pp. 15–84. [Google Scholar]
- Bailey, L.; Gibbs, A.J.; Woods, R.D. Two viruses from adult honey bees (Apis mellifera Linnaeus). Virology 1963, 21, 390–395. [Google Scholar] [CrossRef]
- Maori, E.; Lavi, S.; Mozes-Koch, R.; Gantman, Y.; Peretz, Y.; Edelbaum, O.; Tanne, E.; Sela, I. Isolation and characterization of Israeli acute paralysis virus, a dicistrovirus affecting honeybees in Israel: Evidence for diversity due to intra- and inter-species recombination. J. Gen. Virol. 2007, 88, 3428–3438. [Google Scholar] [CrossRef]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef]
- Bailey, L.; Woods, R.D. Two more small RNA viruses from honey bees and further observations on sacbrood and acute bee-paralysis viruses. J. Gen. Virol. 1977, 37, 175–182. [Google Scholar] [CrossRef]
- Doublet, V.; Labarussias, M.; de Miranda, J.R.; Moritz, R.F.; Paxton, R.J. Bees under stress: Sublethal doses of a neonicotinoid pesticide and pathogens interact to elevate honey bee mortality across the life cycle. Environ. Microbiol. 2015, 17, 969–983. [Google Scholar] [CrossRef] [PubMed]
- Olivier, V.; Blanchard, P.; Chaouch, S.; Lallemand, P.; Schurr, F.; Celle, O.; Dubois, E.; Tordo, N.; Thiéry, R.; Houlgatte, R.; et al. Molecular characterisation and phylogenetic analysis of Chronic bee paralysis virus, a honey bee virus. Virus Res. 2008, 132, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Traynor, K.S.; Rennich, K.; Forsgren, E.; Rose, R.; Pettis, J.; Kunkel, G.; Madella, S.; Evans, J.; Lopez, D.; van Engelsdorp, D. Multiyear survey targeting disease incidence in US honey bees. Apidologie 2016, 47, 325–347. [Google Scholar] [CrossRef]
- Porrini, C.; Mutinelli, F.; Bortolotti, L.; Granato, A.; Laurenson, L.; Roberts, K.; Gallina, A.; Silvester, N.; Medrzycki, P.; Renzi, T.; et al. The Status of Honey Bee Health in Italy: Results from the Nationwide Bee Monitoring Network. PLoS ONE 2016, 11, e0155411. [Google Scholar] [CrossRef]
- Li, B.; Deng, S.; Yang, D.; Hou, C.; Diao, Q. Complete sequences of the RNA 1 and RNA 2 segments of chronic bee paralysis virus strain CBPV-BJ detected in China. Arch. Virol. 2017, 162, 2451–2456. [Google Scholar] [CrossRef]
- Bailey, L. Recent research on honeybee viruses. Bee World 1975, 56, 55–64. [Google Scholar] [CrossRef]
- Ribière, M.; Olivier, V.; Blanchard, P. Chronic bee paralysis: A disease and a virus like no other? J. Invertebr. Pathol. 2010, 103 (Suppl. 1), 120. [Google Scholar] [CrossRef]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103 (Suppl. 1), 48. [Google Scholar] [CrossRef]
- Bailey, L. The multiplication and spread of sacbrood virus of bees. Ann. Appl. Biol. 1969, 63, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.; Gibbs, A.J.; Woods, R.D. Sacbrood virus of the larval honey bee (Apis mellifera linnaeus). Virology 1964, 23, 425–429. [Google Scholar] [CrossRef]
- Chen, Y.P.; Siede, R. Honey bee viruses. Adv. Virus Res. 2007, 70, 33–80. [Google Scholar] [PubMed]
- Wang, D.; Mofller, F.E. The Division of Labor and Queen Attendance Behavior of Nosema-Infected Worker Honey Bees. J. Econ. Entomol. 1970, 63, 1539–1541. [Google Scholar] [CrossRef]
- Bailey, L.; Fernando, E.F.W. Effects of sacbrood virus on adult honey-bees. Ann. Appl. Biol. 1972, 72, 27–35. [Google Scholar] [CrossRef]
- Yue, C.; Genersch, E. RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 2005, 86, 3419–3424. [Google Scholar] [CrossRef]
- Nordström, S. Distribution of deformed wing virus within honey bee (Apis mellifera) brood cells infested with the ectoparasitic mite Varroa destructor. Exp. Appl. Acarol. 2003, 29, 293–302. [Google Scholar] [CrossRef]
- McMahon, D.P.; Natsopoulou, M.E.; Doublet, V.; Fürst, M.; Weging, S.; Brown, M.J.F.; Gogol-Döring, A.; Paxton, R.J. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. Royal Soc. B Biol. Sci. 2016, 283, 20160811. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef]
- Ramsey, S.D.; Ochoa, R.; Bauchan, G.; Gulbronson, C.; Mowery, J.D.; Cohen, A.; Lim, D.; Joklik, J.; Cicero, J.M.; Ellis, J.D.; et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proc. Natl. Acad. Sci. USA 2019, 116, 1792. [Google Scholar] [CrossRef]
- Rosenkranz, P.; Aumeier, P.; Ziegelmann, B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 2009, 103 (Suppl. 1), 96. [Google Scholar] [CrossRef] [PubMed]
- Nazzi, F.; Brown, S.P.; Annoscia, D.; Del Piccolo, F.; Di Prisco, G.; Varricchio, P.; Della Vedova, G.; Cattonaro, F.; Caprio, E.; Pennacchio, F. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog. 2012, 8, e1002735. [Google Scholar] [CrossRef]
- Kang, Y.; Blanco, K.; Davis, T.; Wang, Y.; DeGrandi-Hoffman, G. Disease dynamics of honeybees with Varroa destructor as parasite and virus vector. Math. Biosci. 2016, 275, 71–92. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.; Cordoni, G.; Budge, G. The Acute bee paralysis virus–Kashmir bee virus–Israeli acute paralysis virus complex. J. Invertebr. Pathol. 2009, 103 (Suppl. 1), 30. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, R.S.; Bauchan, G.R.; Murphy, C.A.; Ravoet, J.; de Graaf, D.C.; Evans, J.D. Characterization of Two Species of Trypanosomatidae from the Honey Bee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. gen., n. sp. J. Eukaryot. Microbiol. 2015, 62, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Arismendi, N.; Caro, S.; Castro, M.P.; Vargas, M.; Riveros, G.; Venegas, T. Impact of Mixed Infections of Gut Parasites Lotmaria passim and Nosema ceranae on the Lifespan and Immune-related Biomarkers in Apis mellifera. Insects 2020, 11, 420. [Google Scholar] [CrossRef] [PubMed]
- Langridge, D.F.; McGhee, R.B. Crithidia mellificae n. sp. an acidophilic trypanosomatid of the honey bee Apis mellifera. J. Protozool. 1967, 14, 485–487. [Google Scholar] [CrossRef]
- Runckel, C.; Flenniken, M.L.; Engel, J.C.; Ruby, J.G.; Ganem, D.; Andino, R.; De Risi, J.L. Temporal Analysis of the Honey Bee Microbiome Reveals Four Novel Viruses and Seasonal Prevalence of Known Viruses, Nosema, and Crithidia. PLoS ONE 2011, 6, e20656. [Google Scholar] [CrossRef]
- Cornman, R.S.; Tarpy, D.R.; Chen, Y.; Jeffreys, L.; Lopez, D.; Pettis, J.S.; van Engelsdorp, D.; Evans, J.D. Pathogen Webs in Collapsing Honey Bee Colonies. PLoS ONE 2012, 7, e43562. [Google Scholar] [CrossRef]
- Ravoet, J.; Maharramov, J.; Meeus, I.; De Smet, L.; Wenseleers, T.; Smagghe, G.; de Graaf, D.C. Comprehensive Bee Pathogen Screening in Belgium Reveals Crithidia mellificae as a New Contributory Factor to Winter Mortality. PLoS ONE 2013, 8, e72443. [Google Scholar] [CrossRef]
- Cepero, A.; Martín-Hernández, R.; Bartolomé, C.; Gómez-Moracho, T.; Barrios, L.; Bernal, J.; Teresa Martín, M.; Meana, A.; Higes, M. Passive laboratory surveillance in Spain: Pathogens as risk factors for honey bee colony collapse. J. Apic. Res. 2015, 54, 525–531. [Google Scholar] [CrossRef]
- Schwarz, R.S.; Moran, N.A.; Evans, J.D. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA 2016, 113, 9345. [Google Scholar] [CrossRef] [PubMed]
- Vejnovic, B.; Stevanovic, J.; Schwarz, R.S.; Aleksic, N.; Mirilovic, M.; Jovanovic, N.M.; Stanimirovic, Z. Quantitative PCR assessment of Lotmaria passim in Apis mellifera colonies co-infected naturally with Nosema ceranae. J. Invertebr. Pathol. 2018, 151, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, J.; Schwarz, R.S.; Vejnovic, B.; Evans, J.D.; Irwin, R.E.; Glavinic, U.; Stanimirovic, Z. Species-specific diagnostics of Apis mellifera trypanosomatids: A nine-year survey (2007–2015) for trypanosomatids and microsporidians in Serbian honey bees. J. Invertebr. Pathol. 2016, 139, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Buendía-Abad, M.; Martín-Hernández, R.; Ornosa, C.; Barrios, L.; Bartolomé, C.; Higes, M. Epidemiological study of honeybee pathogens in Europe: The results of Castilla-La Mancha (Spain). Span. J. Agric. Res. 2018, 16, e0502. [Google Scholar] [CrossRef]
- Xu, G.; Palmer-Young, E.; Skyrm, K.; Daly, T.; Sylvia, M.; Averill, A.; Rich, S. Triplex real-time PCR for detection of Crithidia mellificae and Lotmaria passim in honey bees. Parasitol. Res. 2018, 117, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Castelli, L.; Branchiccela, B.; Invernizzi, C.; Tomasco, I.; Basualdo, M.; Rodriguez, M.; Zunino, P.; Antúnez, K. Detection of Lotmaria passim in Africanized and European honey bees from Uruguay, Argentina and Chile. J. Invertebr. Pathol. 2018, 160, 95–97. [Google Scholar] [CrossRef]
- Van Engelsdorp, D.; Lengerich, E.; Spleen, A.; Dainat, B.; Cresswell, J.; Baylis, K.; Nguyen, B.K.; Soroker, V.; Underwood, R.; Human, H.; et al. Standard epidemiological methods to understand and improve Apis mellifera health. J. Apic. Res. 2013, 52, 1–16. [Google Scholar] [CrossRef]
- OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2021. Chapter 9.6. Infestation of Honey Bees with Varroa spp. (Varroosis) Article 9.6.1. Available online: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-code-online-access/?id=169&L=1&htmfile=chapitre_varroa_spp.htm (accessed on 20 January 2022).
- Martín-Hernández, R.; Meana, A.; Prieto, L.; Salvador, A.M.; Garrido-Bailón, E.; Higes, M. Outcome of colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007, 73, 6331–6338. [Google Scholar] [CrossRef]
- Bartolomé, C.; Buendía-Abad, M.; Benito, M.; De la Rua, P.; Ornosa, C.; Martín-Hernández, R.; Higes, M.; Maside, X. A new multiplex PCR protocol to detect mixed trypanosomatid infections in species of Apis and Bombus. J. Invertebr. Pathol. 2018, 154, 37–41. [Google Scholar] [CrossRef]
- Cox-Foster, D.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.; Briese, T.; Hornig, M.; Geiser, D.M. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Carletto, J.; Aurélie, G.; Regnault, J.; Blanchard, P.; Schurr, F.; Ribiere-Chabert, M. Detection of main honey bee pathogens by multiplex PCR. Euroreference 2010, 4, 13–15. [Google Scholar]
- Martinello, M.; Baratto, C.; Manzinello, C.; Piva, E.; Borin, A.; Toson, M.; Granato, A.; Boniotti, M.B.; Gallina, A.; Mutinelli, F. Spring mortality in honey bees in northeastern Italy: Detection of pesticides and viruses in dead honey bees and other matrices. J. Apic. Res. 2017, 56, 239–254. [Google Scholar] [CrossRef]
- Blanchard, P.; Ribière, M.; Celle, O.; Lallemand, P.; Schurr, F.; Olivier, V.; Iscache, A.L.; Faucon, J.P. Evaluation of a real-time two-step RT-PCR assay for quantitation of Chronic bee paralysis virus (CBPV) genome in experimentally-infected bee tissues and in life stages of a symptomatic colony. J. Virol. Methods 2007, 141, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Schurr, F.; Tison, A.; Militano, L.; Cheviron, N.; Sircoulomb, F.; Rivière, M.; Ribière-Chabert, M.; Thiéry, R.; Dubois, E. Validation of quantitative real-time RT-PCR assays for the detection of six honeybee viruses. J. Virol. Methods 2019, 270, 70–78. [Google Scholar] [CrossRef]
- Blanchard, P.; Olivier, V.; Iscache, A.; Celle, O.; Schurr, F.; Lallemand, P.; Ribière, M. Improvement of RT-PCR detection of chronic bee paralysis virus (CBPV) required by the description of genomic variability in French CBPV isolates. J. Invertebr. Pathol. 2008, 97, 182–185. [Google Scholar] [CrossRef]
- Grabensteiner, E.; Ritter, W.; Carter, M.J.; Davison, S.; Pechhacker, H.; Kolodziejek, J.; Boecking, O.; Derakhshifar, I.; Moosbeckhofer, R.; Licek, E.; et al. Sacbrood virus of the honeybee (Apis mellifera): Rapid identification and phylogenetic analysis using reverse transcription-PCR. Clin. Diagn. Lab. Immunol. 2001, 8, 93–104. [Google Scholar] [CrossRef]
- Mutinelli, F.; Costa, C.; Lodesani, M.; Baggio, A.; Medrzycki, P.; Formato, G.; Porrini, C. Honey bee colony losses in Italy. J. Apic. Res. 2010, 49, 119–120. [Google Scholar] [CrossRef]
- Bellucci, V.; Lucci, S.; Bianco, P.; Ubaldi, A.; Felicioli, A.; Porrini, C.; Mutinelli, F.; Battisti, S.; Spallucci, V.; Cersini, A.; et al. Monitoring honey bee healthin five natural protected areas in Italy. Vet. Ital. 2019, 55, 15–25. [Google Scholar] [CrossRef]
- Berényi, O.; Bakonyi, T.; Derakhshifar, I.; Köglberger, H.; Nowotny, N. Occurrence of six honeybee viruses in diseased Austrian apiaries. Appl. Environ. Microbiol. 2006, 72, 2414–2420. [Google Scholar] [CrossRef]
- De Smet, L.; Ravoet, J.; de Miranda, J.R.; Wenseleers, T.; Mueller, M.Y.; Moritz, R.F.A.; de Graaf, D.C. BeeDoctor, a Versatile MLPA-Based Diagnostic Tool for Screening Bee Viruses. PLoS ONE 2012, 7, e47953. [Google Scholar] [CrossRef] [PubMed]
- Matthijs, S.; De Waele, V.; Vandenberge, V.; Verhoeven, B.; Evers, J.; Brunain, M.; Saegerman, C.; De Winter, P.J.J.; Roels, S.; de Graaf, D.C.; et al. Nationwide Screening for Bee Viruses and Parasites in Belgian Honey Bees. Viruses 2020, 12, 890. [Google Scholar] [CrossRef]
- Amiri, E.; Meixner, M.; Nielsen, S.L.; Kryger, P. Four Categories of Viral Infection Describe the Health Status of Honey Bee Colonies. PLoS ONE 2015, 10, e0140272. [Google Scholar] [CrossRef] [PubMed]
- Tentcheva, D.; Gauthier, L.; Zappulla, N.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 2004, 70, 7185–7191. [Google Scholar] [CrossRef] [PubMed]
- Morawetz, L.; Köglberger, H.; Griesbacher, A.; Derakhshifar, I.; Crailsheim, K.; Brodschneider, R.; Moosbeckhofer, R. Health status of honey bee colonies (Apis mellifera) and disease-related risk factors for colony losses in Austria. PLoS ONE 2018, 14, e0219293. [Google Scholar] [CrossRef]
- Blanchard, P.; Guillot, S.; Antùnez, K.; Köglberger, H.; Kryger, P.; de Miranda, J.R.; Franco, S.; Chauzat, M.; Thiéry, R.; Ribière, M. Development and validation of a real-time two-step RT-qPCR TaqMan® assay for quantitation of Sacbrood virus (SBV) and its application to a field survey of symptomatic honey bee colonies. J. Virol. Methods 2014, 197, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.; Ball, B. The incidence and world distribution of honey bee viruses. Bee World 1996, 77, 141–162. [Google Scholar] [CrossRef]
- Bacandritsos, N.; Granato, A.; Budge, G.; Papanastasiou, I.; Roinioti, E.; Caldon, M.; Falcaro, C.; Gallina, A.; Mutinelli, F. Sudden deaths and colony population decline in Greek honey bee colonies. J. Invertebr. Pathol. 2010, 105, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Predictive markers of honey bee colony collapse. PLoS ONE 2012, 7, e32151. [Google Scholar] [CrossRef]
- Mendoza, Y.; Antúnez, K.; Branchiccela, B.; Anido, M.; Santos, E.; Invernizzi, C. Nosema ceranae and RNA viruses in European and Africanized honeybee colonies (Apis mellifera) in Uruguay. Apidologie 2014, 45, 224–234. [Google Scholar] [CrossRef][Green Version]
- Chagas, D.B.; Monteiro, F.L.; Barcelos, L.D.S.; Frühauf, M.I.; Ribeiro, L.C.; Lima, M.D.; Hübner, S.D.O.; Fischer, G. Black queen cell virus and Nosema ceranae coinfection in Africanized honey bees from southern Brazil. Pesqui. Vet. Bras. 2021, 40, 892–897. [Google Scholar] [CrossRef]
- Klee, J.; Besana, A.; Genersch, E.; Gisder, S.; Nanetti, A.; Tam, D.; Chinh, T.; Puerta, F.; Ruz, J.; Kryger, P.; et al. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 2007, 96, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ferroglio, E.; Zanet, S.; Peraldo, N.; Tachis, E.; Trisciuoglio, A.; Laurino, D.; Porporato, M. Nosema ceranae has been infecting honey bees Apis mellifera in Italy since at least 1993. J. Apic. Res. 2013, 52, 60–61. [Google Scholar] [CrossRef]
- Maiolino, P.; Iafigliola, L.; Rinaldi, L.; De Leva, G.; Restucci, B.; Martano, M. Histopathological findings of the midgut in European honey bee (Apis mellifera L.) naturally infected by Nosema spp. Vet. Med. Anim. Sci. 2014, 2, 4. [Google Scholar] [CrossRef]
- Stevanovic, J.; Stanimirovic, Z.; Genersch, E.; Kovacevic, S.R.; Ljubenkovic, J.; Radakovic, M.; Aleksic, N. Dominance of Nosema ceranae in honey bees in the Balkan countries in the absence of symptoms of colony collapse disorder. Apidologie 2011, 42, 49. [Google Scholar] [CrossRef]
- Mráz, P.; Hýbl, M.; Kopecký, M.; Bohatá, A.; Hoštičková, I.; Šipoš, J.; Vočadlová, K.; Čurn, V. Screening of Honey Bee Pathogens in the Czech Republic and Their Prevalence in Various Habitats. Insects 2021, 12, 1051. [Google Scholar] [CrossRef]
- Martínez, J.; Leal, G.; Conget, P. Nosema ceranae an emergent pathogen of Apis mellifera in Chile. Parasitol. Res. 2012, 111, 601–607. [Google Scholar] [CrossRef]
- Morimoto, T.; Kojima, Y.; Yoshiyama, M.; Kimura, K.; Yang, B.; Peng, G.; Kadowaki, T. Molecular detection of protozoan parasites infecting Apis mellifera colonies in Japan. Environ. Microbiol. Rep. 2013, 5, 74–77. [Google Scholar] [CrossRef]
- Emsen, B.; Guzman-Novoa, E.; Hamiduzzaman, M.M.; Eccles, L.; Lacey, B.; Ruiz-Pérez, R.A.; Nasr, M. Higher prevalence and levels of Nosema ceranae than Nosema apis infections in Canadian honey bee colonies. Parasitol. Res. 2016, 115, 175–181. [Google Scholar] [CrossRef]
- Antúnez, K.; Martín-Hernández, R.; Prieto, L.; Meana, A.; Zunino, P.; Higes, M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 2009, 11, 2284–2290. [Google Scholar] [CrossRef]
- Betti, M.I.; Wahl, L.M.; Zamir, M. Effects of infection on honey bee population dynamics: A model. PLoS ONE 2014, 9, e110237. [Google Scholar] [CrossRef] [PubMed]
- Tritschler, M.; Retschnig, G.; Yañez, O.; Williams, G.R.; Neumann, P. Host sharing by the honey bee parasites Lotmaria passim and Nosema ceranae. Ecol. Evol. 2017, 7, 1850–1857. [Google Scholar] [CrossRef] [PubMed]
- Cavigli, I.; Daughenbaugh, K.F.; Martin, M.; Lerch, M.; Banner, K.; Garcia, E.; Brutscher, L.M.; Flenniken, M.L. Pathogen prevalence and abundance in honey bee colonies involved in almond pollination. Apidologie 2016, 47, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Doublet, V.; Natsopoulou, M.E.; Zschiesche, L.; Paxton, R.J. Within-host competition among the honey bees pathogens Nosema ceranae and Deformed wing virus is asymmetric and to the disadvantage of the virus. J. Invertebr. Pathol. 2015, 124, 31–34. [Google Scholar] [CrossRef]
- Wilfert, L.; Long, G.; Leggett, H.C.; Schmid-Hempel, P.; Butlin, R.; Martin, S.; Boots, M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef]
- Mondet, F.; de Miranda, J.R.; Kretzschmar, A.; Le Conte, Y.; Mercer, A.R. On the front line: Quantitative virus dynamics in honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor. PLoS Pathog. 2014, 10, e1004323. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, C.; Wu, P.; Xu, J.; Guo, Y.; Xue, F.; Getachew, A.; Xu, S. Brain metabolomic profiling of eastern honey bee (Apis cerana) infested with the mite Varroa destructor. PLoS ONE 2017, 12, e0175573. [Google Scholar] [CrossRef]
- Shutler, D.; Head, K.; Burgher-MacLellan, K.; Colwell, M.J.; Levitt, A.L.; Ostiguy, N.; Williams, G.R. Honey bee Apis mellifera parasites in the absence of Nosema ceranae fungi and Varroa destructor mites. PLoS ONE 2014, 9, e98599. [Google Scholar] [CrossRef]
- Manley, R.; Temperton, B.; Doyle, T.; Gates, D.; Hedges, S.; Boots, M.; Wilfert, L. Knock-on community impacts of a novel vector: Spillover of emerging DWV-B from Varroa-infested honeybees to wild bumblebees. Ecol. Lett. 2019, 22, 1306–1315. [Google Scholar] [CrossRef]
- Gisder, S.; Möckel, N.; Eisenhardt, D.; Genersch, E. In Vivo evolution of viral virulence: Switching of deformed wing virus between hosts results in virulence changes and sequence shifts. Environ. Microbiol. 2018, 20, 4612–4628. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Dead or alive: Deformed wing virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 2012, 78, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Varroa-Virus interaction in collapsing honey bee colonies. PLoS ONE 2013, 8, e57540. [Google Scholar] [CrossRef] [PubMed]
- Toplak, I.; Rihtarič, D.; Ciglenečki, U.J.; Hostnik, P.; Jenčič, V.; Barlič-Maganja, D. Detection of six honeybee viruses in clinically affected colonies in Carniolan gray bee (Apis mellifera carnica). Slov. Vet. Res. 2012, 49, 89–96. [Google Scholar]
- Tlak Gajger, I.; Šimenc, L.; Toplak, I. The First Detection and Genetic Characterization of Four Different Honeybee Viruses in Wild Bumblebees from Croatia. Pathogens 2021, 10, 808. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.L.; Nicolaisen, M.; Kryger, P. Incidence of acute bee paralysis virus, black queen cell virus, chronic bee paralysis virus, deformed wing virus, Kashmir bee virus and sacbrood virus in honey bees (Apis mellifera) in Denmark. Apidologie 2008, 39, 310–314. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordin, F.; Zulian, L.; Granato, A.; Caldon, M.; Colamonico, R.; Toson, M.; Trevisan, L.; Biasion, L.; Mutinelli, F. Presence of Known and Emerging Honey Bee Pathogens in Apiaries of Veneto Region (Northeast of Italy) during Spring 2020 and 2021. Appl. Sci. 2022, 12, 2134. https://doi.org/10.3390/app12042134
Bordin F, Zulian L, Granato A, Caldon M, Colamonico R, Toson M, Trevisan L, Biasion L, Mutinelli F. Presence of Known and Emerging Honey Bee Pathogens in Apiaries of Veneto Region (Northeast of Italy) during Spring 2020 and 2021. Applied Sciences. 2022; 12(4):2134. https://doi.org/10.3390/app12042134
Chicago/Turabian StyleBordin, Fulvio, Laura Zulian, Anna Granato, Mauro Caldon, Rosa Colamonico, Marica Toson, Laura Trevisan, Laura Biasion, and Franco Mutinelli. 2022. "Presence of Known and Emerging Honey Bee Pathogens in Apiaries of Veneto Region (Northeast of Italy) during Spring 2020 and 2021" Applied Sciences 12, no. 4: 2134. https://doi.org/10.3390/app12042134
APA StyleBordin, F., Zulian, L., Granato, A., Caldon, M., Colamonico, R., Toson, M., Trevisan, L., Biasion, L., & Mutinelli, F. (2022). Presence of Known and Emerging Honey Bee Pathogens in Apiaries of Veneto Region (Northeast of Italy) during Spring 2020 and 2021. Applied Sciences, 12(4), 2134. https://doi.org/10.3390/app12042134







