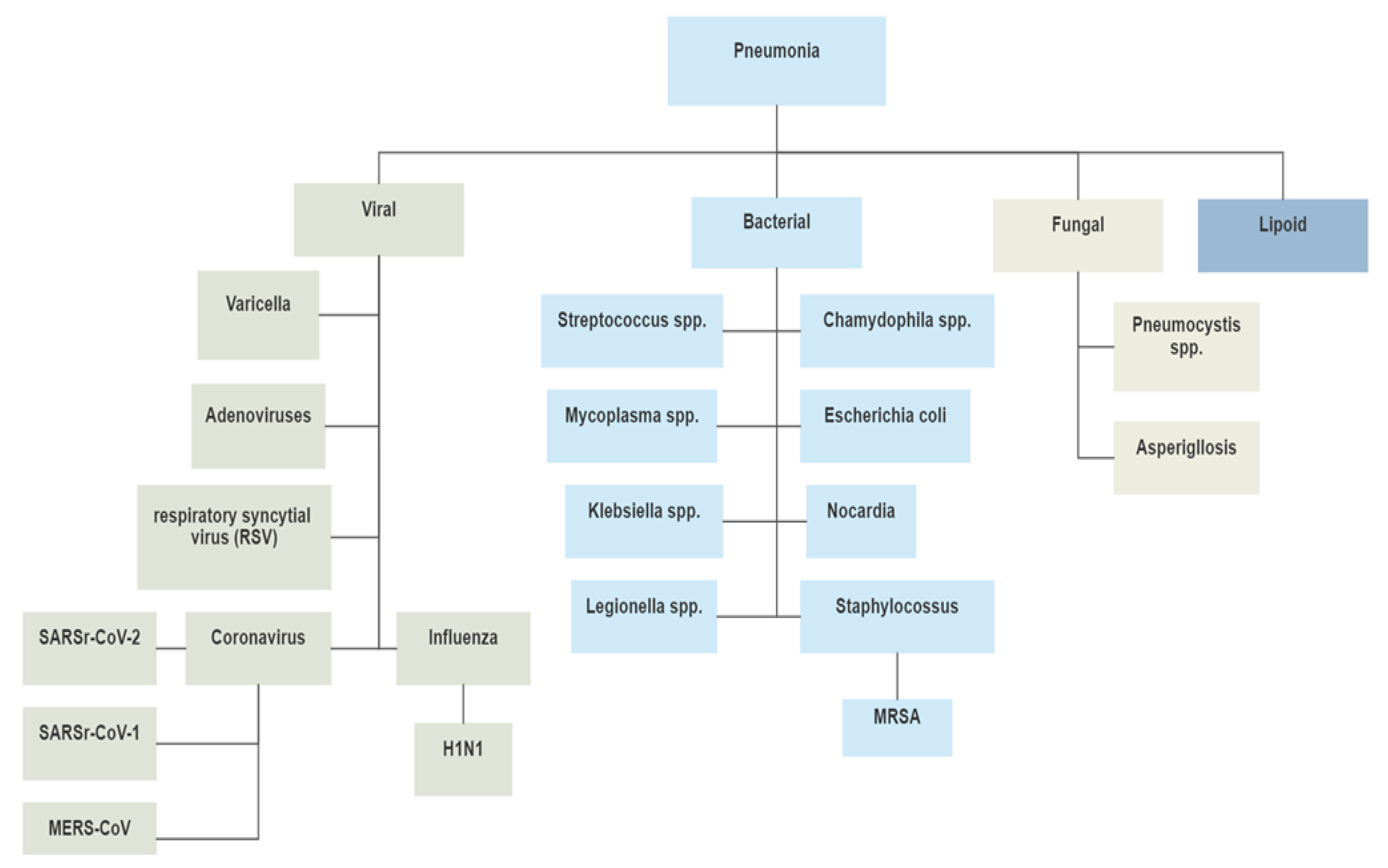
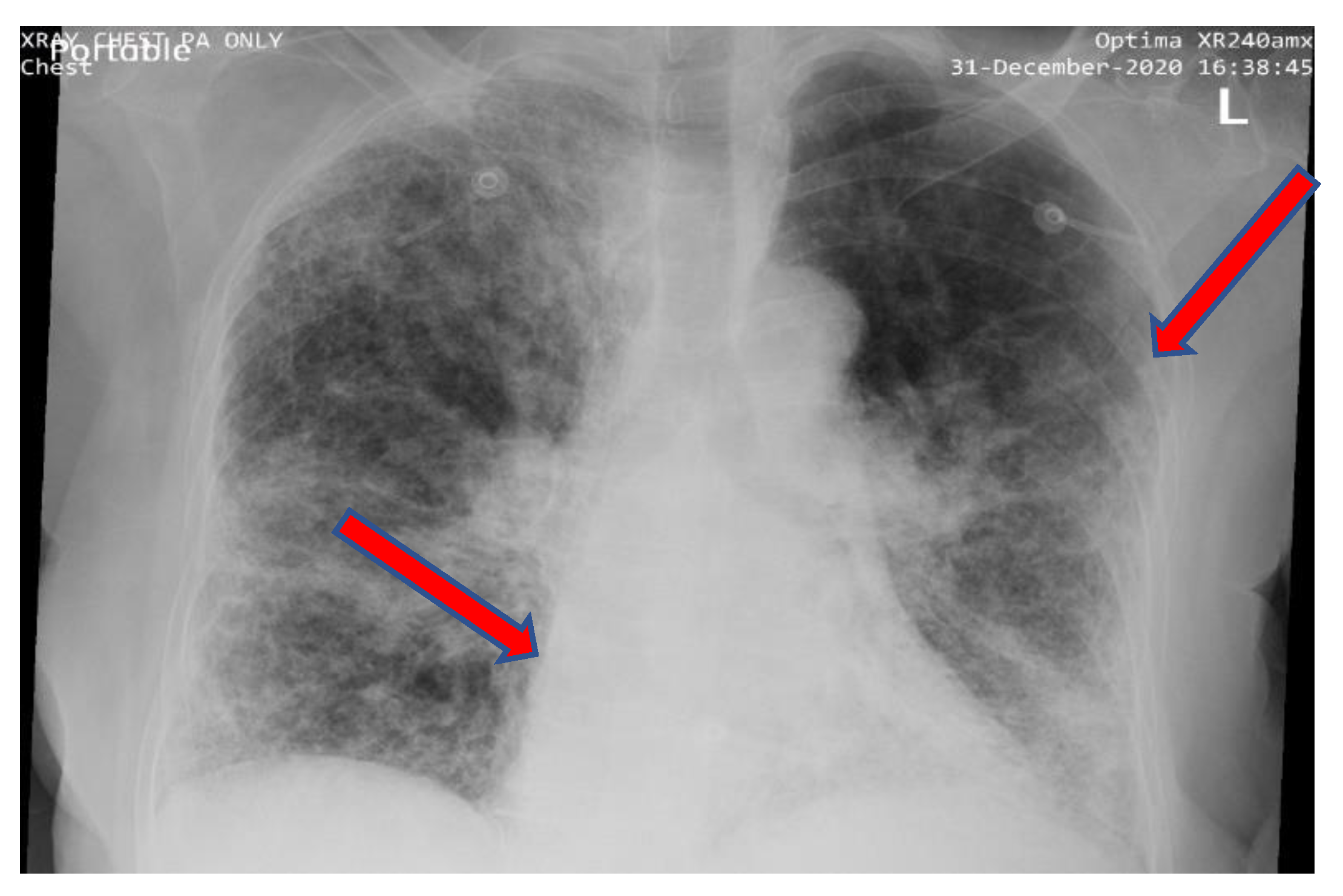
4.1. Identification of COVID-19 from CXR Images using Ensemble of Deep Learning Models
Ensemble learning involves capturing the outputs produced by different classifiers, which helps to ensure a robust prediction and increases the accuracy of deep learning models [
38]. Chouhan et al. [
39] designed a model based on the performance of the following pre-trained deep learning models for the detection of pneumonia: AlexNet, DenseNet121, ResNet18, Inception-v3, and GoogLeNet. The dataset was obtained from the Guangzhou Women and Children’s Medical Center (GWCMC); it consisted of a total of 5232 images labelled with the following classes: viral pneumonia, bacterial pneumonia, and normal. Random distribution of data into training (5232 images) and test (624 images) datasets was provided to avoid bias in performance. Some of the steps for data preprocessing include noise addition, random horizontal flip, and random-resized crop as augmentation techniques, with the images resized to 224 × 224 pixels. The classification results from the pre-trained CNN models were combined into a prediction vector, and majority voting was applied to generate a final prediction output. The proposed deep learning framework of the ensemble model achieved 96.4% testing accuracy.
Khan et al. [
40] compared the performance of deep hybrid learning (COVID-RENet-1 and COVID RENet-2) and deep boosted hybrid learning (COVID-RENet-1 and COVID-RENet-2) with the well-established CNNs (i.e., VGG-16/19, GoogLeNet, InceptionV3, ResNet-18/50, SqueezeNet, DenseNet-201, and Xception). Initially, the dataset images were resized to 224 × 224 pixels, followed by data augmentation with the help of rotation, reflection, and scaling operations. The balanced public CXR dataset from the GitHub and Kaggle repositories was divided into 3224 COVID-19-infected and 3224 healthy cases. In order to reduce bias, the dataset images were randomly distributed into training and validation datasets, with 80% of data for system training and 20% for system validation. Within the deep boosted hybrid learning framework, transfer-learning-based fine-tuned deep COVID-RENet-1 and 2 were used for feature extraction, followed by eigenvector-based transformation using principal component analysis (PCA) and SVM as the final classifier. The authors observed that the deep hybrid learning models outperformed the well-established CNN models, while the deep boosted hybrid learning models performed the most effectively.
Al-Waisy et al. [
41] also proposed an ensemble COVID-19–DeepNet system based on the integration of a deep belief network and a convolutional deep belief network. The purpose of the system was to differentiate between healthy and COVID-19-infected CXR images. A public dataset consisting of 24,000 CXR images was obtained from GitHub, Radiopaedia, the Italian Society of Medical and Interventional Radiology, the Radiological Society of North America, and Kaggle. A balanced dataset with 400 images for COVID-19 and 400 images for normal cases was used to obtain an augmented dataset with around 24,000 images of 128 × 128 pixels in size. Some of the different operations used for data augmentation included random sampling of data into mutually exclusive training, contrast balancing using contrast-limited adaptive histogram equalization (CLAHE), noise removal with the help of a Butterworth bandpass filter, horizontal flipping, and five-degree rotation. To avoid bias in the results, the data were randomly distributed into training (75%), testing (25%), and validation (10%) sets. The results from two different deep learning approaches—namely, the deep belief network and the convolutional deep belief network—were fused together by computing predicted probability scores, and the final decision regarding classification was based on decision-level fusion. The proposed model was compared with the state-of-the-art approaches (e.g., COVID-Net, ResNet 50, COVID-ResNet, and EfficientNet-B3 model), and it outperformed other approaches, with a detection accuracy rate of 99.93%.
In another study, Mazaar et al. [
42] proposed a hybrid model that exploits the potential of deep learning and transfer learning techniques to develop accurate and robust models for detecting COVID-19. The preprocessing function involved transforming the dimensions of the original dataset into 120 × 120 pixels. Three basic blocks were used for building seven different variants. These building blocks included (1) a CNN block, (2) a transfer learning block using VGG16 or VGG19, and (3) a machine learning block. A publicly available dataset obtained from Kaggle was used with a private dataset from Asir Hospital, Abha, Saudi Arabia, which consisted of a total of 4103 CXR images labelled as COVID-19, viral, or normal. To avoid bias in the performance, a random distribution of data into training (3279 images) and validation (820 images) sets was performed. The hybrid model using VGG-16 with transfer learning was able to outperform all other variants, with an accuracy of 97.6%.
Bhowal et al. [
38] applied the Choquet integral for ensemble deep learning models and evaluated the fuzzy measures for each classifier using coalition game theory (Shapley value), information theory, and lambda fuzzy approximation. Some of the preprocessing functions involved downscaling of original images to 512 × 512 pixels using bicubic interpolation and color-space translation to convert the original dataset images from RGB to grayscale. For deep feature extraction, three standard deep learning models with pre-trained weights on the ImageNet dataset were used—namely, VGG-16, Xception, and Inception-v3. The authors obtained a public dataset of CXR images from two repositories (namely, GitHub and Kaggle), which was used to create a single dataset called the Novel COVID-19 Chest X-ray Repository. The dataset consisted of 752 COVID-19-infected, 1584 viral pneumonia, and 1639 normal CXR images. Random sampling of images was used for the training, validation, and testing datasets, with the ratio of training, testing, and validation data as 77%:20%:3%, respectively. Finally, the results were combined to generate a final output as one of the following three classes: COVID-19, pneumonia, or normal. The ensemble model outperformed many recent methods, with an area under the curve (AUC) of 0.97 and a validation accuracy of approximately 98.99%.
Another method used ECG data for the classification of COVID-19 [
43]. The pro-posed automated tool consisted of four steps: ECG trace image preprocessing, deep feature extraction and feature incorporation, hybrid feature selection, and classification. In the preprocessing stage, the dimensionality of the input image was reduced from the original image (ranging between 952 × 1232 and 2213 × 1572 pixels) to 224 × 224 pixels for ResNet-50, ShuffleNet, and MobileNet and 229 × 229 pixels for Inception-v3, Xception, and Inception-ResNet. The dataset consisted of 250 scans of cases with COVID-19, 848 for trace records of cases with a present or former myocardial infarction and irregular heartbeat, and 859 normal images. To avoid the classification bias arising from class imbalances, the number of images per class was kept equal (i.e., 250 images per class) for the binary and multi-class classification. The proposed method for hybrid feature extraction utilized 10 different DL-based approaches. These networks include Inception–ResNet, ResNet-18, ResNet-50, ShuffleNet, Inception-v3, MobileNet, Xception, Dark-Net-19, DarkNet-53, and DenseNet-201. The fully connected deep features obtained from the different networks were followed by hybrid feature selection utilizing a forward search with a random forest classifier. The performance of the proposed system was compared with ResNet-50, ShuffleNet, and MobileNet to reveal the superior performance of the proposed method.
Rajaraman et al. [
44] utilized deep ensemble learning via a proposed network that consisted of two stages of pre-training followed by the actual deep learning architecture developed specifically for the recognition of COVID-19, which provided an input CXR of 256 × 256 pixels in size. The first stage of pre-training was composed of different deep learning models (i.e., ResNet18, VGG-16, VGG-19, Xception, Inception-V3, Dense-Net-121, MobileNet-V2, and NasNet-Mobile) with pre-trained weights on an ImageNet dataset, followed by successive layers for zero-padding, fully convolutional layers, pooling layers, dropout, and softmax activation. The second stage of pre-training was based on the first-stage pre-training model, followed by additional pooling, dropout, and softmax activation layers. The two stages of pre-training were concatenated with additional pooling and activation layers to provide the final recognition results. The ensemble learning leveraged the top three, top five, and top seven results from the proposed models with majority voting, simple averaging, and weighted averaging to obtain the final results. A total of 720 CXRs from the Montreal COVID-19, Twitter-COVID-19, RSNA, CheXpert, and NIH collections were used in this study. These data were randomly split into 80% for training and 20% for validation to avoid bias in the performance of the proposed system. The proposed system with deep ensemble learning was able to achieve an accuracy of 90.97%.
The proposed deep stacked ensemble in [
45] requires the preprocessing stage to reduce the size of the original images to 224 × 224 pixels, which is given as input to the feature extraction module leveraging different deep learning models (i.e., ResNet, Mo-bileNet, Inception, DenseNet, and NasNet). The two models with the best performance are stacked to form a deep ensemble model for COVID-19 prediction. The dataset used in this study contained a total of 2905 images (219 COVID-19 images and 2686 normal class images), which were randomly divided into training (80%) and test sets (20%) by maintaining consistency in class labels at each partition and minimizing classification bias. However, the overall class imbalance was present throughout the training data. The proposed stacked ensemble was able to provide the best accuracy result of 95.1% on the given dataset. The development of COVID-Net and COVID-EnsembleNet was discussed in [
46]. The COVID-Net architecture was based on four pairs of consecutive convolution and maximum-pooling layers, with filters ranging from 16 in the first pair to 128 in the fourth pair. These convolutional layer blocks were followed by the flattening operation and two fully connected layers with 128 and 2 filters, where each fully connected layer employed the softmax regression activation function. The COVID-EnsembleNet was constructed using the proposed COVID-Net architecture along with the existing VGG-16 architecture. The dataset used in this study contained 1281 positive cases and 3269 negative cases, with a random distribution of data into training (3641 images), testing (455 images), and validation (455 images) sets to avoid classification bias. Some of the preprocessing functions included image resizing to 224 × 224 pixels, image normalization for faster network convergence, and data augmentation techniques such as random horizontal and vertical image flipping, followed by a random 72-degree rotation. The proposed ensemble model was able to provide binary accuracy of 99.56% and multiclass accuracy of 97.56%.
A novel ensemble deep learning model for the detection of COVID-19 from CT images was developed in [
47], utilizing 2500 lung CT images from COVID-19 patients, along with 2500 CT images of lung tumors and 2500 CT images of normal lungs from a hospital. These images were randomly distributed into 6000 images for training and 1500 images for validation. Transfer learning was used for model parameter initialization, followed by deep feature extraction using three pre-trained deep convolutional neural network models, namely, AlexNet, GoogLeNet, and ResNet. The ensemble classifier EDL-COVID was obtained via relative majority voting of the aforementioned individual classifiers. With an average accuracy of 99.1%, precision of 99.1%, and recall of 99.6%, the ensemble classifier was able to outperform the results of the individual classifiers.
Taken together, the results of these previous studies strongly indicate that deep learning models built using multiple learning algorithms and, in particular, with ensemble learning classifiers, can benefit from improved performance. Nevertheless, it is worth mentioning that these studies have some limitations, not least because building ensemble models based on a single modality (in this case, mainly CT over CXR) leads to clinical challenges because, practically speaking, it is not preferable to expose the patient to CT radiation, and CXR imaging is not sufficient for the diagnosis of COVID-19 without any additional data [
48].
4.2. Identification of COVID-19 in CXR Images Using Deep Learning Models
A substantial body of research has been published recently on models for the detection and classification of COVID-19 using binary/multi-class classifiers for CXR images with off-the-shelf networks. In this review, we chose not to address studies with solutions involving binary classification. This is because the ability of AI systems to differentiate between the different classes has considerably improved, as these systems are able to learn from diverse data belonging to different classes [
49]. Abdelsamea et al. [
50] proposed the use of a CNN called Decompose, Transfer, and Compose (DeTraC); this method helps to deal with any irregularities and the limited availability of annotated CXR images. Their study used data from different sources—80 cases of normal CXR images with 4020 × 4892 pixels from the Japanese Society of Radiological Technology (JSRT), along with 105 and 11 cases of COVID-19 and SARS, respectively, with 4248 × 3480 pixels. Data augmentation techniques were used to generate 1764 samples from the original limited dataset. The different augmentation techniques included random up/down and right/left flipping, random translation, and rotation using five different angles. A histogram modification technique was applied to the augmented images to enhance the contrast of the images. The augmented dataset was randomly divided into 70% training and 30% validation sets to minimize classification bias. An AlexNet network based on shallow learning was used for the class decomposition layer, and different ImageNet pre-trained CNN networks were used for the transfer learning stage. The high-dimensional feature space was substantially reduced using PCA. The highest accuracy was achieved by VGG19 in DeTraC. The accuracy rate after applying the model was 93.1%.
Brunese et al. [
51] implemented a deep learning model using a dataset containing 6523 CXR images collected from three different CXR image sources. The dataset was labelled with 250 COVID-19 images, 2753 images belonging to patients with other pulmonary diseases, and 3520 normal patients. The preprocessing stage allowed the reduction of the image dimensions to 224 × 224 pixels as well as random distribution into training (2000 images), testing (1100 images), and validation (803 images) sets. Data augmentation was performed via random clockwise and counterclockwise rotation by 15 degrees. The proposed approach is based on a threefold method: Firstly, a process for the detection of any type of pneumonia in the CXR image is conducted. Secondly, if the lungs are not normal, then the system tries to classify between COVID-19 and other pneumonia. Finally, in the event of COVID-19 classification, the images are used to identify the area in the CXR that indicates the presence of COVID-19. The researchers applied the VGG-16 CNN model with 16 layers, which yielded an accuracy of 98% for the detection of COVID 19.
The limited number of CXR images that exist for COVID-19 research constituted the focus of the study undertaken by Loey et al. [
52]. The dataset used in this research was created by Dr. Joseph Cohen from the University of Montreal; it consists of total 307 CXR images, including 69 from COVID-19 patients, while the remainder belong to normal, bacterial, and viral pneumonia patients. The proposed model consists of two stages: In the first stage, a generative adversarial network (GAN) is used to generate additional images to increase the size of the existing limited dataset. In the second stage, deep transfer learning is used in the training, validation, and testing phases of the proposed model. For their investigation, the researchers selected the following deep transfer learning models: AlexNet, GoogLeNet, and ResNet18. Each of these networks can take an input image of 512 × 512 pixels in size. The choice of the models was due to their architectures, which contain a small number of layers, thereby reducing the processing time, the memory consumed, and the proposed model’s complexity. The highest test accuracy (80.6%) for a scenario that included all four classes was achieved by the GoogLeNet framework.
Oh et al. [
53] proposed a batch-based CNN approach with a probabilistic Grad-CAM saliency map that was compatible with a batch-based approach. This approach considered the limited availability of CXR images for the classification of COVID-19. The researchers used a public dataset containing 502 CXR images, including 180 COVID-19 images, 191 normal images and 113 belonging to three other classes: viral pneumonia, bacterial pneumonia, and tuberculosis. To reduce classification bias in the system, the data were randomly distributed into training (354 images), validation (49 images), and test (99 images) sets. The CXR images were first preprocessed for data normalization, and the images were resized to 224 × 224 pixels to obtain the preprocessed data. The preprocessed data were then fed into a segmentation network, from which lung areas could be extracted for network training and classification using patch-by-patch training and inference. The final decision regarding the network classification was based on majority voting. The experimental results were stable with a small dataset and achieved 88.9% accuracy using the proposed batch-based CNN approach. The effects of patch size, different segmentation methods (e.g., U-Net, FC-DenseNet63, FC-DenseNet-103), and training dataset size were also evaluated in relation to the overall performance of the proposed system.
An attention-based VGG-16 model was used for the classification of COVID-19 in [
54]. A total of 4901 image data were used in this study from three different datasets with their own sets of unique challenges and limitations. The original image size was reduced in dimensions to 224 × 224 pixels. To reduce the classification bias, data from each dataset were randomly divided into 70% training and 30% validation sets. The proposed attention-based method was based on four main building blocks: an attention module, a convolution module, FC layers, and a softmax classifier. The attention module was used to capture the spatial relationships of visual clues in the COVID-19 CXR images. The output from the attention module was given as an input to both maximum pooling and average pooling on the input tensor, which was the fourth pooling layer of the VGG-16 model in the proposed method. After that, these two resultant tensors (maximum-pooled 2D tensor and average-pooled 2D tensor) were concatenated to one another to perform a convolution, followed by the fully connected layers and the softmax classifier to give the final output. Based on the inherent characteristics and limitations of each dataset, the performance of the proposed approach varied, with the accuracy ranging between 80% and 87%.
A deep learning pipeline for the diagnosis and discrimination of viral, non-viral, and COVID-19 pneumonia CXR images was developed in [
55]. The dataset used in this study included data from two public datasets: CheXpert and CC-CXRI. The total CXR images included 1571 COVID-19 images, 5656 viral pneumonia images, 11,591 other pneumonia images, and 10,477 normal images. The CXR images were resized to 512 × 512 pixels. The common thoracic disease detection module classified the standardized CXR images into 14 different classes. Multiple external validations were performed, with an average ratio of random training, validation, and testing data distribution amounting to 80%, 10%, and 10%, respectively. The following three modules provide the main functionality of the proposed deep learning pipeline: (1) a CXR standardization module, (2) a common thoracic disease detection module, and (3) a final pneumonia analysis module. The pneumonia analysis module consists of a lung-lesion segmentation model and a final classification model to estimate the subtype of pneumonia and the severity of COVID-19. The lung-lesion segmentation training was based on 1016 CXR images that were manually segmented into four categories and common lesions to develop a model that could differentiate between COVID-19 and other types of pneumonia. The final classification model was developed based on the DenseNet-121 architecture, which was able to perform lung-lesion segmentation and pneumonia diagnosis. The results showed that the proposed deep learning pipeline was able to predict COVID-19 pneumonia with an AUC of 86.8%, along with a recall of 80.65% and precision of 82.05%.
Fusion Module–Hand-Crafted Features–Deep Learning Features (FM–HCF–DLF) is another model for COVID-19 CXR classification given in [
56]. The study made use of an imbalanced dataset containing 220 images for COVID-19, 27 for normal lungs, 11 for SARS, and 15 for pneumonia. In the preprocessing part of the system, a 1D Gaussian operator was used for noise removal and image smoothing for the input images, followed by resizing the original images down to 299 × 299 pixels. The FM model incorporates the fusion of hand-crafted features with the help of local binary patterns (LBPs) and deep learning. The deep learning (DL) features are computed using the convolutional neural network (CNN)-based Inception-v3 framework, followed by a multilayer perceptron (MLP) to provide the final output classification. The proposed method’s performance was compared with that of traditional ML algorithms to highlight the superior performance of the proposed model, which achieved 94.08% accuracy.
CVDNet is a deep convolutional neural network (CNN) model to distinguish COVID-19 infection from normal lungs and other pneumonia cases using chest X-ray images, as presented in [
57]. The proposed architecture is based on a residual neural network and is constructed by using two parallel levels with different kernel sizes to capture the local and global features of the inputs. This model is trained on a publicly available dataset containing a combination of 219 COVID-19, 1341 normal, and 1345 viral pneumonia CXR images. The images are randomly distributed into 70% for training, 10% for validation, and 20% for testing to reduce classification bias. Some of the preprocessing functions include cropping and resizing the original images to provide input images of 256 × 256 pixels in size. Two streams with four parallel residual blocks are used for deep feature extraction, followed by feature concatenation leading to a final residual block, and ending with a fully connected layer and a softmax classifier. The proposed system can provide an accuracy of 96.69%, which is comparable to state-of-the-art methods for the classification of COVID-19.
Azemin et al. [
58] used a ResNet-101-based deep learning model. A total of 10,359 images were used in this study, of which 154 were from COVID-19. Despite the over-whelming imbalance in the dataset used in this study, the authors failed to provide an adequate strategy for mitigating the effects of data imbalance on the overall system performance. The input data were randomly distributed into training (3063 images), validation (1313 images), and test (5828 images) sets to evaluate the performance of the proposed system. The best accuracy provided by the proposed system was 71.9%, which is considerably lower than that achieved in many of the studies discussed in this section.
Despite the findings of these studies, there are notable limitations in terms of small sample sizes, the use of too few pneumonia classes, and dependence on one modality. These limitations have implications for the potential use of these research findings in real-world healthcare applications.
4.3. Identification of COVID-19 in Chest CT Images Using Deep Learning Models
Multiple studies have used chest CT scans to detect and classify COVID-19 in im-ages. CT images are chosen due to the ease of detecting abnormal regions in infected lungs, along with their accuracy in extracting features of pneumonia. However, in practice, CT scans are not the most suitable option for COVID-19 diagnosis due to the limited availability of the necessary equipment and the high cost associated with the process. In addition, both the American College of Radiology and the Italian Society of Radiology (SIRM) do not recommend chest CT scans as a screening tool for COVID-19 [
59]. Based on these guidelines, many studies refrain from using CT images as the input modality for the classification of COVID-19. Consequently, this subsection of the paper considers studies that ultimately decided to rely on CT scans as the primary modality for data collection for the development of their deep-learning-based models for the classification of COVID-19.
Amyar et al. [
60] designed a slice-level classification model for three learning tasks—segmentation, classification, and image reconstruction—for CT scan images. A CNN model was used, the architecture of which consists of a common encoder and two decoders based on U-Net. A common encoder module was used for the three tasks, taking a CT scan as its input, and its output was then used for image reconstruction through the first decoder module, followed by the segmentation task completed by the second decoder module and multi-class classification for COVID-19 (and other lung diseases) performed by the multilayer perceptron. Different state-of-the-art models were compared to this classification model; the authors used AlexNet, VGG-16, VGG- 19, ResNet-50, 169-layer DenseNet, InceptionV3, Inception-ResNet v2, and EfficientNet. The dataset was collected from three hospitals and contained 1369 images divided into three classes: 425 for normal cases, 449 for COVID-19 cases, and 495 for other infections. In order to avoid classification bias, the original balanced dataset was randomly distributed into training (1069 images), validation (150 images), and test (150 images) sets for performance evaluation. The preprocessing stage involved the conversion of original images down to 256 × 256 × 3 pixels and 512 × 512 × 3 pixels, and these two sizes were used as inputs to the proposed models, with the smaller images providing a comparatively better performance. The proposed model outperformed state-of-the-art methods for the classification and segmentation of COVID-19, achieving an accuracy of 94.67% for the classification task and a dice coefficient of 88.0% for the segmentation task.
Li et al. [
61] developed a 3D deep learning framework using the architecture of COVNet to identify COVID-19 from CT scan images. The initial input data of 3D CT scans were preprocessed to reduce the original dimensions to 512 × 512 × 3 pixels, followed by the extraction of the region of interest (i.e., lungs) using a U-Net-based segmentation model. The preprocessed image was given as an input to COVNet to extract visual features from 2D local and 3D global images. The COVNet framework leverages ResNet-50 as the backbone model for deep feature extraction from CT slices, which are combined using a maximum pooling operation, followed by a fully connected layer and softmax activation function to generate a probability score for classification (COVID-19, CAP, and non-pneumonia). The study used a dataset obtained from six hospitals, amounting to 4356 CT scan images divided into three classes: 1296 for COVID-19, 1735 for CAP, and 1325 for non-pneumonia. To remove classification bias, the original balanced dataset was randomly distributed into 3918 CT images for model training and 434 CT images for model testing. The proposed framework achieved an AUC of 96% for the identification of COVID-19 and 95% for CAP on CT scan images.
Another study proposed a DL-based pipeline for CT images called CoviWavNet for the automatic diagnosis of COVID-19 [
62]. In the preprocessing phase of the proposed system, data from two different datasets are concatenated and augmented to increase the size of each of the datasets and reduce overfitting. Some of the augmentation processes performed include scaling, shearing, rotation, flipping on the x- and y-axes, and random translation in the x- and y-directions. Finally, the augmented images are resized to 227 × 227 × 3 pixels. The proposed CoviWavNet uses multilevel discrete wavelet transform (DWT) and heatmaps of the approximation levels to train three different ResNet models for classification. To examine the effect of the combination of spatial and spectral–temporal information on diagnostic accuracy, deep spectral–temporal features are generated from ResNet using transfer learning and integrated with deep spatial features extracted from ResNet models trained with the original CT slices. To reduce the dimensionality, the most valuable feature is selected using the minimum-redundancy–maximum-relevance (mRMR) technique and used as inputs to three support-vector machine (SVM) classifiers. The performance of the proposed system achieves accuracy of 98.62%, precision of 99.54%, an F1-score of 99.62%, and recall of 99.69%.
Alshazly et al. [
63] applied two separate CT datasets for developing a deep-learning-based system for the classification of COVID-19 using the most advanced networks, such as SqueezeNet, Inception, ResNet, ResNeXt, Xception, ShuffleNet, and DenseNet. The original images from the two datasets were randomly distributed into 60% for training and 40% for testing, and were resized to 253 × 349 × 3 pixels. Data augmentation methods were implemented to effectively increase the number of training samples for improved generalization. Some of these methods included random horizontal flipping, normalization, cropping, blurring, Gaussian noise addition, and brightness and contrast improvement. To assess the performance of the proposed models, stratified K-fold (K = 5) cross-validation was used to ensure class-level consistency in each of the five folds. The proposed model achieved high accuracy, with 99.4%, and an F1-score of 99.4%.
Another study proposed a combination of radiomics and artificial intelligence for the analysis of medical images using a CAD framework with four phases [
64]: image preprocessing, feature extraction, feature fusion, and classification. In the first phase, the images are analyzed using two texture-based radiomic approaches: gray-level co-variance matrix (GLCM), and discrete wavelet transform (DWT). The radiomics and original CT images are resized to 227 × 227 × 3 pixels before they are given as inputs for the feature generation and extraction phase. Different data augmentation techniques are performed on the radiomics and original CT data, including shearing, scaling, random translation, and rotation. In the second phase of the proposed framework, three residual networks (ResNets) are used for deep feature extraction. In the third phase, these features are fused together using discrete cosine transform (DCT). In the fourth phase, three machine learning classifiers are used to perform the classification procedure. The dataset used in this study was a benchmark 2D CT dataset containing a total of 2482 CT images, with 1252 for COVID-19, while the remaining 1230 were non-COVID-19. The proposed framework was able to provide accuracy of 99.6% and an F1-score of 99.6%.
The uAI Intelligent Assistant Analysis System (IAAS) is a deep-learning-based software platform developed by United Imaging Medical Technology Company Limited (Shanghai, China) for the classification of COVID-19 [
65]. The IAAS software has an underlying deep learning architecture consisting of a modified 3D convolutional neural network and a combined V-Net with bottleneck structures. A total of 2460 images were used in this study (2215 for COVID-19, while the rest were normal). The authors failed to provide details regarding the data imbalance issues with respect to the distribution of the training and validation sets. At the same time, detailed information regarding preprocessing procedures and final performance was also not provided. One potential reason could be that the main purpose of the study was to assess the feasibility of utilizing the uAI IAAS as a diagnostic tool for COVID-19 from CT images.
Shah et al. [
66] utilized different deep learning frameworks to differentiate between the CT images of COVID-19 and non-COVID-19 patients. CTnet-10 was designed for the diagnosis of COVID-19, developed using six successive layers of convolution and maximum pooling, followed by flattening and dropout layers, and ending in a softmax classification layer. Some of the other models that were tested included DenseNet-169, VGG-16, ResNet-50, InceptionV3, and VGG 19. The performance of the different deep learning models was assessed to highlight the most suitable option for the classification of COVID-19. For the CT-net model, the input image size was 128 × 128 pixels. For the VGG-19 model, the image dimensions used were 224 × 224 × 3 pixels. The images were randomly distributed into 80% for training, 10% for validation, and 10% for testing. CT-net provided an accuracy of 82.1%, while the VGG-19 model was able to provide an accuracy of 94.5%.
Wang et al. [
67] developed a deep-learning-based framework for CT scans of COVID-19 cases. The dataset used in this study was based on 1065 CT images of COVID-19 patients as well as patients who had a prior history of typical viral pneumonia. Unfortunately, the authors did not mention the class-wise distribution of data for the different lung diseases and COVID-19, limiting the ability to assess class balance for this study. The dataset was randomly divided into one training subset (320 CT im-ages), one internal validation subset (455 CT images), and one external validation cohort (290 CT images). The proposed architecture consisted of the preprocessing module, deep feature extraction module, and final classification module. The preprocessing module involved the conversion of the original images into grayscale, followed by grayscale binarization, background area filling, color reversal, and ROI selection. The preprocessed images were rescaled to 299 × 299 × 3 pixels before being used for deep feature extraction. The transfer learning process involved training with a predefined model, which in this study was the GoogLeNet–Inception-v3 deep learning architecture. After feature extraction, the final step was to provide multi-class classification using an ensemble of classifiers to improve performance. The proposed model was able to provide an accuracy of 89.5% on internal validation and 79.3% on external validation, which is significantly lower than the performance provided by prior studies discussed in this section.
As this review’s findings indicate, many studies have been published that rely on CT scans to diagnose COVID-19. However, these studies not only suffer from limitations, but also produce models that cannot be applied practically and economically in routine clinical practice. This is especially due to the cost and limited availability of CT scans, as well as the circumstances of the ongoing pandemic. In comparison, CXR has better availability and ease of execution, and minimizes in-hospital transmission; it is neither time-consuming with lengthy waiting times, such as the CT scan procedure, nor does it require wearing a special suit. In addition, CT scans are often optimally applied in critical cases where the infected lungs are very clear for radiologists.
4.4. Identification of COVID-19 from CXR Images using Deep Learning Models in a Hierarchical Classification Scenario
One of the earliest studies that applied hierarchical image classification using deep learning was carried out in 2015 [
68]. A hierarchical deep CNN (HD-CNNs) model was proposed by embedding deep CNNs into a two-level category hierarchy, where the easily distinguishable classes were classified using a coarse-category classifier and difficult classes were classified using fine-category classifiers. Hierarchical classification models have a noticeable impact on reducing classification errors.
Despite their advantages in classifying COVID-19 due to the similarity in symptoms and physical features shared with many other diseases (e.g., viral pneumonia, bacterial pneumonia, tuberculosis), only one study by Pereira et al. [
69] has used a deep hierarchical model for the classification of COVID-19. The study leveraged deep learning with a pre-trained CNN model to classify 1144 CXR images for 7 flat labels and 14 hierarchical labels of multi-class pneumonia classification, including COVID-19 and healthy lungs. To reduce classification bias, the dataset was randomly distributed into training and validation sets of 70% and 30%, respectively. To reduce the effect of imbalanced data on the final performance of the classifier, different resampling algorithms were employed (i.e., ADASYN, SMOTE, SMOTE-B1, SMOTE-B2, Al-lKNN, ENN, RENN, TomekLinks (TL), and SMOTE + TL). In the feature extraction phase, different hand-crafted features were used, including oriented basic image features (oBIFs), locally encoded transform feature histograms (LETRISTs), and local directional numbers (LDNs), using local phase quantization (LPQ) and deep learning (Inception-v3) methods. Both early and late fusion techniques were employed, followed by data resampling and multi-class and hierarchical classifiers. For multi-class classification, different traditional ML methods were employed, namely, support-vector machines, multilayer perceptrons, random forests, and decision trees. For the hierarchical classification, Clus-HMC—an unsupervised predictive clustering algorithm—was selected as the hierarchical classifier. The experiment proved that the hierarchical structure for the classification of COVID-19, which was tested on the RYDLS-20 dataset, achieved a higher F1-score (89%) with early fusion and BSIF, EQP, and LPQ features, compared to a flat structure for classification (83%). However, these findings are limited due to the way in which the authors classified pneumonia, as well as the extraction of features from a single modality, which were applied on a small sample size.
4.5. Identification of COVID-19 in Chest CT and CXR Images Using Multimodal Deep Learning Models
Models based on data from diverse radiological imaging modalities can achieve a higher accuracy rate compared to models that use only one imaging modality. However, analyzing CXR and CT images for the same patient in the context of the COVID-19 pandemic is impractical in clinical practice and is considered a significant gap in prior studies.
Horry et al. [
70] proposed multimodal imaging data to detect COVID-19 by combining CXR, ultrasound, and CT scans using a VGG-19 model. A publicly accessible dataset gathered from different data sources, containing 1118 COVID-19 images, 996 pneumonia images, and 60,533 images with no findings, was used in the experiment. To prevent bias in classification, the original dataset was randomly distributed into 20% for validation and 80% for training. To enhance different features in the original images, the contrastive limited adaptive histogram equalization (CLAHE) method was used. Since the different deep learning models have different limitations in terms of image size, the original image dataset had to be modified due to the utilization of different deep learning models, and the input image sized varied from 224 × 224 pixels for VGG variants to 299 × 299 pixels for Inception-v3. The performance of the proposed approach varied based on the type of data modality being used for system training, such that the highest performance was provided by the proposed models trained on ultrasound data and the lowest performance was provided for the proposed models trained on CT scan data. The results indicate that ultrasound images were more accurate compared to CXR and CT scans, with a precision of 100% compared to 86% and 84%, respectively.
Vinod et al. [
71] demonstrated the possibility of integrating CXR images with CT scans in deep learning models to diagnose COVID-19 patients. A deep COVIX-Net model was used to classify CXR and CT images into one of the following three classes: normal, COVID-19, and pneumonia. The dataset was obtained from the Kaggle and GitHub repositories, consisting of 9000 CXR images (3000 pneumonia, 3000 COVID-19, and 3000 normal) and 6000 CT scans (3000 pneumonia and 3000 COVID-19). The original dataset was randomly distributed between 80% training and 20% validation subsets; this ensured that the developed system was able to minimize the classification bias. The images from the original dataset were converted to 224 × 224 pixels, and these images were subjected to different techniques for feature extraction. The features were extracted from the medical images using the following techniques: texture, gray-level co-occurrence matrix (GLCM), gray-level difference method (GLDM), fast Fourier transform (FFT), and discrete wavelet transform (DWT). From the different feature extraction techniques used, some of the different statistical features used for model training included average, skewness, kurtosis, energy, average deviation, dimension, RMS, consistency, average gradient, minimum, and median. Unfortunately, there is no further information regarding the structure of deep COVIX-Net shown in the paper. The outputs from feature extraction from different techniques were given as inputs to a random forest classifier to provide the final classification output. The proposed model achieved promising outcomes, with 96.8% accuracy for the CXR images and 97% for CT scans.
Yadav et al. [
72] proposed a deep unsupervised framework (Lung-GANs) to classify lung diseases based on chest CT scans and CXR images using unlabeled data. The proposed method involved the use of six large, publicly available datasets consisting of 38,155 CXR and CT scan images from healthy, sick, tuberculosis (TB), viral pneumonia, COVID-19, and non-COVID-19 classes. A number of different image preprocessing procedures were leveraged for modifying the original dataset images, such as color mode conversion, image resizing (original images were converted to 512 × 512 pixels), and image normalization. After that, the preprocessed images were randomly distributed into training (70%) and validation (30%) sets to reduce classification bias. The generator module in Lung-GAN takes in a 100-dimensional vector as its input and outputs a single 512 × 512 image. The discriminator module in Lung-GAN is a CNN architecture that can differentiate between real and synthesized images of 512 × 512 pixels in size as inputs, and its output is given as a probability value specifying whether the image is real or not. The authors developed an ensemble of classifiers, with linear support-vector classification (SVC) and random forests serving as the base classifiers, which were combined with predictions from each classifier to produce the final result. The CNN architecture was used for both models. The performance of a GAN-based single framework for all binary classifications on all datasets achieved higher accuracy compared to other state-of-the-art unsupervised models in this area. The breakdown of the performance of Lung-GAN for the different datasets showed diverse results, which varied considerably from one dataset to another. The accuracy values ranged between 94% and 99.5%, with an average accuracy of 97.6%. This shows that the proposed system is sensitive to variations in dataset characteristics (i.e., noise, image dimensions, quality and quantity of data, metadata).
Kalaiselvi et al. [
73] designed three artificial intelligence models: sANN ML, using machine learning; pVGG TL, using transfer learning; and pCNN DL, using deep learn-ing. The purpose of each model was to detect COVID-19 from CXR and CT scan images. Each model used the ReLU and E-Tanh activation functions. The public dataset was collected from different research and medical centers, and contained a total of 650 CXR images and 746 CT images divided into positive and negative COVID-19 cases. The training and validation sets contained a total of 625 images for training and 10 images for validation. Despite the considerable data imbalance, the study failed to shed light on the techniques to be used for mitigating the underlying issues. For the different models highlighted above, a VGG-16 network, ANNs with different activation functions, and CNNs with different activation functions were used to provide the backbone for classification. The sANN ML and pVGG TL models achieved high accuracy (100%) in the detection of COVID-19 from CXR images. However, pCNN DL did not perform well, which could be attributed to the small size of the dataset. Notably, each model had a low detection performance for CT scan images compared to CXR images. In addition, the E-Tanh activation function yielded positive results for CXR images.
Another study made use of 8879 CXR and 3724 CT images for the training and development of the proposed deep learning model [
74]. The authors did not specify the distribution of the original data between training, test, and validation sets. In the preprocessing stage of the proposed model, contrast-limited adaptive histogram equalization (CLAHE) was used for contrast and feature enhancement. The use of different data augmentation strategies, such as random transformations (i.e., rotation, horizontal and vertical translations, zooming, and shearing), ensured that the system could be generalized well to unknown data. All of the data were resized to 512 × 512 pixels before being used in the system. In the first stage of the proposed model, an In-ception-v3 deep model was trained for the recognition of COVID-19 using multimodal learning by leveraging data from both modalities, i.e., X-ray and CT scan. The second stage was based on a convolutional neural network architecture for recognizing three types of lung disease. The third stage was based on transfer learning from pulmonary nodule segmentation in CT scans to produce binary masks for segmenting similar regions in the given data. Ultimately, this method showed an accuracy of 99.4%, precision of 99.5%, recall of 99.1%, and F1-score of 99.3%.
Ibrahim et al. [
75] examined the effects of four deep learning models for the classification of COVID-19 using multimodal data consisting of CXR and CT images. The first stage of the proposed system is responsible for performing different image preprocessing functions, such as resizing, image augmentation (i.e., flipping, rotation, and skewing), and random data distribution of the total dataset (75,000 images) into training (70%) and validation (30%) subsets. Unfortunately, the authors did not provide the distribution of the dataset, limiting our ability to assess the level of balance in the dataset between the individual classes. The input images were resized to 224 × 224 pixels, which is a standard size that is suitable for input to the different deep learning models used in this study. The second and third stages are responsible for deep feature extraction and image classification using the following four networks: VGG19-CNN, ResNet152-v2, ResNet152-v2 + gated recurrent unit (GRU), and ResNet152-v2 + bidirectional GRU (Bi-GRU). Of the four networks mentioned, the best results were provided by ResNet152-v2 + bidirectional GRU, with an accuracy of 98%, precision of 99.5%, recall of 98%, and F1-score of 98.24%.
Sharma et al. [
76] used the VGG-16 deep learning model for the classification of COVID-19. Before identifying the different lung infections, different preprocessing functions and data augmentation operations were performed to minimize the classification bias, enhance the system’s generalizability, and improve the quantity and diversity of the data. Open-source data were acquired for this study, namely, the COVIDx CT-2A dataset, which includes 194,922 images from 3745 patients. From the open-source data, original data were used and randomly distributed into 80% for training and 20% for testing. The authors of this paper failed to highlight the class-wise distribution of the dataset used in their study, limiting our ability to assess level of class imbalance. Augmented images were converted into 512 × 512 pixels before being used for model training and validation. For the classification of COVID-19, the proposed model was able to provide the best performance, with 99.2% accuracy, 99.6% precision, and 99.8% recall.
A deep transfer learning algorithm was proposed in another study [
77] to provide a rapid-response-based system for the classification of COVID-19 using multimodal data for CXR and CT images. Data from different publicly available sources were consolidated and utilized in this study. For example, a total of 6111 CXR images were acquired from two separate sources. Similarly, another data source was used for acquiring a total of 1252 CT scans. In the preprocessing stage, the original images were converted into 512 × 512 pixels. The data were randomly split into training, validation, and testing subsets of 64%, 20%, and 16%, respectively. VGG-19 was used for transfer learning in this study; this deep learning model is based on 16 convolution layers and 3 fully connected layers, followed by 5 maximum-pooling layers and a softmax layer. The VGG-19 model is followed by the Grad-CAM model, which takes an image input to provide improved visualization output for detecting regions of interest. Once the predicted label has been calculated using the VGG-19 model, Grad-CAM is applied to the last convolutional layer of the VGG-19 model. Based on the different experiments performed, the best results obtained by the proposed algorithm provided an accuracy of 95.61%, precision of 88%, recall of 97%, and F1-score of 92%.
In another study [
78], two separate deep learning models were used for the classification of COVID-19. The major difference between the two models was in terms of the data modalities used: the first model used CNN with CT and X-ray images separately, whereas the second model used CNN with both CT and X-ray images simultaneously. The dataset used in this study contained CXR and CT images divided into three separate classes: COVID-19, normal lungs, and pneumonia. The data were randomly distributed into training (3135 images) and testing (900 images) subsets. In the preprocessing stage, the sizes of the CXR and CT images were reduced to 299 × 299 pixels and 512 × 512 pixels, respectively. In the first model, the architecture consisted of different convolutional layers, pooling layers, dropout layers, and a softmax classification layer. In the second model architecture, the data from two modalities were provided to two separate architectures (where each architecture was based on the first model), followed by the concatenation of data from two models after the flattening layer and softmax classification layer. The deep learning model trained on multimodal data was able to outperform models trained on single data modalities, with accuracy, precision, and recall of 99% each.
4.6. Identification of COVID-19 in Chest CT or CXR Images with Clinical/lab Test Features Using Multimodal Deep Learning Models
The use of deep learning for multimodal data fusion has a substantial effect on medical applications. The EMIXER model is the only model that has been proposed in the literature to utilize CXR images along with radiologists’ reports to classify CXR images and generate diagnostic reports [
79]. The study was based on end-to-end multimodal data fusion that combined CXR images and corresponding text reports. CNN and recurrent neural network (RNN) models were used with the MIMIC-CXR dataset, which contains CXR images with associated reports. EMIXER is composed of five different modules, namely, the image generator (used to synthesize X-ray images from a prior noise distribution conditioned on label information), image-to-report decoder (used to provide an output text report when an X-ray image is given as an input), image discriminator (used to differentiate between real and synthetic X-ray images), text discriminator (used to differentiate between real and synthetic X-ray reports), and joint discriminator (used to combine X-ray images and text to discriminate between real and synthetic data). The COVID-19 classification task dataset contained 14 different classes that were resized to 128 × 128 pixels. Overall, EMIXER used 100,000 real images as well as 300,000 synthetic images; the addition of synthetic images led to a considerable improvement in results, with a maximum accuracy value of 92.4%. The researchers observed that EMIXER improved the classification of COVID-19 from CXR images. Unfortunately, the authors did not specify the class-level information or the training/validation split for the dataset used, limiting our ability to assess class-level imbalance and associated biases in performance.
Mei et al. [
80] suggested that developing a model based only on CT scans may lead to limited negative predictive power. Therefore, the authors proposed the use of chest CT images along with clinical symptoms (e.g., fever, cough, and cough with sputum), laboratory testing (e.g., white blood cells, neutrophils, percentage neutrophils, lymphocytes, and percentage lymphocytes), and exposure history. The dataset was collected from 905 patients across 18 healthcare centers in China, where 419 cases were positive and 486 were negative, to provide a balanced dataset, with an image size of 512 × 512 × 3 pixels. The original data were randomly distributed into 60% training, 10% validation, and 30% testing sets. The Inception–ResNet-v2 model was used to process the CT images, while support-vector machine (SVM), random forest, and MLP classifiers were used for clinical information. The joint model achieved better performance (AUC = 0.92) compared to the CNN model trained only on CT scans (AUC = 0.86); furthermore, it outperformed the MLP model trained on clinical information (AUC = 0.80).
Chen et al. [
81] also found that the diagnostic model composed of radiological semantic features with clinical features was significantly different. The dataset used in the experiment (CT scans and clinical information) was collected from five independent hospitals in China. A total of 136 cases were labelled as COVID-19 or non-COVID-19. The researchers identified 18 radiological semantic features and 17 clinical features, including demographic information, daily body temperature, blood pressure, heart rate, clinical symptoms, history of exposure to epidemic centers, total white blood cell (WBC) counts, lymphocyte counts, lymphocyte ratios, neutrophil count, neutrophil ratios, procalcitonin (PCT), C-reactive protein (CRP) levels, and erythrocyte sedimentation rates (ESR). To compare diagnostic performance, the authors developed three models: one for clinical features, a second for CT scan features, and a third for the proposed model that combined them. The proposed model outperformed the others, with an AUC of 0.986, while the AUC values for the clinical and CT scan feature models were 0.952 and 0.969, respectively.
The combined use of CT scan images and clinical findings also occurred in another study [
82]. Notably, this research also achieved promising diagnostic results for COVID-19. The study’s dataset consisted of 168 patients, including 88 COVID-19-positive and 80 COVID-19-negative patients. The latter category included patients with bacterial infection, Mycobacterium tuberculosis complex, influenza virus A, influenza virus B, influenza virus B and mycoplasma, and mycoplasma. The total dataset was randomly distributed into a training subset (118 patients) and a testing subset (50 patients). The data from continuous variables within the clinical information were categorized into different classes using means, medians, and standard deviations. The categorical data were analyzed using chi-squared or Fisher’s tests. Using 10 variables, logistic regression analysis was performed with ROC curves for analyzing the performance. Data from CT scans allowed the separation of visual features (i.e., the number of affected lobes and segments, segments with peripheral GGO, consolidation, air bronchograms, crazy-paving patterns, subpleural curvilinear lines, bronchiectasis, and patchy lesions), which were used to categorize between COVID-19 and non-COVID-19 patients. The predictive model utilizing clinical information with CT scan features was able to achieve good results, with an AUC of approximately 0.91.
In addition, Chen et al. [
83] noted that there is a lack of awareness of the importance of biomedical features and choosing the right technical approach to diagnose COVID-19. Therefore, they proposed a late fusion deep learning–machine learning multimodal diagnostic approach to classify 214 patients with non-severe COVID-19, 148 patients with severe COVID-19, 129 patients with other viral infections, and 198 uninfected individuals. The data for 689 patients were collected from different hospitals in China, and the healthy cases (control group) were selected from patients who made up a regular annual physical examination cohort. For the preprocessing stage, the original image dataset was reduced to 512 × 512 pixels. The data were randomly distributed into 80% training and 20% validation subsets. The features used in the model were clinical (23), lab testing (10), and CT scan features. A customized ResNet CNN model was used for CT scan images and applied with three different ML models—random forests, SVM, and k-nearest neighbors (kNN)—for clinical findings and lab testing. The best performance with regard to all metrics was achieved when integrating SVM with ResNet, with an overall multimodal classification accuracy of 99.8%. This was a higher result compared to the use of a single modality (unimodality), which achieved 75.5% accuracy on clinical features alone, 67.7% accuracy on lab test features alone, and 90.8% accuracy on CT scan data alone.
A number of different deep learning models were used in another study, and their performance was compared to highlight the most suitable alternative for the classification of COVID-19 [
84]. The private dataset was collected from King Fahad University Hospital, Dammam, KSA, and consisted of 270 cases. The preprocessing step was able to reduce the image size to 224 × 224 pixels (for case 2) and 300 × 300 pixels (for case 3), followed by random data distribution into 80% training and 20% validation sets. Different data augmentation techniques were employed, such as flipping (both horizontal and vertical), rotation, shifting, cropping, blurring, zooming, rescaling, and shearing. Three different cases were used in this study for selecting the most suitable deep learning model for the classification of COVID-19. Case 1 used clinical data for the training and validation of a 13-layer deep learning model. Case 2 used CXR data for training and validation on a diverse range of CNN architectures (i.e., ResNet, DenseNet, VGG19, EfficientNet). Case 3 used multimodal data (i.e., clinical and CXR images) with transfer learning to learn weights for EfficientNet, which was used as a backbone architecture for the classification of COVID-19. For case 3, which used multimodal data, the proposed system was able to provide the best performance (accuracy of 97%, recall of 98.6%, precision of 97.8%, and F1-score of 98.2%).
Similarly, Attaullah et al. [
85] used multimodal data of symptoms and CXR images for the development of a deep-learning-based model for the classification of COVID-19. The public dataset contained a total of five classes: bacterial, COVID-19, non-COVID-19 viral, initial-stage COVID-19, and normal. The total dataset was subjected to random splitting into an 80% training set and 20% testing set. The images in the publicly available dataset were converted into 150 × 150 pixels as one of the main preprocessing steps. For the symptom data, the preprocessing step involved the removal of duplicate rows and null values and the application of resampling techniques to address the class imbalance issues. This step was followed by the training and validation of the preprocessed data using logistic regression and CNN models. For image data, different data augmentation techniques—zooming, rotation, and translation—were performed during CNN training. The CNN model was trained on the transformed images, and the decision tree model was trained on the labeled results of the previous two trained models to provide the final output, with an accuracy of 78.88%.
A fully automated hybrid framework based on capsule networks (CT-CAPS) and random forest classifiers was used in another study for the classification of COVID-19 using chest CT images and clinical/demographic data [
86]. Private CT scans and the associated clinical/demographic data were collected for a total of 312 patients (176 COVID-19 patients, 60 pneumonia patients, and 76 normal cases) in this study. The dataset was randomly split into training (60%), validation (10%), and testing (30%) subsets. A capsule-network-based framework—namely, CT-CAPS—was used in this study, consisting of a stack of convolutional, pooling, batch normalization, and capsule network layers to extract slice-level feature maps from CT images in the first stage of the proposed model. The second stage of the proposed model leveraged the maximum pooling output of the first stage, followed by a conventional multilayer perceptron for final classification. The proposed model was able to provide 90.8% accuracy, 94.5% precision, 86% recall, and an AUC of 0.92.
Jiao et al. [
87] leveraged deep learning models for the classification and severity prediction of COVID-19; a total of 1834 patients’ CXR images and clinical data were used. The data were randomly distributed into 70% for training, 10% for validation, and 20% for testing. The deep learning features extracted from the model and the clinical data were used to predict the risk of COVID-19 progression. All images and masks were resized to 512 × 512 pixels in size and were normalized before being given as inputs to the segmentation model (i.e., U-Net). For the severity prediction model, the CXR image data were segmented using a pre-trained U-Net architecture, followed by feature extraction using a VGG-13 model with five encoder and five decoder blocks to learn the transformation from input images and binary masks. The final results for disease progression were computed based on the combined weighted sum of the individual image and clinical data scores. The model using combined data was able to provide better prediction performance on internal and external testing.
For the development of a deep-learning-based system for the classification of COVID-19 in [
88], data from 654 patients with a total of 5645 CXR images were acquired. Imaging and clinical data were used to train five longitudinal transformer-based networks, applying fivefold cross-validation. In the preprocessing stage, some of the different functions used to modify the images included inversion, padding, resizing (512 × 512 pixels), pixel value normalization, and scaling. The data were randomly divided into 80% for the training subset and 20% for the validation subset. The extracted features from CXRs were combined using global average and global maximum pooling operations, followed by two fully connected layers and a softmax layer to provide risk probability. The deep learning model with the combined data modalities was able to provide the best performance, with 73.2% accuracy and a 70.7% F1-score.
Despite the superior results that these studies have achieved by using multimodal models, the existing systems still suffer from some gaps. These include the fact that the radiologist’s report in the EMIXER model is generated from the CXR image only, without using any clinical information that may support the report’s findings. Moreover, relying on a small sample size of CT images to diagnose COVID-19 patients using multimodal deep learning is another limitation.
Table 1.
Summary of the data extracted for each paper included in our review.
Table 1.
Summary of the data extracted for each paper included in our review.
| References | Data Used | Sample Size | Dataset
Balance | Balance Strategy | Model Type | Classification Method | Performance |
|---|
| Chauhan et al. [39] | CXR | 5232 images | No | Augmentation techniques | Ensemble | Multi-class | Accuracy = 96% |
| Khan et al. [40] | CXR | 3224 positive,
3224 negative | Yes | - | Ensemble | Binary | Accuracy = 98%
F-score = 98% |
| Al-Waisy et al. [41] | CXR | 400 negative,
400 positive | Yes | - | Ensemble | Binary | Accuracy = 99% |
| Bhowal et al. [38] | CXR | 752 COVID-19
1584 viral, 1639 normal | No | Augmentation techniques | Ensemble | Multi-class | AUC = 97%
Accuracy = 99% |
| Mazaar et al. [42] | CXR | 219 COVID-19,
1345 viral pneumonia,
1341 normal | No | Augmentation
techniques | Ensemble | Multi-class | Accuracy = 97.8% |
| Attallah et al. [43] | ECG records and images | 250 COVID-19,
859 normal
848 others | Yes | - | Ensemble | Multi-class | Accuracy = 91.6%
Precision = 91.8%
Recall = 91.6% |
| Rajaraman et al. [44] | CXR | 360 normal
360 COVID-19 | Yes | - | Ensemble | Binary | Accuracy = 90.97%
AUC = 95.08
Precision = 93.94
F1 = 90.91 |
| Bharadwaj et al. [45] | CXR | 219 COVID-19,
2686 normal | No | Not mentioned | Ensemble | Binary | Accuracy = 95.1%
Precision = 100%
Recall = 97% |
| Al-Mansur et al. [46] | CXR | 1281 COVID-19,
3269 healthy lungs | No | Augmentation
techniques | Ensemble | Binary | Accuracy = 97.56% |
| Tao et al. [47] | CT-scan | 2500 COVID-19,
2500 normal,
2500 lung tumors | Yes | - | Ensemble | Multi-class | Accuracy = 99.1%
Precision = 99.1%
Recall = 99.6% |
| Abdelsamea et al. [50] | CXR | 105 COVID-19,
80 normal, 11 SARS | No | Augmentation
techniques | Single modality | Multi-class | Accuracy = 93% |
| Brunese et al. [51] | CXR | 250 COVID-19,
6273 other pulmonary diseases and normal | No | Augmentation
techniques | Single modality | Multi-class | Accuracy = 98% |
| Loey et al. [52] | CXR | 69 COVID-19,
79 normal,
79 bacterial,
79 viral | Yes | - | Single modality | Multi-class | Accuracy = 81% |
| Oh et al. [53] | CXR | 180 COVID-19,
191 normal,
113 others | Yes | - | Single modality | Multi-class | Accuracy = 89% |
| Sitaula et al. [54] | CXR | 4901 total images | No | Not mentioned | Single modality | Multi-class | Accuracy = 80–87%
Precision = 91–96%
Recall = 77–95%
F1-score = 83–93% |
| Wang et al. [55] | CXR | 1571 COVID-19,
5656 viral pneumonia,
11,591 other pneumonia,
10,477 normal | No | Augmentation
techniques | Single modality | Multi-class | AUC = 86.8%,
Recall = 80.65%,
Precision = 82.05% |
| Shankar et al. [56] | CXR | 220 COVID-19,
27 normal,
11 SARS,
15 pneumonia | No | Not mentioned | Single modality | Multi-class | Accuracy = 94.08%,
Precision = 94.85%,
F1-score = 93.2% |
| Ouchica et al. [57] | CXR | 219 COVID-19,
1341 normal,
1345 viral | No | Not mentioned | Single modality | Multi-class | Accuracy = 96.69%
Precision = 96.72%
Recall = 96.84%
F1-score = 96.68% |
| Azemin et al. [58] | CXR | 154 COVID-19,
5828 no findings,
2166 opacity,
2210 no opacity | No | Not mentioned | Single modality | Binary | Accuracy = 71.9%
Precision = 77.3%
Recall = 71.8% |
| Amyar et al. [60] | CT-Scan | 449 COVID-19,
425 normal,
495 others | Yes | - | Single modality | Multi-class | Accuracy = 95% |
| Li et al. [61] | CT-Scan | 1296 COVID-19,
1735 CAP,
1325 non-CAP | Yes | - | Single modality | Multi-class | Accuracy = 96% |
| Attallah et al. [62] | CT-scan | 7264 positive,
6382 normal | Yes | Augmentation
techniques | Single modality | Binary | Accuracy = 98.62%
Precision = 99.54%
F1-score = 99.62%
Recall = 99.69% |
| Alshazly et al. [63] | CT-scan | 2517 COVID-19,
758 normal,
1644 others | No | Augmentation
techniques | Single modality | Multi-class | Accuracy = 99.4%
Precision = 99.6%
Recall = 99.1%
F1-score = 99.4% |
| Attallah et al. [64] | CT-scan | 1252 COVID-19,
1230 non-COVID-19 | Yes | Augmentation
techniques | Single modality | Binary | Accuracy = 99.6%
Precision = 99.72%
Recall = 99.47%
F1-score = 99.6% |
| Zhang et al. [65] | CT-scan | 2215 COVID-19,
245 normal | No | Not mentioned | Single modality | Binary | Not mentioned |
| Shah et al. [66] | CT-scan | 349 positive,
463 negative | No | Augmentation
techniques | Single modality | Binary | Accuracy = 94.5% |
| Wang et al. [67] | CT-scan | Total 1065 images including COVID-19 | N/A | Not mentioned | Single modality | Binary | Accuracy = 89.5% |
| Periera et al. [69] | CXR | 90 COVID-19,
1000 normal,
54 others | No | Resampling algorithms | Single modality | Multi-class | F-score = 89% |
| Horry et al. [70] | CXR/CT-Scan/Ultrasound | CXR:
115 COVID-19,
322 pneumonia,
60,361 no finding,
CT-scan:
349 COVID-19,
397 non-COVID-19
Ultrasound:
654 COVID-19,
277 Pneumonia,
172 no finding. | No | - | Multi-modal | Multi-class | Precision = 100% for Ultrasound
Precision = 86% for CXR
Precision = 84% for CT-Scan |
| Vinod et al. [71] | CXR/CT-Scan | 3000 CT-scan, COVID-19,
3000 CT-scan pneumonia,
3000 CXR COVID-19, 3000 CXR pneumonia,
3000 CXR normal | Yes | - | Multi-modal | Multi-class | Accuracy = 96% for CXR
Accuracy = 97% for CT-Scan |
| Yadav et al. [72] | CXR/CT-Scan | 38,155 CXR and CT-scan | No | Not mentioned | Multi-modal | Multi-class | Accuracy = 96% for CXR,
97% for CT-scan. |
| Kalaiselvi et al. [73] | CXR/CT-Scan | 650 CXR,
746 CT-Scan | No | Not mentioned | Multi-modal | Binary | Accuracy = 100% for CXR |
| El-Banaa et al. [74] | CXR/CT-scan | 5719 COVID-19,
2485 normal,
2122 bacterial,
2277 viral | No | Augmentation
techniques | Multi-modal | Multi-class | Accuracy = 99.4%
Precision = 99.5%
Recall = 99.1%
F1-score = 99.3% |
| Ibrahim et al. [75] | CXR/CT-scan | 75,000 for COVID-19, normal, pneumonia,
lung cancer | No | Augmentation
techniques | Multi-modal | Multi-class | Accuracy = 98%
Recall = 98%
Precision = 99.5%
F1-score = 98.24% |
| Sharma et al. [76] | CXR/CT-scan | Total images = 194,922 | No | Augmentation
techniques | Multi-modal | Multi-class | Precision = 99%
Recall = 91%
F1-score = 89% |
| Panwar et al. [77] | CXR/CT-scan | 526 CXR and CT-scan for COVID-19,
1252 CT-scan for COVID-19,
1230 CT-scan for other,
5856 CXR for normal and pneumonia | No | Not mentioned | Multi-modal | Multi-class | Accuracy = 95.61% |
| Ouahab et al. [78] | CXR/CT-scan | 1345
CXR COVID-19,
1345
CXR normal, 1345
CXR pneumonia,
1345
CT COVID-19,
1345
CT normal,
1345
CT pneumonia | Yes | - | Multi-modal | Multi-class | Accuracy = 99%
Recall = 99%
Precision = 99% |
| Biswal et al. [79] | CXR/Radiologist report | 377,110 CXR | N/A | Not mentioned | Multi-modal | Multi-class | Accuracy = 92%
AUC = 90% |
| Mei et al. [80] | CT-Scan/Symptoms/Lab Tests/Exposure history to COVID-19 | 415 COVID-19,
486 negative | Yes | - | Multi-modal | Binary | AUC = 92% |
| Chen et al. [81] | CT-Scan/Clinical information | 70 COVID-19,
66 non-COVID-19, | Yes | - | Multi-modal | Binary | AUC = 98.6% |
| Yang et al. [82] | CT-Scan/Clinical information/Lab Tests | 88 COVID-19,
80 other pneumonias | Yes | - | Multi-modal | Multi-class | AUC = 91% |
| Xu et al. [83] | CT-Scan/Clinical information/Lab Tests | 689 cases | Yes | - | Multi-modal | Multi-class | Accuracy = 99%
|
| Khan et al. [84] | CXR/Clinical data | 222 COVID-19,
48 normal | No | Augmentation
techniques | Multi-modal | Binary | Accuracy = 97%
Recall = 98.6%
Precision = 97.8%
F-Score = 98.2% |
| Attah Ullah et al. [85] | CXR/Symptoms | 200 bacterial,
290 COVID-19,
180 viral,
130 normal | No | Resampling
techniques/Augmentation
techniques | Multi-modal | Multi-class | Accuracy = 78.88% |
| Afshar et al. [86] | CT-scan/Clinical data | 176 COVID-19,
76 normal,
60 CAP. | No | Augmentation
techniques | Multi-modal | Multi-class | Accuracy = 90.8% |
| Jiao et al. [87] | CXR/Clinical data | Total data = 1834 patients | N/A | Not mentioned | Multi-modal | Binary | C-index = 0.805 |
| Cheng et al. [88] | CXR/Clinical data | Total data = 5645 cases | N/A | Not mentioned | Multi-modal | Binary | Accuracy = 73.2%
Recall = 70.7%
Precision = 71.4%
F1-score = 74.6% |