Clustering of Heart Failure Phenotypes in Johannesburg Using Unsupervised Machine Learning
Abstract
1. Introduction
2. Methods
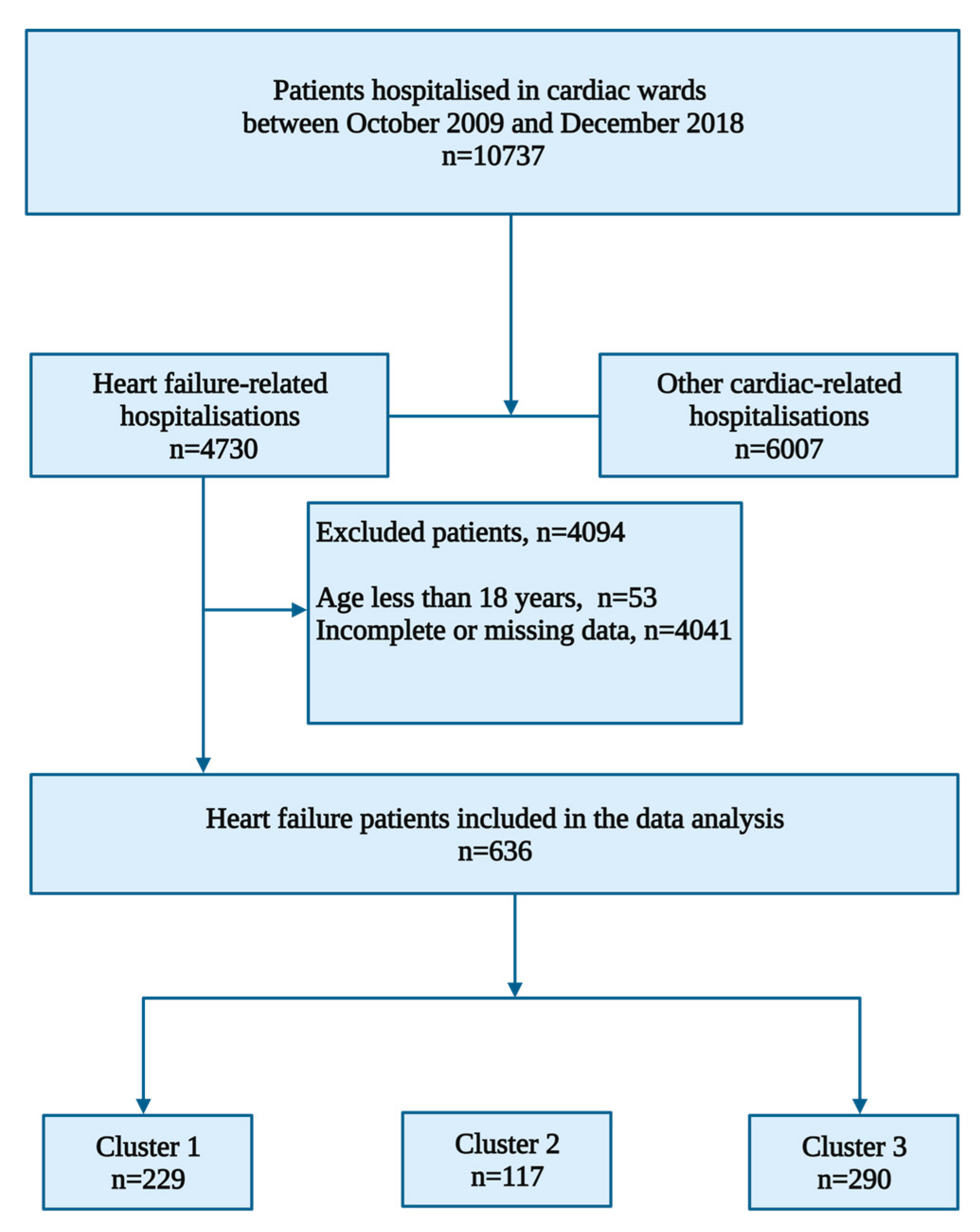
2.1. Study Design and Participants
2.2. Descriptive Statistics
2.3. Data Pre-Processing for Clustering of Heart Failure Patients
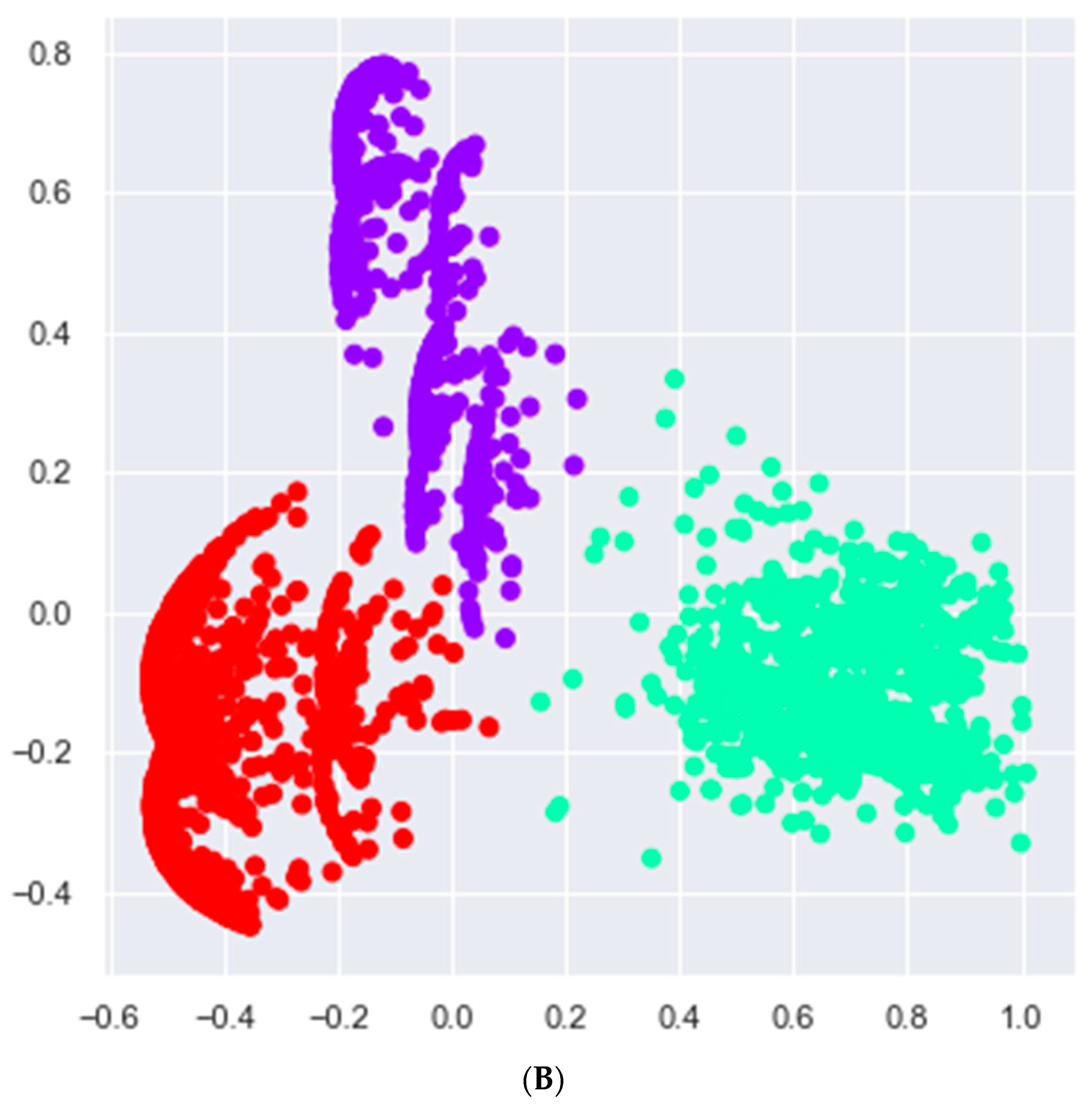
2.4. Cluster Analysis
3. Results
3.1. Baseline Demographics and Clinical Characteristics
3.2. Cluster Analysis
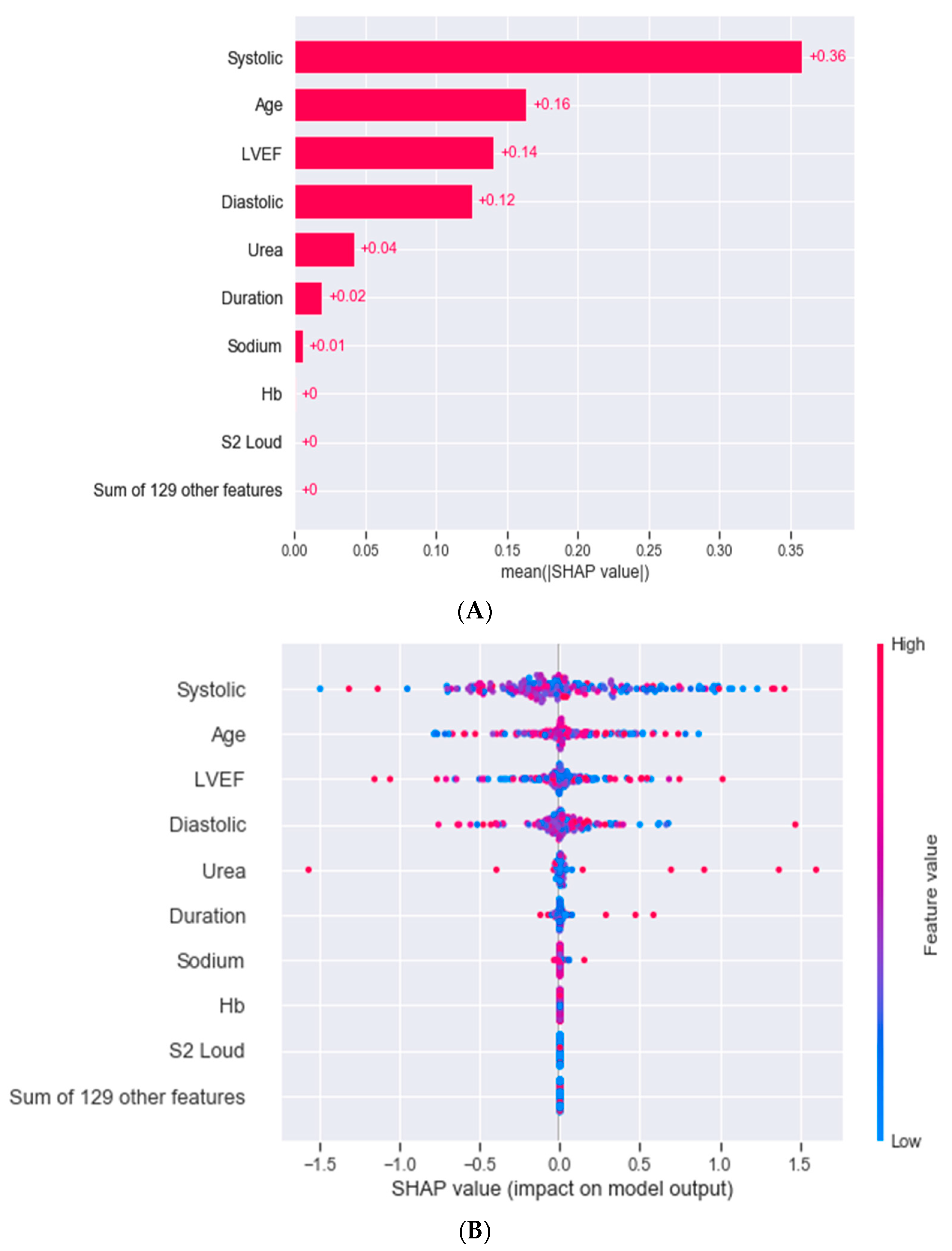
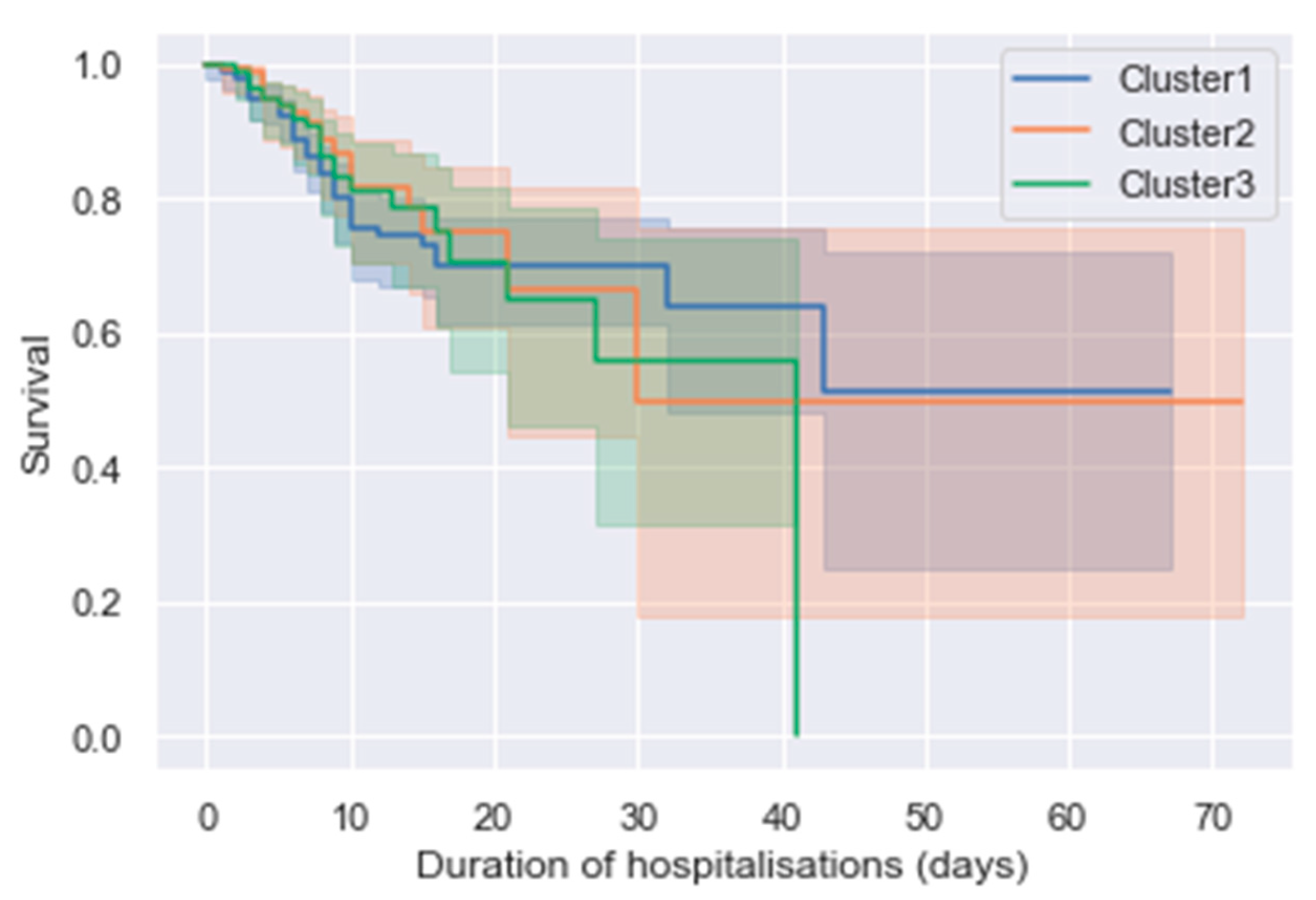
3.3. Outcomes
3.4. Cluster Analysis on a Dataset without the Left Ventricular Ejection Fraction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jovel, J.; Greiner, R. An Introduction to Machine Learning Approaches for Biomedical Research. Front. Med. 2021, 8, 771607. [Google Scholar] [CrossRef] [PubMed]
- Ponikowski, P.; Anker, S.D.; AlHabib, K.F.; Cowie, M.R.; Force, T.L.; Hu, S.; Jaarsma, T.; Krum, H.; Rastogi, V.; Rohde, L.E.; et al. Heart failure: Preventing disease and death worldwide. ESC Heart Fail. 2014, 1, 4–25. [Google Scholar] [CrossRef] [PubMed]
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.; Coats, A.J.; Falk, V.; González-Juanatey, J.R.; Harjola, V.-P.; Jankowska, E.A.; et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. J. Heart Fail. 2016, 18, 891–975. [Google Scholar] [CrossRef] [PubMed]
- Kosaraju, A.; Goyal, A.; Grigorova, Y.; Makaryus, A.N. Left Ventricular Ejection Fraction. In StatPearls. Treasure Island (FL); StatPearls Publishing: Online, January 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK459131/ (accessed on 1 July 2022).
- Jasinska-Piadlo, A.; Bond, R.; Biglarbeigi, P.; Brisk, R.; Campbell, P.; McEneaneny, D. What can machines learn about heart failure? A systematic literature review. Int. J. Data Sci. Anal. 2022, 13, 163–183. [Google Scholar] [CrossRef]
- Banerjee, A.; Chen, S.; Fatemifar, G.; Zeina, M.; Lumbers, R.T.; Mielke, J.; Gill, S.; Kotecha, D.; Freitag, D.F.; Denaxas, S.; et al. Machine learning for subtype definition and risk prediction in heart failure, acute coronary syndromes and atrial fibrillation: Systematic review of validity and clinical utility. BMC Med. 2021, 19, 85. [Google Scholar] [CrossRef] [PubMed]
- Dokainish, H.; Teo, K.; Zhu, J.; Roy, A.; AlHabib, K.F.; ElSayed, A.; Palileo-Villaneuva, L.; Lopez-Jaramillo, P.; Karaye, K.; Yusoff, K.; et al. Heart Failure in Africa, Asia, the Middle East and South America: The INTER-CHF study. Int. J. Cardiol. 2016, 204, 133–141. [Google Scholar] [CrossRef]
- Ajayi, A.A.; Sofowora, G.G.; Ladipo, G.O. Explaining Heart Failure Hyper-mortality in Sub Saharan Africa: Global Genomic and Environmental Contribution Review. J. Natl. Med. Assoc. 2020, 112, 141–157. [Google Scholar] [CrossRef]
- Van Diepen, S.; Katz, J.N.; Albert, N.M.; Henry, T.D.; Jacobs, A.K.; Kapur, N.K.; Kilic, A.; Menon, V.; Ohman, E.M.; Sweitzer, N.K.; et al. Contemporary Management of Cardiogenic Shock: A Scientific Statement from the American Heart Association. Circulation 2017, 136, e232–e268. [Google Scholar] [CrossRef]
- Kwon, J.M.; Kim, K.H.; Jeon, K.H.; Park, J. Deep learning for predicting in-hospital mortality among heart disease patients based on echocardiography. Echocardiography 2019, 36, 213–218. [Google Scholar] [CrossRef]
- Segar, M.W.; Jaeger, B.C.; Patel, K.V.; Nambi, V.; Ndumele, C.E.; Correa, A.; Butler, J.; Chandra, A.; Ayers, C.; Rao, S.; et al. Development and Validation of Machine Learning-Based Race-Specific Models to Predict 10-Year Risk of Heart Failure: A Multi-Cohort Analysis. Circulation 2021, 143, 2370–2383. [Google Scholar] [CrossRef]
- Su, Z.; Brecht, T.; O’Donovan, F.; Boussios, C.; Menon, V.; Gliklich, R.; Fonarow, G. Machine Learning Enhanced Predictions of Hospital Readmission or Death in Heart Failure. Circulation 2017, 136, A16941. [Google Scholar]
- Ahmad, T.; Lund, L.H.; Rao, P.; Ghosh, R.; Warier, P.; Vaccaro, B.; Dahlström, U.; O’Connor, C.M.; Felker, G.M.; Desai, N.R. Machine Learning Methods Improve Prognostication, Identify Clinically Distinct Phenotypes, and Detect Heterogeneity in Response to Therapy in a Large Cohort of Heart Failure Patients. J. Am. Heart Assoc. 2018, 7, e008081. [Google Scholar] [CrossRef]
- Shameer, K.; Johnson, K.W.; Yahi, A.; Miotto, R.; Li, L.I.; Ricks, D.; Jebakaran, J.; Kovatch, P.; Sengupta, P.P.; Gelijns, S.; et al. Predictive modeling of hospital readmission rates using electronic medical record-wide machine learning: A case-study using mount sinai heart failure cohort. Pac. Symp. Biocomput. 2017, 22, 276–287. [Google Scholar] [CrossRef]
- Golas, S.B.; Shibahara, T.; Agboola, S.; Otaki, H.; Sato, J.; Nakae, T.; Hisamitsu, T.; Kojima, G.; Felsted, J.; Kakarmath, S.; et al. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: A retrospective analysis of electronic medical records data. BMC Med. Inform. Decis. Mak. 2018, 18, 44. [Google Scholar] [CrossRef]
- Miao, F.; Cai, Y.P.; Zhang, Y.X.; Fan, X.M.; Li, Y. Predictive modeling of hospital mortality for patients with heart failure by using an improved random survival forest. IEEE Access 2018, 6, 7244–7253. [Google Scholar] [CrossRef]
- Meng, F.; Zhang, Z.; Hou, X.; Qian, Z.; Wang, Y.; Chen, Y.; Wang, Y.; Zhou, Y.; Chen, Z.; Zhang, X.; et al. Machine learning for prediction of sudden cardiac death in heart failure patients with low left ventricular ejection fraction: Study protocol for a retroprospective multicentre registry in China. BMJ Open 2019, 9, e023724. [Google Scholar] [CrossRef]
- Liu, W.; Stansbury, C.; Singh, K.; Ryan, A.M.; Sukul, D.; Mahmoudi, E.; Waljee, A.; Zhu, J.; Nallamothu, B.K. Predicting 30-day hospital readmissions using artificial neural networks with medical code embedding. PLoS ONE 2020, 15, e0221606. [Google Scholar] [CrossRef]
- Ahmad, T.; Pencina, M.J.; Schulte, P.J.; O’Brien, E.; Whellan, D.J.; Pina, I.L.; Kitzman, D.W.; Lee, K.L.; O’Connor, C.M.; Felker, G.M. Clinical implications of chronic heart failure phenotypes defined by cluster analysis. J. Am. Coll. Cardiol. 2014, 64, 1765–1774. [Google Scholar] [CrossRef]
- Ieva, F.; Paganoni, A.M.; Pietrabissa, T. Dynamic clustering of hazard functions: An application to disease progression in chronic heart failure. Health Care Manag. Sci. 2017, 20, 353–364. [Google Scholar] [CrossRef]
- Moser, D.K.; Lee, K.S.; Wu, J.-R.; Mudd-Martin, G.; Jaarsma, T.; Huang, T.-Y.; Fan, X.; Strömberg, A.; Lennie, T.A.; Riegel, B. Identification of symptom clusters among patients with heart failure: An international observational study. Int. J. Nurs. Stud. 2014, 51, 1366–1372. [Google Scholar] [CrossRef]
- Bose, E.; Radhakrishnan, K. Using Unsupervised Machine Learning to Identify Subgroups among Home Health Patients with Heart Failure Using Telehealth. Comput. Inform. Nurs. 2018, 36, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Segar, M.W.; Patel, K.V.; Ayers, C.; Basit, M.; Tang, W.H.W.; Willett, D.; Berry, J.; Grodin, J.L.; Pandey, A. Phenomapping of patients with heart failure with preserved ejection fraction using machine learning-based unsupervised cluster analysis. Eur. J. Heart Fail. 2020, 22, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Nouraei, H.; Nouraei, H.; Rabkin, S.W. Comparison of Unsupervised Machine Learning Approaches for Cluster Analysis to Define Subgroups of Heart Failure with Preserved Ejection Fraction with Different Outcomes. Bioengineering 2022, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Perry, A.; Loh, F.; Adamo, L.; Zhang, K.W.; Deych, E.; Foraker, R.; Mann, D.L. Unsupervised cluster analysis of patients with recovered left ventricular ejection fraction identifies unique clinical phenotypes. PLoS ONE 2021, 16, e0248317. [Google Scholar] [CrossRef]
- Harada, D.; Asanoi, H.; Noto, T.; Takagawa, J. Different Pathophysiology and Outcomes of Heart Failure with Preserved Ejection Fraction Stratified by K-Means Clustering. Front. Cardiovasc. Med. 2020, 7, 607760. [Google Scholar] [CrossRef]

| Algorithm | 2 Clusters | 3 Clusters | 4 Clusters | 5 Clusters | 6 Clusters |
|---|---|---|---|---|---|
| K-prototype | 0.580 | 0.100 | 0.120 | 0.120 | 0.120 |
| K-means | 0.036 | 0.042 | 0.048 | 0.048 | 0.048 |
| Agglomerative | 0.640 | 0.720 | 0.680 | 0.540 | 0.580 |
| BIRCH | 0.030 | 0.020 | 0.022 | 0.023 | 0.026 |
| Gaussian mixture model | 0.580 | 0.580 | 0.570 | 0.550 | 0.460 |
| Clinical Parameters | All Patients (n = 636) | HFrEF (n = 409) | HFmEF (n = 91) | HFpEF (n = 136) | p-Value |
|---|---|---|---|---|---|
| Age (years), mean | 55.6 ± 16.9 | 54.6 ± 16.9 | 58.0 ± 17.1 | 56.9 ± 16.4 | 0.1183 |
| Male sex, n (%) | 352 (55.3) | 218 (61.9) | 53 (15.1) | 81 (23.0) | 0.3720 |
| Ethnicity, n (%) | |||||
| African | 380 (59.7) | 251 (66.1) | 49 (12.9) | 80(21.1) | 0.4038 |
| Caucasian | 154 (24.2) | 93 (60.4) | 31 (20.1) | 30 (19.5) | 0.0595 |
| Indian | 72 (11.3) | 46 (63.9) | 8 (11.1) | 18 (25.0) | 0.5831 |
| Mixed ancestry | 29 (4.6) | 19 (65.5) | 3 (10.3) | 7 (24.1) | 0.7992 |
| Clinical examination | |||||
| Lung crepitations | 392 (61.6) | 251 (64.0) | 60 (15.3) | 81 (20.7) | 0.6153 |
| Ascites | 144 (22.6) | 90 (62.5) | 20 (13.9) | 34 (23.6) | 0.7597 |
| Cyanosed | 51 (8.0) | 32 (62.5) | 2 (13.9) | 17 (23.6) | 0.7597 |
| Hepatomegaly | 249 (39.2) | 173 (69.5) | 36 (14.5) | 40 (16.1) | 0.0284 |
| Peripheral oedema | 436 (68.6) | 282 (64.7) | 58 (13.3) | 96 (22.0) | 0.5298 |
| S3 | 324 (50.9) | 227 (70.1) | 32 (9.9) | 65 (20.1) | 0.0015 |
| JVP elevated | 493(77.5) | 326(66.1) | 69(14.0) | 98(19.9) | 0.1653 |
| Pulse | 100 ± 24.3 | 102 ± 25.1 | 101 ± 16.8 | 94 ± 25.1 | 0.1324 |
| Systolic BP, mmHg | 128 ± 30.1 | 125 ± 28.8 | 136 ± 35.7 | 135 ± 27.5 | 0.0002 |
| Diastolic BP, mmHg | 82 ± 19.7 | 83 ± 20.4 | 85 ± 20.5 | 78 ± 16.1 | 0.0318 |
| Laboratory tests | |||||
| Haemoglobin, g/dL | 12.9 ± 2.5 | 13.3 ± 2.4 | 12.4 ± 2.9 | 12.5 ± 2.8 | 0.0016 |
| Sodium, mmol/L | 139 ± 5.4 | 139 ± 5.5 | 138 ± 5.1 | 140 ± 5.2 | 0.1415 |
| Potassium, mmol/L | 4.4 ± 0.7 | 4.4 ± 0.7 | 4.3 ± 0.9 | 4.4 ± 0.8 | 0.5181 |
| Urea, mmol/L, IQR | 8.5 (6.0–12.5) | 12.1 ± 16.9 | 16.5 ± 43.3 | 13.2 ± 20.8 | 0.2692 |
| eGFR, mL/min, IQR | 66.1 (41.2–85.0) | 66.7 ± 27.8 | 73.5 ± 35.5 | 63.7 ± 37.3 | 0.4604 |
| Echocardiography | |||||
| LVIDd, cm | 5.6 ± 1.1 | 6.0 ± 0.9 | 5.3 ± 1.1 | 4.5 ± 1.1 | <0.001 |
| LVIDs, cm | 4.6 ± 1.3 | 5.2 ± 1.0 | 4.3 ± 0.8 | 3.1 ± 1.3 | <0.001 |
| Clinical Parameters | All Patients (n = 636) | Cluster 1 Uraemic (n = 229) | Cluster 2 Hypotensive (n = 117) | Cluster 3 Congestive (n = 290) | p-Value |
|---|---|---|---|---|---|
| Age (years), mean | 55.6 ± 16.9 | 56.2 ± 17.2 | 54.3 ± 15.9 | 55.6 ± 16.9 | 0.7642 |
| Male sex, n (%) | 352 (55.3) | 128 (36.4) | 66 (18.8) | 158 (44.9) | 0.9189 |
| Ethnicity, n (%) | |||||
| African | 380 (59.7) | 132 (34.7) | 68 (17.9) | 180 (47.4) | 0.5485 |
| Caucasian | 154 (24.2) | 57 (37.0) | 25 (16.2) | 72 (46.8) | 0.7286 |
| Indian | 72 (11.3) | 28 (38.9) | 17 (23.6) | 27 (37.5) | 0.2787 |
| Mixed Ancestry | 29 (4.6) | 12 (41.4) | 7 (24.1) | 10 (34.5) | 0.4468 |
| Clinical examination | |||||
| Lung crepitations | 392 (61.6) | 122 (31.1) | 67 (17.1) | 203 (51.8) | 0.0002 |
| Ascites | 144 (22.6) | 23 (16.0) | 28 (19.4) | 93 (64.6) | <0.0001 |
| Cyanosed | 51 (8.0) | 9 (17.6) | 10 (19.6) | 32 (62.7) | 0.0122 |
| Hepatomegaly | 249 (39.2) | 65 (26.1) | 44 (17.7) | 140 (56.2) | <0.0001 |
| Peripheral oedema | 436 (68.6) | 115 (26.4) | 82 (18.8) | 239 (54.8) | <0.0001 |
| S3 | 324 (50.9) | 102 (31.5) | 66 (20.4) | 156 (48.1) | 0.0474 |
| JVP elevated | 493 (77.5) | 148 (30.0) | 94 (19.1) | 251 (50.9) | <0.001 |
| Systolic BP, mmHg | 128 ± 30.1 | 135 ± 28.6 | 117 ± 26.5 | 128 ± 26.2 | 0.0135 |
| Diastolic BP, mmHg | 82 ± 19.7 | 83 ± 17.3 | 76 ± 16.8 | 84 ± 18.8 | 0.7012 |
| Laboratory tests | |||||
| Haemoglobin, g/dL | 12.9 ± 2.5 | 12.7 ± 2.4 | 13.2 ± 2.3 | 13.1 ± 2.4 | 0.0986 |
| Sodium, mmol/L | 139 ± 5.4 | 139 ± 4.7 | 138 ± 4.9 | 140 ± 5.8 | 0.4697 |
| Potassium, mmol/L | 4.4 ± 0.7 | 4.3 ± 0.6 | 4.5 ± 0.8 | 4.4 ± 0.8 | 0.1063 |
| Urea, mmol/L, IQR | 8.5 (6.0–12.5) | 14.5 ± 31.3 | 14.3 ± 16.0 | 11.2 ± 16.4 | 0.0917 |
| Echocardiography | |||||
| LVEF, % | 32 (25–45) | 45 ± 17.6 | 31 ±13.8 | 33 ± 14.7 | <0.001 |
| LVIDd, cm | 5.6 ± 1.1 | 5.2 ± 1.1 | 5.8 ± 1.1 | 5.9 ± 1.0 | <0.001 |
| LVIDs, cm | 4.6 ± 1.3 | 4.1 ± 1.3 | 5.0 ± 1.0 | 4.9 ± 1.1 | <0.001 |
| Clinical Parameters | All Patients (n = 636) | HFrEF vs. HFmEF | HFrEF vs. HFpEF | HFmEF vs. HFpEF |
|---|---|---|---|---|
| Systolic BP, mmHg | 128 ± 30.1 | 0.0049 | 0.0022 | 0.9000 |
| Diastolic BP, mmHg | 82 ± 19.7 | 0.6592 | 0.0626 | 0.0468 |
| Haemoglobin, g/dL | 12.9 ± 2.5 | 0.0142 | 0.0156 | 0.9000 |
| LVIDd, cm | 5.6 ± 1.1 | 0.0010 | 0.0010 | 0.0010 |
| LVIDs, cm | 4.6 ± 1.3 | 0.0010 | 0.0010 | 0.0010 |
| Clinical Parameters | All Patients (n = 636) | Cluster 1 vs. Cluster 2 | Cluster 1 vs. Cluster 3 | Cluster 2 vs. Cluster 3 |
|---|---|---|---|---|
| Systolic BP, mmHg | 128 ± 30.1 | 0.001 | 0.0152 | 0.001 |
| LVEF, % | 32 (25–45) | 0.001 | 0.001 | 0.5583 |
| LVIDd, cm | 5.6 ± 1.1 | 0.001 | 0.001 | 0.7457 |
| LVIDs, cm | 4.6 ± 1.3 | 0.001 | 0.001 | 0.9000 |
| Clinical Parameters | All Patients (n = 636) | Cluster 1 (n = 38) | Cluster 2 (n = 56) | Cluster 3 (n = 118) | Cluster 4 (n = 134) | Cluster 5 (n = 49) | Cluster 6 (n = 123) | Cluster 7 (n = 118) | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| Age (years), mean | 55.6 ± 16.9 | 53.3 ± 19.4 | 50.9 ± 16.3 | 53.7 ± 17.6 | 57.3 ± 16.9 | 58.0 ± 13.9 | 56.0 ± 15.7 | 56.9 ± 17.4 | 0.0283 |
| Male sex, n (%) | 352 (55.3) | 15 (39.5) | 30 (53.6) | 69 (58.5) | 74 (55.2) | 32 (65.3) | 70 (56.9) | 62 (52.5) | 0.3323 |
| Ethnicity, n (%) | |||||||||
| African | 380 (59.7) | 27 (71.1) | 33 (58.9) | 69 (58.5) | 82 (61.2) | 30 (61.2) | 75 (61.0) | 64 (54.2) | 0.6979 |
| Caucasian | 154 (24.2) | 5 (13.2) | 12 (21.4) | 32 (27.1) | 32 (23.9) | 14 (28.6) | 26 (21.1) | 33 (28.0) | 0.4979 |
| Indian | 72 (11.3) | 5 (13.2) | 9 (16.1) | 6 (5.1) | 16 (11.9) | 4 (8.2) | 17 (13.8) | 15 (12.7) | 0.2779 |
| Mixed Ancestry | 29 (4.6) | 1 (2.6) | 2 (3.6) | 11 (9.3) | 3 (2.2) | 1 (2.0) | 5 (4.1) | 6 (5.1) | 0.1671 |
| Clinical examination | |||||||||
| Lung crepitations | 392 (61.6) | 32 (84.2) | 31 (55.4) | 92 (78.0) | 85 (63.4) | 21 (42.9) | 77 (62.6) | 54 (45.8) | <0.0001 |
| Ascites | 144 (22.6) | 12 (31.6) | 11 (19.6) | 37 (31.4) | 40 (29.9) | 2 (4.1) | 34 (27.6) | 8 (6.8) | <0.0001 |
| Cyanosed | 51 (8.0) | 2 (5.3) | 0 (0.0) | 9 (7.6) | 16 (11.9) | 2 (4.1) | 17 (13.8) | 5 (4.2) | 0.0092 |
| Hepatomegaly | 249 (39.2) | 17 (44.7) | 9 (16.1) | 61 (51.7) | 65 (48.5) | 10 (20.4) | 58 (47.2) | 29 (24.6) | <0.0001 |
| Peripheral oedema | 436 (68.6) | 34 (89.5) | 31 (55.4) | 95 (80.5) | 104 (77.6) | 18 (36.7) | 105 (85.4) | 49 (41.5) | <0.0001 |
| S3 | 324 (50.9) | 20 (52.6) | 37 (66.1) | 77 (65.3) | 70 (52.2) | 23 (46.9) | 54 (43.9) | 43 (36.4) | 0.0001 |
| JVP elevated | 493 (77.5) | 38 (100.0) | 40 (71.4) | 102 (86.4) | 108 (80.6) | 33 (67.3) | 108 (87.8) | 64 (54.2) | <0.001 |
| Systolic BP, mmHg | 128 ± 30.1 | 131 ± 23.8 | 120 ± 27.3 | 136 ± 28.7 | 129 ± 25.9 | 136 ± 29.8 | 119 ± 26.4 | 132 ± 27.5 | 0.4155 |
| Diastolic BP, mmHg | 82 ± 19.7 | 86 ± 17.0 | 76 ± 16.1 | 86 ± 18.8 | 85 ± 19.3 | 86 ± 15.5 | 76 ± 17.5 | 82 ± 17.0 | 0.0759 |
| Laboratory tests | |||||||||
| Haemoglobin, g/dL | 12.9 ± 2.5 | 13.4 ± 2.2 | 13.1 ± 2.2 | 12.3 ± 2.6 | 13.1 ± 2.5 | 12.8 ± 2.4 | 13.4 ± 2.4 | 13.0 ± 2.3 | 0.2322 |
| Sodium, mmol/L | 139 ± 5.4 | 140 ± 4.1 | 138 ± 4.4 | 140 ± 5.3 | 138 ± 5.8 | 140 ± 4.2 | 138 ± 5.8 | 139 ± 5.0 | 0.0147 |
| Potassium, mmol/L | 4.4 ± 0.7 | 4.3 ± 0.6 | 4.3 ± 0.6 | 4.3 ± 0.6 | 4.4 ± 0.7 | 4.3 ± 0.7 | 4.6 ± 0.9 | 4.3 ± 0.6 | 0.1096 |
| Urea, mmol/L | 8.5 (6.0–12.5) | 8.5 ± 4.2 | 13.9 ± 17.4 | 11.3 ± 16.5 | 10.2 ± 5.4 | 12.0 ± 14.7 | 15.2 ± 25.5 | 16.8 ± 39.8 | 0.0194 |
| Echocardiography | |||||||||
| LVIDd, cm | 5.6 ± 1.1 | 6.1 ± 0.9 | 5.6 ± 1.2 | 5.4 ± 0.9 | 5.8 ± 1.1 | 5.2 ± 1.1 | 5.9 ± 1.0 | 5.3 ± 1.2 | 0.1748 |
| LVIDs, cm | 4.6 ± 1.3 | 5.2 ± 0.9 | 4.8 ± 1.1 | 4.4 ± 1.1 | 4.8 ± 1.2 | 3.9 ± 1.4 | 5.0 ± 1.0 | 4.3 ± 1.5 | 0.0536 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mpanya, D.; Celik, T.; Klug, E.; Ntsinjana, H. Clustering of Heart Failure Phenotypes in Johannesburg Using Unsupervised Machine Learning. Appl. Sci. 2023, 13, 1509. https://doi.org/10.3390/app13031509
Mpanya D, Celik T, Klug E, Ntsinjana H. Clustering of Heart Failure Phenotypes in Johannesburg Using Unsupervised Machine Learning. Applied Sciences. 2023; 13(3):1509. https://doi.org/10.3390/app13031509
Chicago/Turabian StyleMpanya, Dineo, Turgay Celik, Eric Klug, and Hopewell Ntsinjana. 2023. "Clustering of Heart Failure Phenotypes in Johannesburg Using Unsupervised Machine Learning" Applied Sciences 13, no. 3: 1509. https://doi.org/10.3390/app13031509
APA StyleMpanya, D., Celik, T., Klug, E., & Ntsinjana, H. (2023). Clustering of Heart Failure Phenotypes in Johannesburg Using Unsupervised Machine Learning. Applied Sciences, 13(3), 1509. https://doi.org/10.3390/app13031509







