Sequential Data Fusion Techniques for the Authentication of the P.G.I. Senise (“Crusco”) Bell Pepper
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
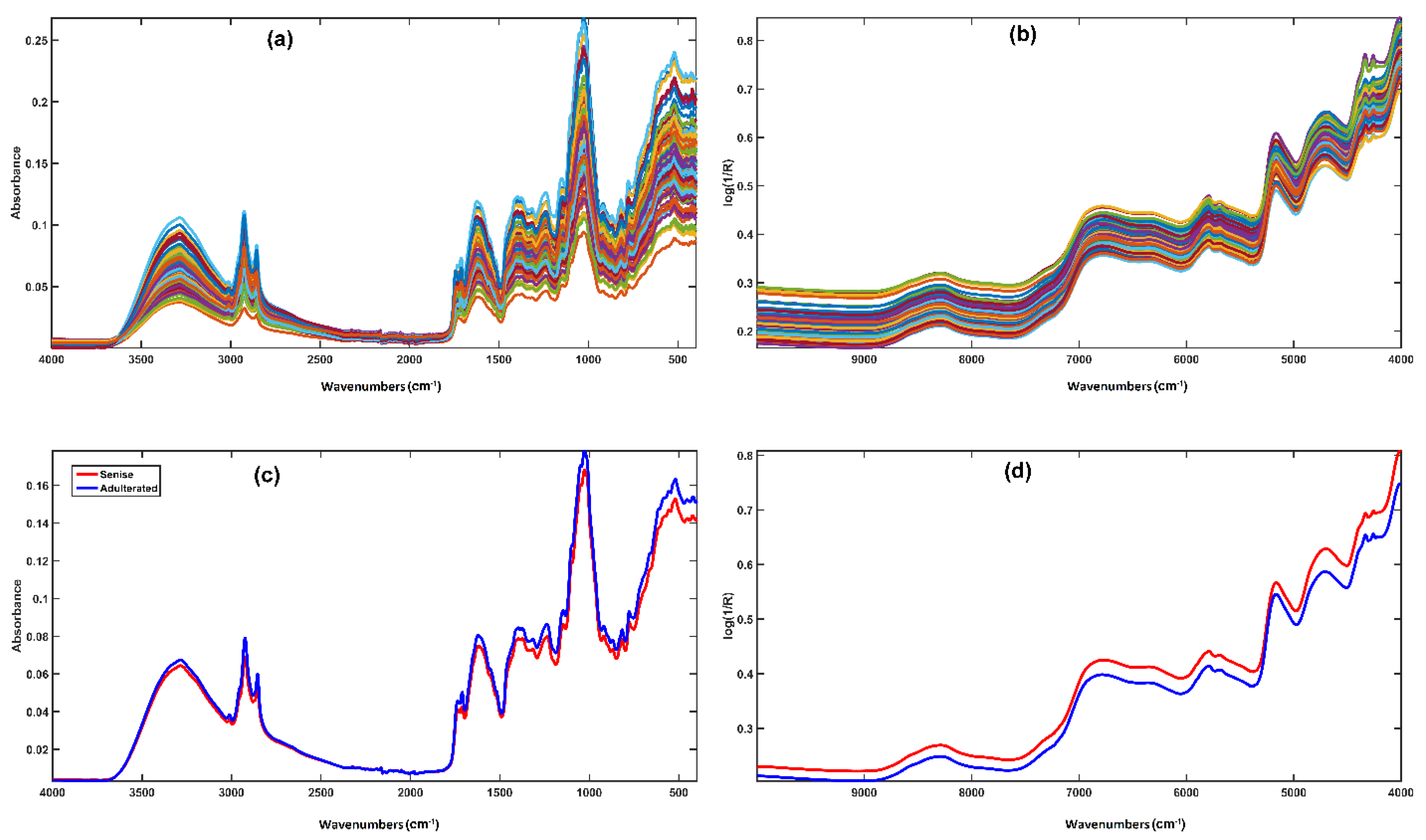
2.1.1. NIR Measurements
2.1.2. MIR Measurements
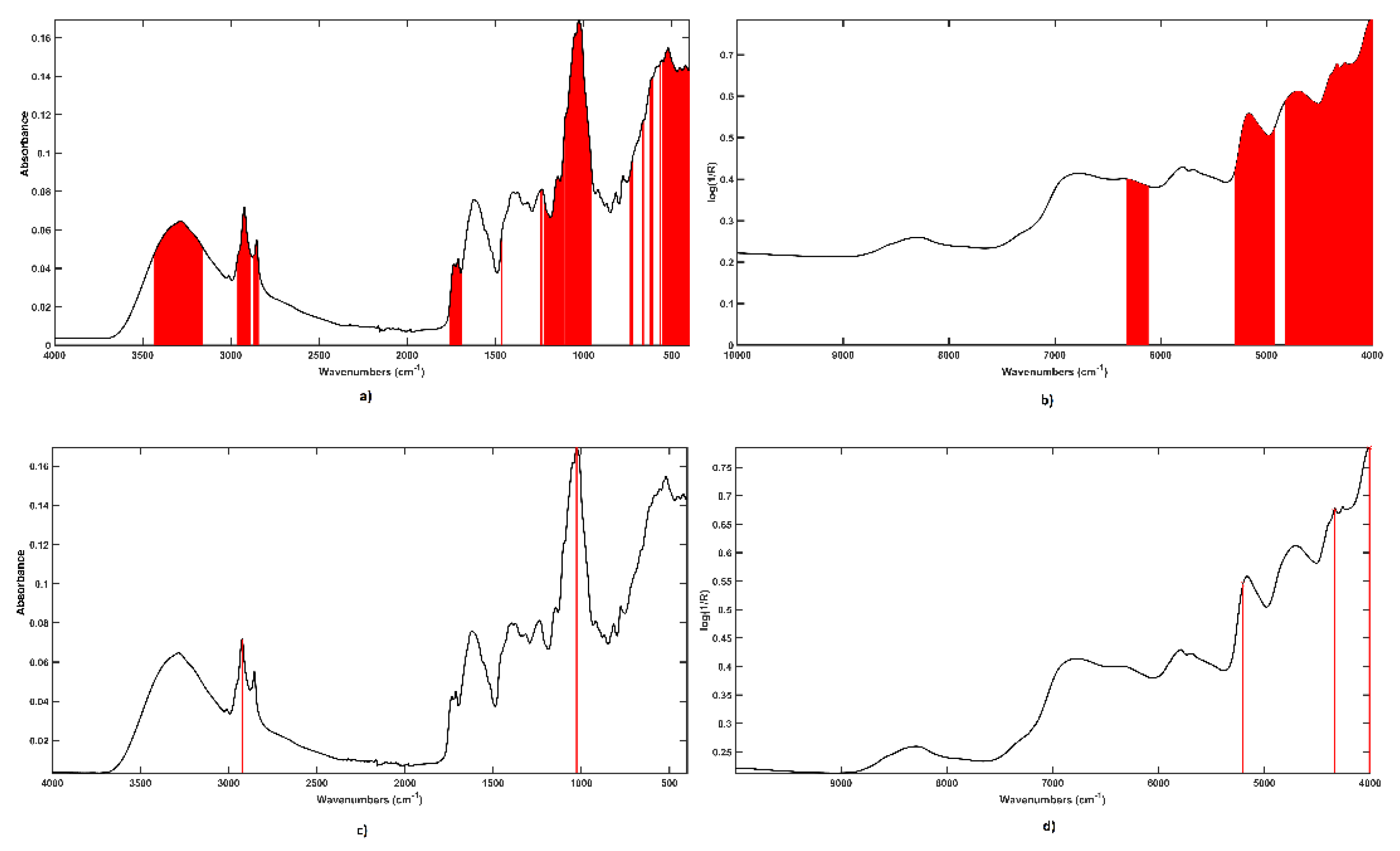
2.2. Data Fusion Approaches: Sequential and Orthogonalized Partial Least Squares-Linear Discriminant Analysis (SO-PLS-LDA) and Sequential and Orthogonalized Covariance Selection-Linear Discriminant Analysis (SO-CovSel-LDA)
- (a)
- is fitted to by PLS regression: .
- (b)
- is orthogonalized with respect to the -scores estimated in (a), obtaining
- (c)
- is fitted to the residuals from step (a) by partial least squares (PLS) regression.
- (d)
- The overall regression model is obtained by combining the outcomes of (a) and (c), and can be expressed as: , where the hat (^) indicates model predictions and and are the regression coefficient matrices.
- (a)
- A set of -variables are selected by means of CovSel (obtaining ).
- (b)
- is fitted to by means of ordinary least squares (OLS).
- (c)
- is orthogonalized with respect to (obtaining ).
- (d)
- A set of -variables are selected by means of CovSel (obtaining ).
- (e)
- The -residuals from step (a) are fitted to by means of OLS.
- (f)
- The overall regression model is obtained by combining the outcomes of steps (b) and (e):.
3. Results
3.1. Classification Models Built on Individual Spectroscopic Blocks
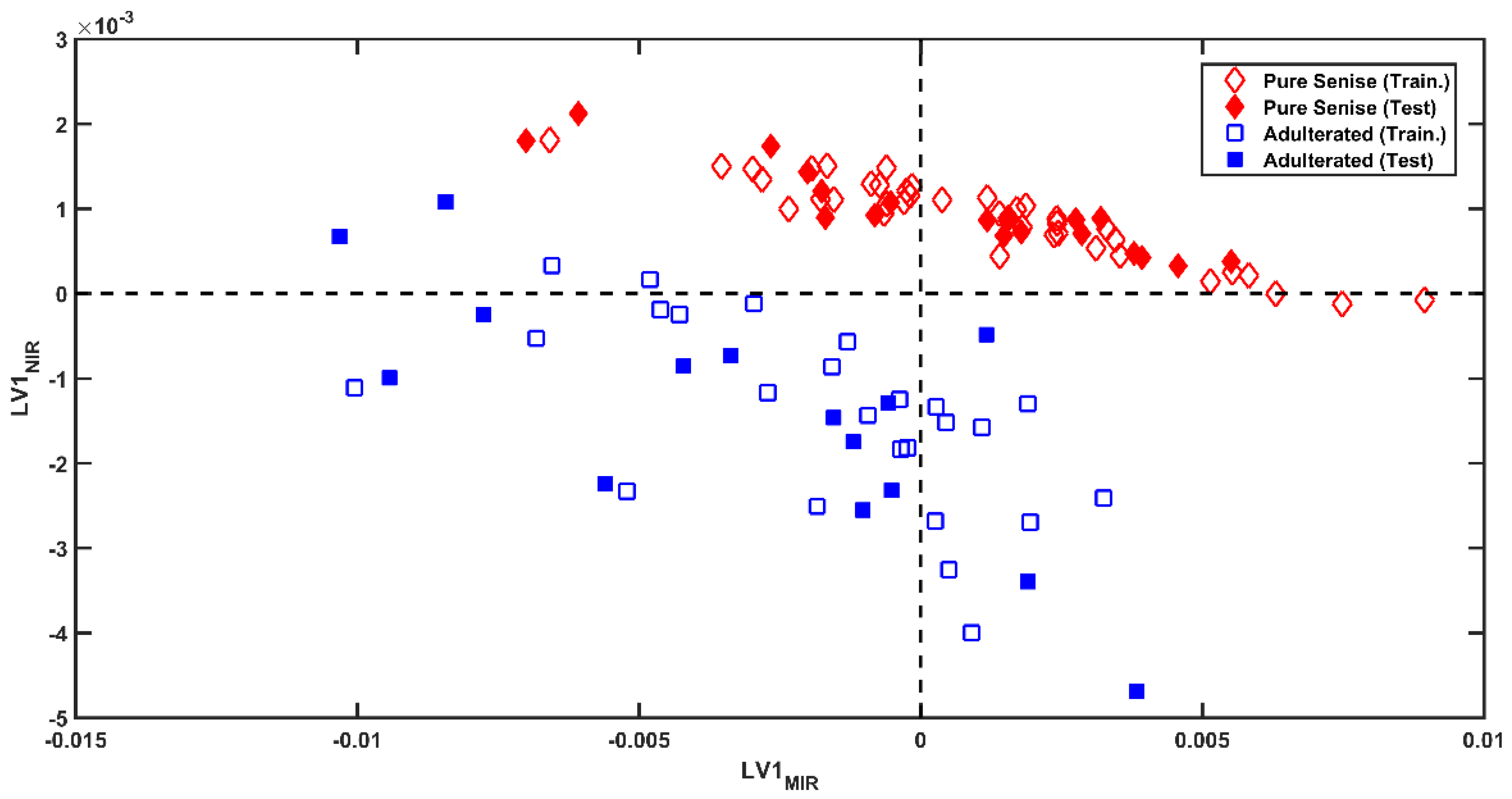
3.2. Multi-Block Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement.
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McClure, W.F. Near-Infrared Spectroscopy: The Giant is Running Strong. Anal. Chem. 1994, 66, 43A–53A. [Google Scholar] [CrossRef]
- Caballero, B.; Finglas, P.; Toldrá, F. Peppers and chilies. In Encyclopedia of Food and Health; Caballero, B., Finglas, P., Toldra, F., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 301–306. [Google Scholar]
- Gregory, G.K.; Chen, T.-S.; Philip, T. Quantitative Analysis of Carotenoids and Carotenoid Esters in Fruits by HPLC: Red Bell Peppers. J. Food Sci. 1987, 52, 1071–1073. [Google Scholar] [CrossRef]
- Gentili, A.; Dal Bosco, C.; Fanali, S.; Fanali, C. Large-scale profiling of carotenoids by using non aqueous reversed phase liquid chromatography–photodiode array detection–triple quadrupole linear ion trap mass spectrometry: Application to some varieties of sweet pepper (Capsicum annuum L.). J. Pharm. Biomed. Anal. 2019, 164, 759–767. [Google Scholar] [CrossRef]
- Sora, G.T.S.; Haminiuk, C.W.I.; da Silva, M.V.; Zielinski, A.A.F.; Gonçalves, G.A.; Bracht, A.; Peralta, R.M. A comparative study of the capsaicinoid and phenolic contents and in vitro antioxidant activities of the peppers of the genus Capsicum: An application of chemometrics. J. Food Sci. Technol. 2015, 52, 8086–8094. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.-T.; Torres, I.; de la Haba, M.-J.; Chamorro, A.; Garrido-Varo, A.; Pérez-Marín, D. Rapid, simultaneous, and in situ authentication and quality assessment of intact bell peppers using near-infrared spectroscopy technology. J. Sci. Food Agric. 2019, 99, 1613–1622. [Google Scholar] [CrossRef] [PubMed]
- Amirvaresi, A.; Nikounezhad, N.; Amirahmadi, M.; Daraei, B.; Parastar, H. Comparison of near-infrared (NIR) and mid-infrared (MIR) spectroscopy based on chemometrics for saffron authentication and adulteration detection. Food Chem. 2021, 344. [Google Scholar] [CrossRef] [PubMed]
- Aykas, D.P.; Menevseoglu, A. A rapid method to detect green pea and peanut adulteration in pistachio by using portable FT-MIR and FT-NIR spectroscopy combined with chemometrics. Food Control 2021, 121. [Google Scholar] [CrossRef]
- Di Donato, F.; Di Cecco, V.; Torricelli, R.; D’Archivio, A.A.; Di Santo, M.; Albertini, E.; Veronesi, F.; Garramone, R.; Aversano, R.; Marcantonio, G.; et al. Discrimination of potato (solanum tuberosum l.) accessions collected in majella national park (abruzzo, Italy) using mid-infrared spectroscopy and chemometrics combined with morphological and molecular analysis. Appl. Sci. 2020, 10, 1630. [Google Scholar] [CrossRef]
- Firmani, P.; Bucci, R.; Marini, F.; Biancolillo, A. Authentication of “Avola almonds” by near infrared (NIR) spectroscopy and chemometrics. J. Food Compos. Anal. 2019, 82, 103235. [Google Scholar] [CrossRef]
- Firmani, P.; La Piscopia, G.; Bucci, R.; Marini, F.; Biancolillo, A. Authentication of P.G.I. Gragnano pasta by near infrared (NIR) spectroscopy and chemometrics. Microchem. J. 2020, 152, 104339. [Google Scholar] [CrossRef]
- Biancolillo, A.; Boqué, R.; Cocchi, M.; Marini, F. Data Fusion Strategies in Food Analysis. Data Handl. Sci. Technol. 2019, 31, 271–310. [Google Scholar]
- Frank, I.E.; Kowalski, B.R. prediction of wine quality and geographic origin from chemical measurements by parital least-squares regression modeling. Anal. Chim. Acta 1984, 162, 241–251. [Google Scholar] [CrossRef]
- Skov, T.; Honoré, A.H.; Jensen, H.M.; Næs, T.; Engelsen, S.B. Chemometrics in foodomics: Handling data structures from multiple analytical platforms. TrAC Trends Anal. Chem. 2014, 60, 71–79. [Google Scholar] [CrossRef]
- Biancolillo, A.; Næs, T. The Sequential and Orthogonalized PLS Regression for Multiblock Regression: Theory, Examples, and Extensions. Data Handl. Sci. Technol. 2019, 31, 157–177. [Google Scholar] [CrossRef]
- Biancolillo, A.; Preys, S.; Gaci, B.; Le-Quere, J.-L.; Laboure, H.; Deuscher, Z.; Cheynier, V.; Sommerer, N.; Fayeulle, N.; Costet, P.; et al. Multi-block classification of chocolate and cocoa samples into sensory poles. Food Chem. 2021, 340. [Google Scholar] [CrossRef] [PubMed]
- Biancolillo, A.; Marini, F.; D’Archivio, A.A. Geographical discrimination of red garlic (Allium sativum L.) using fast and non-invasive Attenuated Total Reflectance-Fourier Transformed Infrared (ATR-FTIR) spectroscopy combined with chemometrics. J. Food Compos. Anal. 2020, 86, 103351. [Google Scholar] [CrossRef]
- Firmani, P.; Nardecchia, A.; Nocente, F.; Gazza, L.; Marini, F.; Biancolillo, A. Multi-block classification of Italian semolina based on Near Infrared Spectroscopy (NIR) analysis and alveographic indices. Food Chem. 2020, 309, 125677. [Google Scholar] [CrossRef] [PubMed]
- Giannetti, V.; Mariani, M.B.; Marini, F.; Torrelli, P.; Biancolillo, A. Grappa and Italian spirits: Multi-platform investigation based on GC–MS, MIR and NIR spectroscopies for the authentication of the Geographical Indication. Microchem. J. 2020, 157, 104896. [Google Scholar] [CrossRef]
- Schiavone, S.; Marchionni, B.; Bucci, R.; Marini, F.; Biancolillo, A. Authentication of Grappa (Italian grape marc spirit) by Mid and Near Infrared spectroscopies coupled with chemometrics. Vib. Spectrosc. 2020, 107, 103040. [Google Scholar] [CrossRef]
- Tao, L.; Via, B.; Wu, Y.; Xiao, W.; Liu, X. NIR and MIR spectral data fusion for rapid detection of Lonicera japonica and Artemisia annua by liquid extraction process. Vib. Spectrosc. 2019, 102, 31–38. [Google Scholar] [CrossRef]
- Næs, T.; Tomic, O.; Mevik, B.-H.; Martens, H. Path modelling by sequential PLS regression. J. Chemom. 2011, 25, 28–40. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936. [Google Scholar] [CrossRef]
- Biancolillo, A.; Måge, I.; Næs, T. Combining SO-PLS and linear discriminant analysis for multi-block classification. Chemom. Intell. Lab. Syst. 2015, 141, 58–67. [Google Scholar] [CrossRef]
- Biancolillo, A.; Marini, F.; Roger, J.-M. SO-CovSel: A novel method for variable selection in a multiblock framework. J. Chemom. 2020, 34, e3120. [Google Scholar] [CrossRef]
- Roger, J.M.; Palagos, B.; Bertrand, D.; Fernandez-Ahumada, E. CovSel: Variable selection for highly multivariate and multi-response calibration. Application to IR spectroscopy. Chemom. Intell. Lab. Syst. 2011. [Google Scholar] [CrossRef]
- Snee, R.D. Validation of Regression Models: Methods and Examples. Technometrics 1977, 19, 415–428. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003. [Google Scholar] [CrossRef]
- Roger, J.-M.; Biancolillo, A.; Marini, F. Sequential preprocessing through ORThogonalization (SPORT) and its application to near infrared spectroscopy. Chemom. Intell. Lab. Syst. 2020, 199. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964. [Google Scholar] [CrossRef]
- Biancolillo, A.; Liland, K.H.; Måge, I.; Næs, T.; Bro, R. Variable selection in multi-block regression. Chemom. Intell. Lab. Syst. 2016, 156. [Google Scholar] [CrossRef]
- Cortés-Estrada, C.E.; Gallardo-Velázquez, T.; Osorio-Revilla, G.; Castañeda-Pérez, E.; Meza-Márquez, O.G.; López-Cortez, M.D.S.; Hernández-Martínez, D.M. Prediction of total phenolics, ascorbic acid, antioxidant capacities, and total soluble solids of Capsicum annuum L. (bell pepper) juice by FT-MIR and multivariate analysis. LWT 2020, 126. [Google Scholar] [CrossRef]
| Sample # | Adulterant (%) | Sample # | Adulterant (%) | Sample # | Adulterant (%) | Sample # | Adulterant (%) |
|---|---|---|---|---|---|---|---|
| 1 | 2 | 11 | 6 | 21 | 10.5 | 31 | 29 |
| 2 | 2.5 | 12 | 6.5 | 22 | 11 | 32 | 31 |
| 3 | 2.8 | 13 | 7 | 23 | 13 | 33 | 33 |
| 4 | 3 | 14 | 7.5 | 24 | 15 | 34 | 35 |
| 5 | 3.5 | 15 | 8 | 25 | 17 | 35 | 37 |
| 6 | 4 | 16 | 8.5 | 26 | 19 | 36 | 39 |
| 7 | 4.5 | 17 | 9 | 27 | 20 | 37 | 43 |
| 8 | 5 | 18 | 9.5 | 28 | 23 | 38 | 46 |
| 9 | 5.5 | 19 | 9.7 | 29 | 25 | 39 | 48 |
| 10 | 5.7 | 20 | 10 | 30 | 27 | 40 | 50 |
| Method | Data Set | Optimal Number of LVs | Correct Classification Rate (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Raw | SNV | 1st Der. | 2nd Der. | Training Set (Cross-Validation) | Test Set (Prediction) | ||||
| Senise | Adulterated | Senise | Adulterated | ||||||
| PLS-DA | MIR | 3 | 80.0 | 72.0 | |||||
| 4 | 80.0 | 76.0 | |||||||
| 3 | 80.0 | 84.0 | |||||||
| 3 | 87.5 | 80.0 | 75.0 | 93.3 | |||||
| PLS-DA | NIR | 2 | 100.0 | 92.0 | |||||
| 3 | 100.0 | 92.0 | |||||||
| 1 | 100.0 | 100.0 | 100.0 | 86.7 | |||||
| 2 | 100.0 | 100.0 | |||||||
| SPORT | MIR | 1 | 0 | 0 | 4 | 92.5 | 92.0 | 75.0 | 93.3 |
| NIR | 0 | 0 | 1 | 0 | 100.0 | 100.0 | 100.0 | 86.7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biancolillo, A.; Di Donato, F.; Merola, F.; Marini, F.; D’Archivio, A.A. Sequential Data Fusion Techniques for the Authentication of the P.G.I. Senise (“Crusco”) Bell Pepper. Appl. Sci. 2021, 11, 1709. https://doi.org/10.3390/app11041709
Biancolillo A, Di Donato F, Merola F, Marini F, D’Archivio AA. Sequential Data Fusion Techniques for the Authentication of the P.G.I. Senise (“Crusco”) Bell Pepper. Applied Sciences. 2021; 11(4):1709. https://doi.org/10.3390/app11041709
Chicago/Turabian StyleBiancolillo, Alessandra, Francesca Di Donato, Francesco Merola, Federico Marini, and Angelo Antonio D’Archivio. 2021. "Sequential Data Fusion Techniques for the Authentication of the P.G.I. Senise (“Crusco”) Bell Pepper" Applied Sciences 11, no. 4: 1709. https://doi.org/10.3390/app11041709
APA StyleBiancolillo, A., Di Donato, F., Merola, F., Marini, F., & D’Archivio, A. A. (2021). Sequential Data Fusion Techniques for the Authentication of the P.G.I. Senise (“Crusco”) Bell Pepper. Applied Sciences, 11(4), 1709. https://doi.org/10.3390/app11041709









