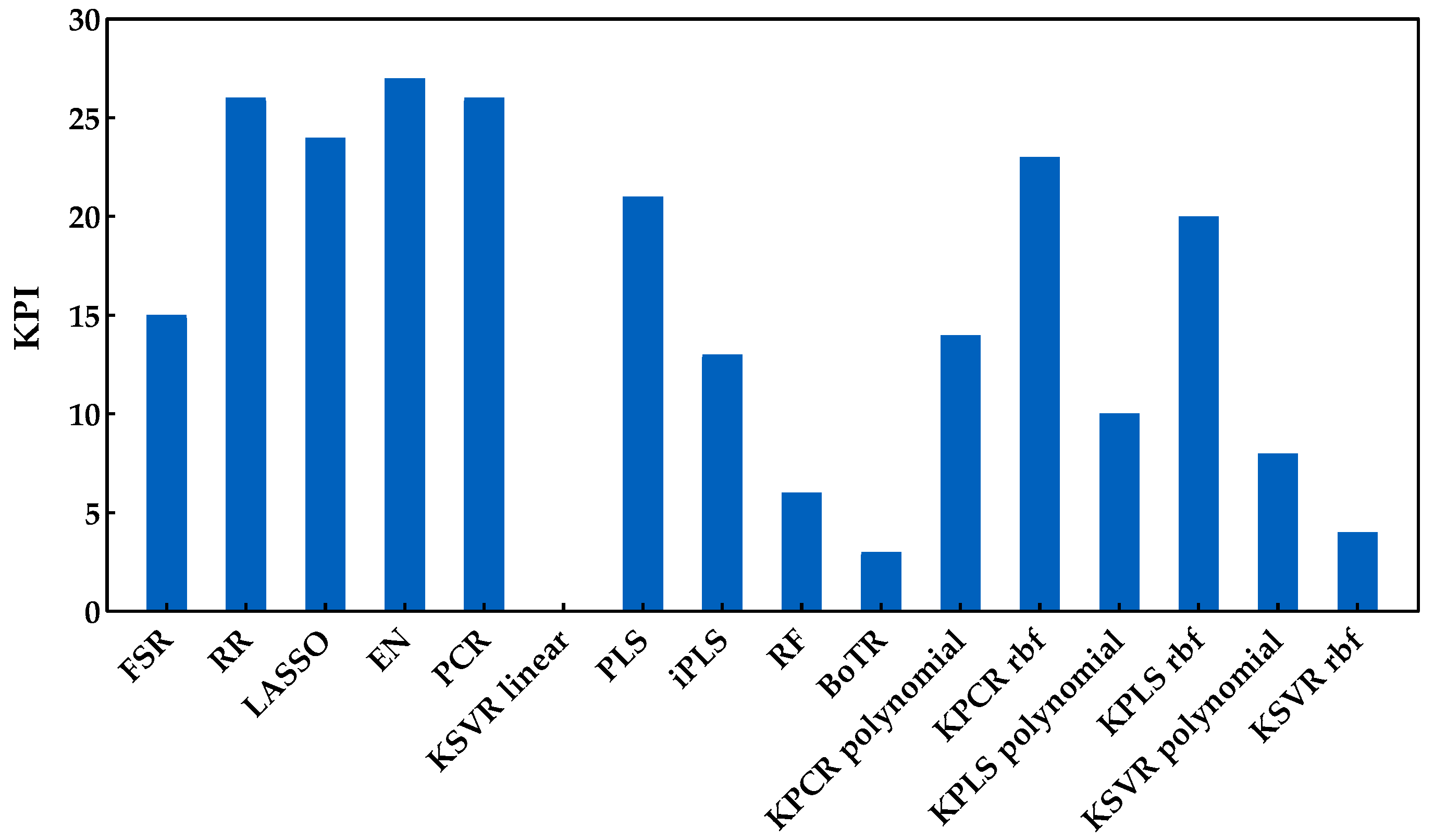Abstract
This paper presents an extended comparison study between 16 different linear and non-linear regression methods to predict the sugar, pH, and anthocyanin contents of grapes through hyperspectral imaging (HIS). Despite the numerous studies on this subject that can be found in the literature, they often rely on the application of one or a very limited set of predictive methods. The literature on multivariate regression methods is quite extensive, so the analytical domain explored is too narrow to guarantee that the best solution has been found. Therefore, we developed an integrated linear and non-linear predictive analytics comparison framework (L&NL-PAC), fully integrated with five preprocessing techniques and five different classes of regression methods, for an effective and robust comparison of all alternatives through a robust Monte Carlo double cross-validation stratified data splitting scheme. L&NLPAC allowed for the identification of the most promising preprocessing approaches, best regression methods, and wavelengths most contributing to explaining the variability of each enological parameter for the target dataset, providing important insights for the development of precision viticulture technology, based on the HSI of grape. Overall, the results suggest that the combination of the Savitzky−Golay first derivative and ridge regression can be a good choice for the prediction of the three enological parameters.
1. Introduction
Wine quality is intrinsically linked to the quality and geographical origin of the grapes utilized as the raw material, along with the success of post-harvest winemaking techniques. These factors need to be properly controlled to achieve the desired wine properties and quality standards. In particular, the search for the optimal grape berries’ maturity stage is a permanent concern of producers who need to make better decisions regarding the best moment for harvesting, as well as to select grapes according to their quality features to accomplish the desired wine consistency and quality. This can be attained through monitoring the enological parameters, such as the sugar content, pH, and anthocyanin concentration. Usually, these enological parameters are assessed along with the grape maturation stage by conventional physical and chemical techniques, which have the disadvantage of being limited to a certain number of samples, as well as being destructive, time-consuming, and expensive. In order to overcome these disadvantages, there have been extensive research efforts for faster, non-destructive, and less expensive ways to assess the enological parameters, with hyperspectral imaging (HSI) emerging as a very promising alternative [1,2,3,4]. This technology has the benefit of merging the features of both imaging and spectroscopy that, in the reflectance mode, allows for the collection of information about the intensity of the light reflected by grapes as a function of their wavelengths [5,6]. However, the large amount of data generated by hyperspectral imaging poses significant challenges for data-driven modelling, requiring the use of suitable data analytic tools to properly deal with the complex spatial-wavelength structure and to extract the relevant information and the underlying patterns.
In this context, supervised learning methods have been used to predict the value of a variety of output variables from the available predictors. However, the number of regression methods currently available is large, and selecting a suitable method is a cumbersome task with practitioners often relying on their preferred method and ignoring others that may present predictive advantages. In fact, most of the studies focusing on grape ripeness assessment (see Table 1) are still based on partial least squares regression (PLS) [7,8,9,10,11]. Nevertheless, some authors have also implemented support vector machines (SVM) [12] and artificial neural networks (ANN) [13,14]. Thus, and given the plethora of methods available, the selection of the most suitable methods requires conducting extensive comparison studies, which represent an unbiased and effective approach to assess the performance of different regression methods in predicting the response variable of interest. As previously published works tend to focus on one, or a very limited set of predictive methods, overlooking entire classes of approaches, there is currently a gap of studies in the literature that compare, in a fair and unbiased way, a wide variety of methods over the same dataset in order to find the most adequate ones, also extracting insights from their combined analysis. Therefore, the present work reports the development and comparison of distinct linear and non-linear regression methods using hyperspectral imaging data collected in reflectance mode. The major novelty relies on putting together a rich variety of carefully chosen regression methods, arising from different classes of machine learning, statistical, and artificial intelligence domains, to predict sugar, pH, and anthocyanin contents in wine grape berries for the target dataset. To drive the comparison, an integrated linear and non-linear predictive analytics comparison framework (L&NL-PAC) was developed to assess the prediction performance of different classes of regression methods, covering the main classes of machine learning methods that are fully integrated with the most common spectral preprocessing approaches. Thus, the broad goal of this study was to identify the most suitable regression and preprocessing approaches, and also to extract insights into the characteristics of the relationship between the hyperspectral imaging of grapes and their enological parameters. ANNs are not included in this work because they have been employed before on the same data set (see [7,13]), and the results obtained here will be compared with those. Furthermore, the current comparison framework does not comprise the deep learning class due to the large amount of data necessary to properly train deep learning algorithms; however, they will be considered in future work when more data are accumulated.

Table 1.
Summary of most published works for predicting enological parameters using spectroscopic measurements in reflectance mode.
2. Dataset Description
The wine grape berries considered in the present work are from a Portuguese native variety, Touriga Franca (Vitis vinifera L.), which is widely used to produce Port wine in one of the oldest appellation regions of the world, the Portuguese Douro region. This variety was chosen due to its high importance for our industrial partner, Symington Family Estates (www.symington.com), which is one of the world’s largest producers of Port wine. A total of 240 bunches, 24 per day, were harvested from Quinta do Bomfim, Pinhão-Portugal, between the beginning of veraison (end of July) and maturity (end of September). The 24 bunches were collected at three different locations inside the vineyard with small, medium, and large vigor and at two different sun exposition levels. Then, line-scan hyperspectral image acquisition was performed in our laboratory-based imaging system using the fresh grape samples. Each sample measured by hyperspectral imaging was composed of six grape berries randomly collected from a single bunch with their pedicel attached, resulting in a total of 240 samples. After imaging and before conventional analysis, the samples were frozen at −18 °C.
2.1. VIS-NIR Spectral Data Acquisition
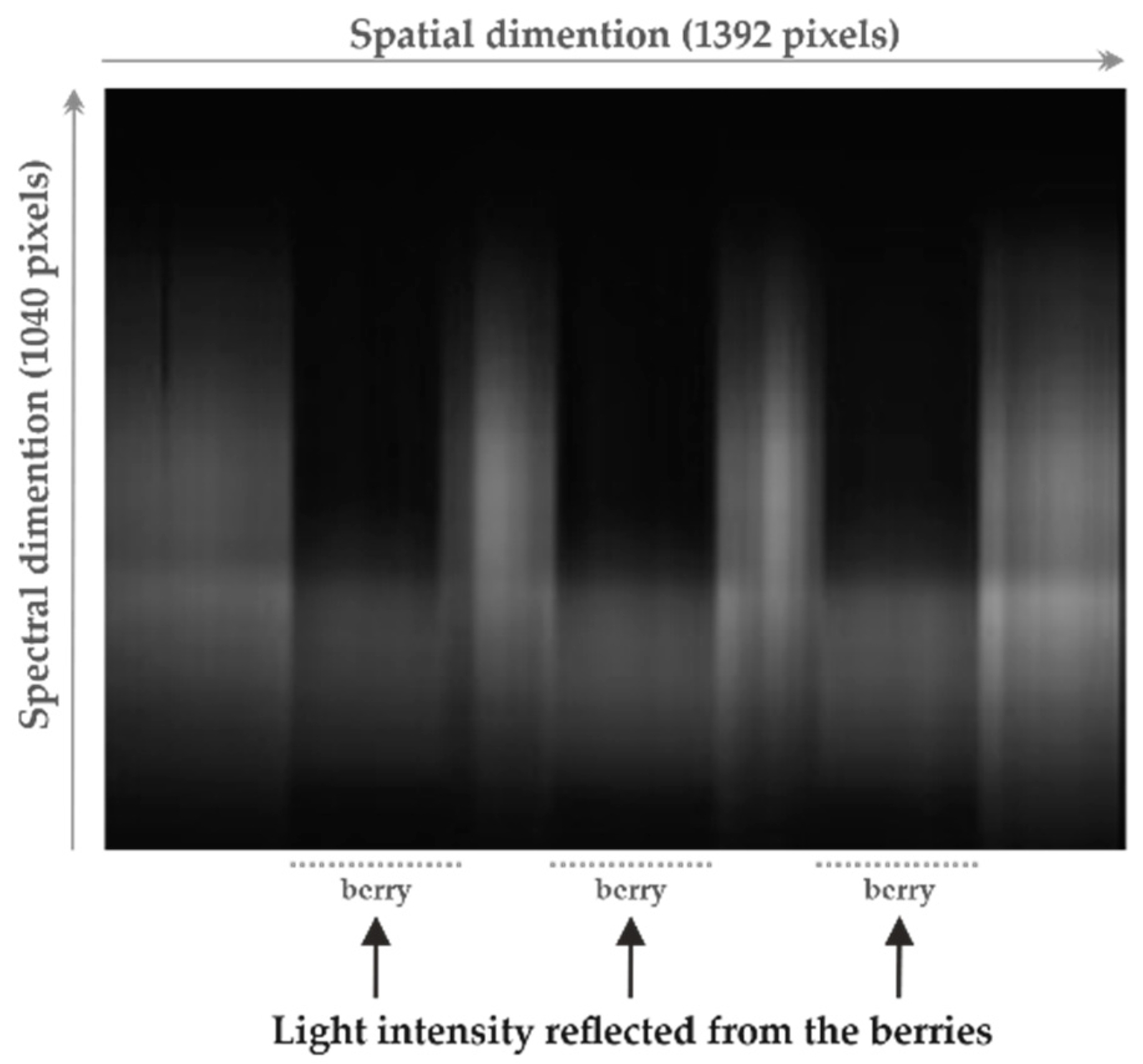
The experimental setup used to acquire the spectral data (line-scan hyperspectral imaging), as well as the procedure to compute the reflectance previously described by the authors in [7,12,13]. Thus, the reader is directed to additional references for a more detailed description. In summary, the system consisted of a hyperspectral camera, composed of a black and white camera and spectrograph, and a lighting source comprising a lamp holder to hold four 20 W, 12 V halogen lamps and two 40 W, 220 V blue reflector lamps. The acquired line-scan hyperspectral images had 1040 × 1392 pixels, in which the 1040 pixels were related to the measured wavelength channels that had a width of approximately 0.6 nm (ranging from 380 to 1028 nm), and the 1392 pixels denoted the spatial dimension (one line over the sample) with a width of approximately 110 nm [7,12,13]. After imaging, a threshold-based segmentation method was applied to identify and extract the grape berries from the complete image. Figure 1 displays an example of a line-scan hyperspectral image acquired by the described setup and for three berries measured simultaneously.
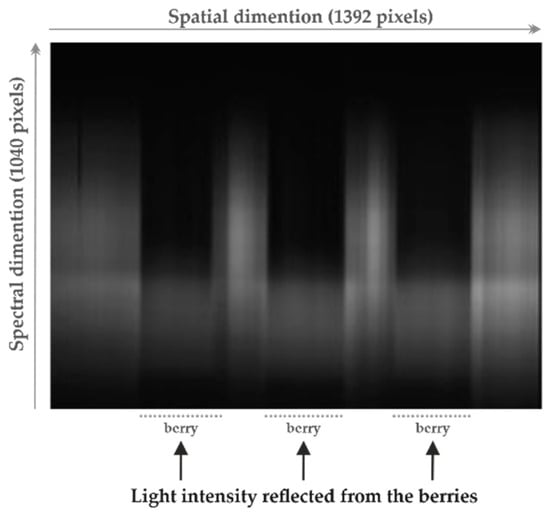
Figure 1.
Example of a line-scan hyperspectral image acquired before segmentation, considering three grape berries simultaneously imaged.
For a certain wavelength range, λ, and position, x, the reflectance values were obtained according to the following:
where GI is the intensity of light reflected by the grapes; SI is the intensity of light coming from a white reflectance target (Spectralon) that reflects almost all the light reaching its surface in the ultraviolet, visible, and infra-red wavelengths; and DI is the dark current signal (electronic noise) measured by keeping the camera shutter closed. This electronic noise is independent of the object being imaged, and must be subtracted from the grape berries and the Spectralon in order to avoid tampering in the determination of the reflectance values.
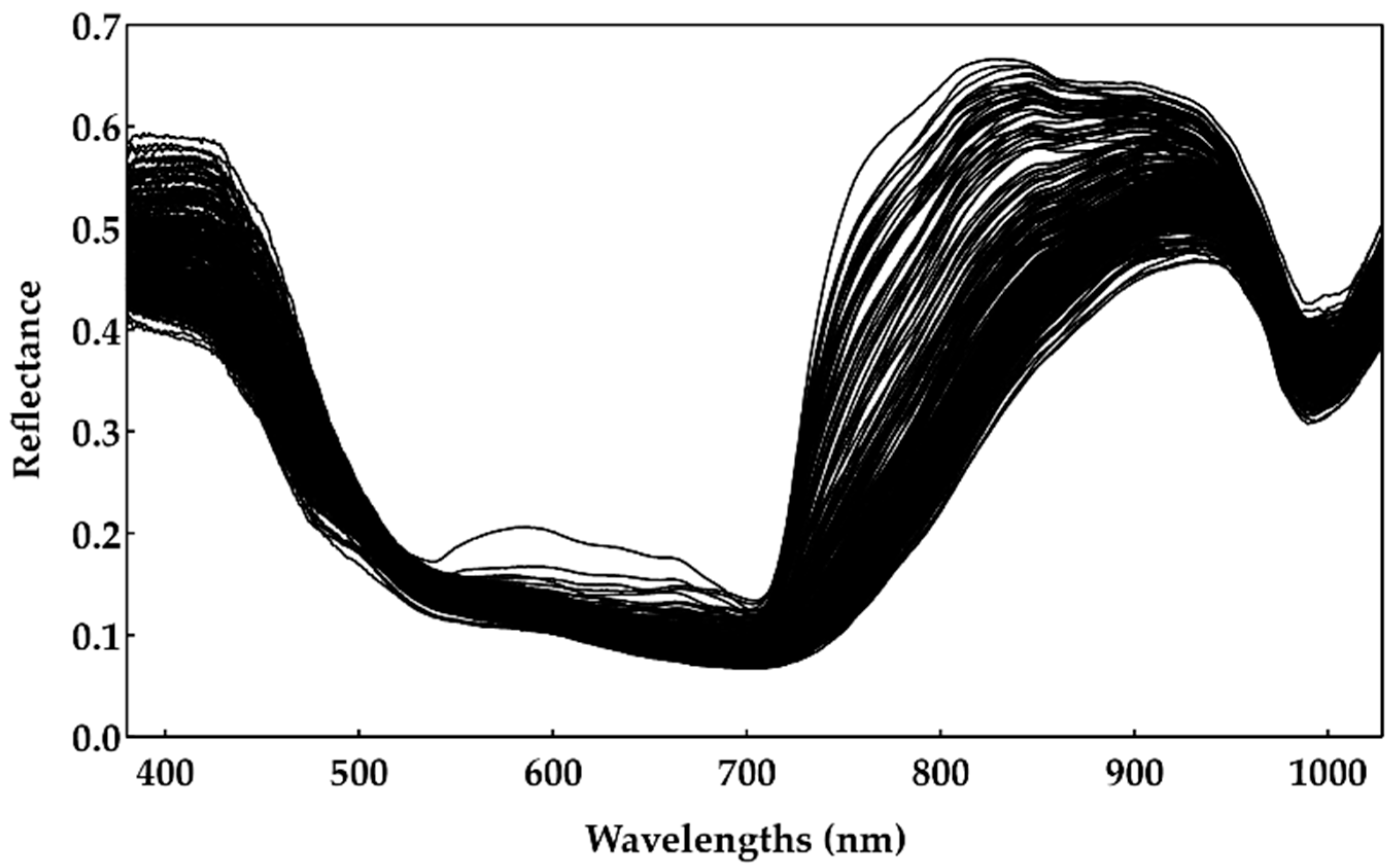
Each hyperspectral image was acquired over the berry’s equator, considering the pedicel as the pole, and for three different positions of the berries, corresponding approximately to 120° rotation between positions [7,12,13,14]. In order to minimize the measurement noise, 32 hyperspectral images were acquired. The final hyperspectral images were obtained by averaging the 32 images and, after identifying the grape berries, the reflectance measurements were computed. Finally, to create a unique reflectance spectrum for each sample, all berries’ points were averaged over the spatial dimension and across all positions. All of the reflectance spectra gathered for the Touriga Franca variety are illustrated in Figure 2.
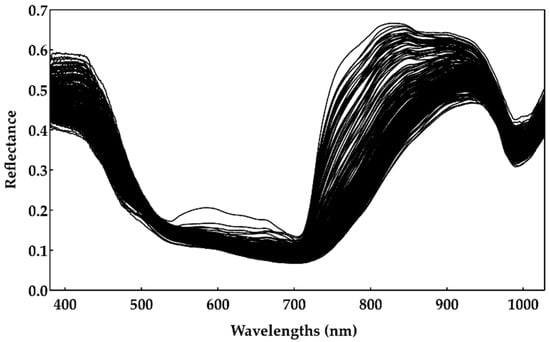
Figure 2.
Reflectance spectrum of 240 samples of the Touriga Franca variety (hyperspectral collected data).
2.2. Analytical Determination
In order to obtain the reference responses for establishing the training set required to derive the predictive models, the contents of the sugar, anthocyanin, and pH were quantified by conventional chemical analysis, as previously described in [13]. Briefly, each set of six grapes was defrosted at room temperature and then crushed. The juice released was analyzed for °Brix and pH according to validated standard methods [19]. The total anthocyanin concentration was determined photometrically by the SO2 bleaching method [20] using a UV/Vis spectrophotometer (Shimadzu) and 1 cm path length disposable cells for spectral measurements at 520 nm. Pigment content, expressed in mg·L−1, was calculated from a calibration curve of malvidin-3-glucoside and all determinations were performed in duplicate.
After the analytical determination of the enological parameters, each reflectance spectrum was paired with the sugar, pH, and anthocyanin reference values to assemble the final dataset.
3. Linear and Non-Linear Predictive Analytics Comparison Framework: L&NL-PAC
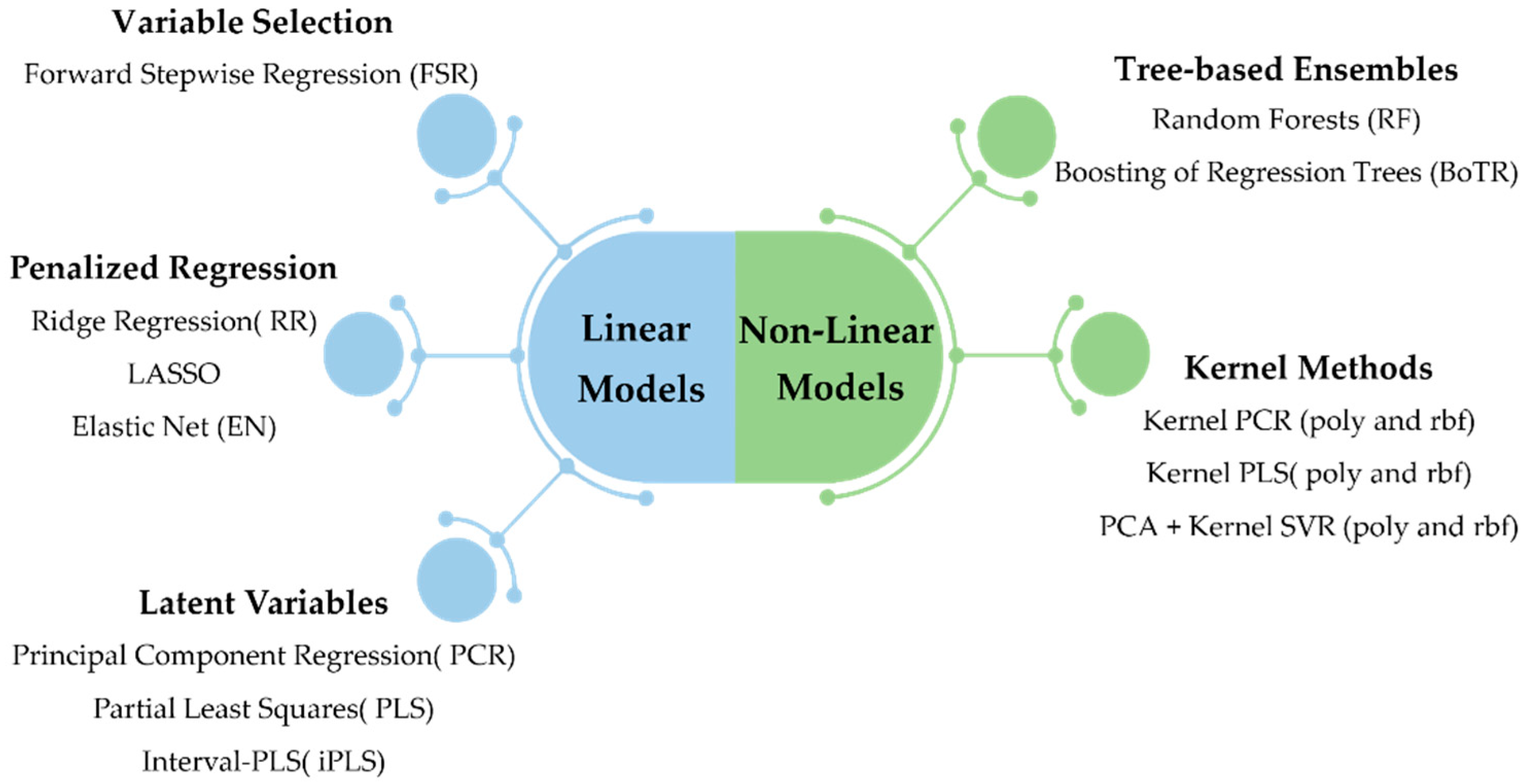
The methodology employed in this work encompasses the simultaneous and integrated consideration of a variety of preprocessing approaches and regression methods, which were submitted to a systematic comparison scheme, whose outcomes were summarized by a results reporting engine. All of these components were assembled and combined in an integrated framework, called L&NL-PAC, which facilitates the identification of the most promising preprocessing approaches and best regression methods for the target dataset. The regression methods included cover both linear and non-linear classes of approaches. Linear methods were grouped into three subclasses: variable selection methods, latent variables methods, and penalized regression methods. Non-linear models were grouped into tree-based ensembles and kernel methods. Each regression method presents different a priori assumptions regarding the nature of the relationships between the predictors and the response variable(s), which may lead to different levels of prediction accuracy for the case study under analysis. A brief description of the designated preprocessing and regression methods is presented in Section 3.1 and Section 3.2, respectively. Finally, the comparison framework procedure is detailed in Section 3.3.
3.1. Preprocessing Approaches
Preprocessing of the spectral data is an integral part of the development of parsimonious and stable predictive models. The purpose of this task is to mitigate/remove physical phenomena in the spectra unrelated to the target responses, including, in this work, the size and curvature of the grape berry [7,13,14]. The methods adopted in the preprocessing step belong to the reference-independent class of preprocessing methods, as they strictly involve the spectra data. The literature on preprocessing methods is extensive and various techniques have been applied for spectroscopic data in food/fruit analyses [21,22,23,24,25]. Thus, representatives of the most well-known spectra reference-independent preprocessing techniques were considered, which can be allocated into two categories: scatter correction methods and spectral derivatives. For the current purpose, multiplicative scatter correction (MSC), standard normal variate (SNV), and normalization techniques (auto-scaling) were selected from the first category, while Savitzky−Golay (SG) with first and second derivatives were chosen from the second group. SG employs a second order polynomial with a window size of fifteen points. The rationale for choosing these preprocessing methods was based on the fact that, according to the scientific literature, these have been the most commonly applied approaches for predicting the enological parameters of grape berries from spectroscopic data (see Table 1), as well as for the treatment of spectroscopic data to assess the quality of various fruits [21,22,23,24,25]. In addition, normalization (auto-scaling) of the spectra was selected due to the similarity with SNV. Mathematically, auto-scaling performs a column-wise normalization with the column mean and standard deviation, whereas SNV performs the same operation row-wise. Details on the spectra preprocessing techniques are available in the literature [26,27,28,29,30,31,32,33,34].
3.2. Predictive Methods
The literature on predictive regression methods is extensive, and to take advantage of the many developments in the field, a careful selection of the potentially effective methods was conducted in this work. The selected methods include linear and non-linear approaches, and cover a wide range of classes of regression methods (see Figure 3). These classes contain different a priori assumptions regarding the distribution of predictors, response variables, and their relationship. Thus, they provide a suitable pool of methods to infer, from the data, which class/method leads to a superior prediction performance. Overall, 16 regression methods were included in this study and compared using L&NL-PAC. This set contains the most popular methods that have found more success in spectroscopic applications, as well as other relevant methods from the general field of regression modeling that have different assumptions regarding the underlying data structure (namely regarding the presence of effect sparsity, collinearity, non-linearity, etc.). Although additional methods could always be considered, the current pool of methodologies provides a comprehensive modeling basis to support the use of hyperspectral imaging for predicting the enological properties of interest. A summary of the regression methods within each class is provided in the following subsections. However, the reader is directed to additional references for a detailed description of the linear- and tree-based ensemble methods adopted [35], as well as for the kernel methods [36,37,38].
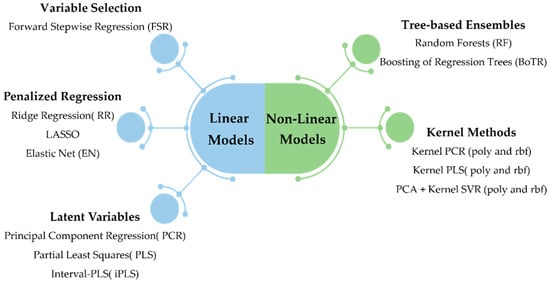
Figure 3.
Linear and non-linear regression methods considered in the comparison study. Kernel methods include the polynomial (poly) kernel and the radial basis function (rbf) kernel.
3.2.1. Variable Selection Methods
The methods belonging to the class of variable selection have the implicit assumption that, although many predictors are measured, some are expected to be irrelevant or too noisy. Thus, various strategies are employed to select the important predictors and remove the noisy or irrelevant ones. In this class, forward stepwise regression (FSR) was selected as a representative method. FSR sequentially builds a model by including and excluding predictors based on the p-values of the partial-F test. The process starts by including the predictor with the lowest p-value (more significant contribution to explain the Y-variability). Then, the importance of all other variables is assessed (given that one predictor is already included), and the one with the smallest p-value is added to the model, as long as the p-value is below a threshold (pin). After this inclusion step, the predictors are assessed, and the one with the highest p-value is excluded from the model (provided that the p-value is above a threshold (pout)). This iterative process continues until no predictor can be added or removed from the model. The regression coefficients are then obtained by multiple linear regression, considering only the variables that were selected in the iterative process.
3.2.2. Penalized Regression Methods
The class of penalized regression methods is characterized by the fact that a penalty term is employed for the magnitude of the regression coefficients, constraining their magnitude to be small. The penalty serves as a model regularization term and helps to mitigate the effects of collinearity and overfitting, and to improve model robustness. In this class, three methods were considered: ridge regression (RR), least absolute shrinkage and selector operator (LASSO), and the elastic net (EN). EN is a more general method and contains RR and LASSO as particular cases. Equation (2) presents the objective function used to obtain the EN model:
where α
is a hyperparameter that weights the relative contributions of the different types of penalization to the magnitude of the coefficients (the L1-norm and the L2-norm penalization), and γ controls the bias−variance trade-off, by weighting the contribution of the classical least-squares term with the penalization term for the regression coefficients size.
3.2.3. Latent Variable Methods
The latent variable methods estimate an underlying latent structure where the influence of the unobserved sources of variability are estimated. Although hundreds or thousands of predictors can be collected, many predictors are correlated and constitute manifestations of a few unmeasured sources of variability. Thus, these methods estimate the underlying sources of variability and, in turn, compress the dimensionality of the dataset. Three methods were considered from this class: principal component regression (PCR), partial least squares (PLS), and interval PLS (iPLS).
3.2.4. Tree-Based Ensembles
The class of tree-based ensembles contains non-linear methods whose basic building blocks are regression trees. A regression tree is a particular model that approximates the relationship between predictors and response variables by a piece-wise constant function. Furthermore, regression trees are very flexible, which often lead to a high variance in their predictions (i.e., small changes in the training set could lead to significant differences in the predicted values). A common solution is then to use ensembles of regression trees, in order to decrease the variance by aggregating the pools of models.
In the class of tree-based ensembles, two approaches were selected: random forests (RF) and boosting of regression trees (BoRT). In RF, for each regression tree in the ensemble, only a small percentage of randomly chosen predictors are selected for the model instead of utilizing all the predictors available. On the other hand, BoRT is based on the boosting idea, where models are fit to residuals from previous models in order to improve the prediction ability. Both RF and BoRT have several tunable parameters (also called hyperparameters, such as the number of trees in the ensemble and the minimum number of samples in each leaf). For RF, the number of trees in the ensemble (TRF) is often the most important hyperparameter for controlling overfitting, thus, it is optimized by k-fold cross-validation. The other parameters are left at their default values. For BoRT, two parameters are often more relevant: the number of trees (TRF) and the learning rate (u). As they are inversely related (lower learning rates require more trees and vice-versa), we opted to set a low value for the learning rate (u = 0.02) and to optimize the number of trees in the ensemble.
3.2.5. Kernel Methods
The last class of regression methods included in this work are kernel methods. Kernel methods are non-linear approaches that implicitly project samples to a high-dimensional space (also called a feature space) where the model is developed. Thus, they are suitable for identifying and approximating non-linear relationships between predictors and response variable(s). In this work, two commonly used kernels were included: the polynomial kernel and the radial basis function (rbf). The former is more suitable for scenarios where the nonlinearity follows a polynomial relationship (quadratic, cubic, etc.), while the latter addresses other more general types of non-linearities. Kernel versions of PCR and PLS were included in the model comparison framework (L&NL-PAC) to enable modelling different types of non-linearities besides those described by tree-based methods (tree ensembles are more suitable when a step-wise relationship exists between the predictors and the response variable).
Kernel PCR starts by constructing a kernel matrix K (with dimensions ) between all pairs of samples in the training set. The kernel matrix represents the projection of the samples in a non-linear space, where the traditional linear PCA algorithm can be applied. This implies that the relationship modelled in the original data space is non-linear. The application of PCA provides a low-dimensional scores matrix that can be regressed to the response variable. Details for kernel PCR model building and for data scaling are readily available in the literature [36].
Kernel PLS is a natural extension of PLS that uses the kernel trick to model non-linear relationships. The starting point is the construction of a kernel matrix K, containing all the similarity/dissimilarity measures between all pairs of samples. The model is then built by applying the PLS algorithm to the matrix K and the response y. Details for kernel PLS model building and preprocessing the kernel matrices are available in the literature [36].
Additionally, two alternatives based on PCA and kernel SVR were included in this work. Initially, PCA was applied and the first principal components were extracted. These principal components constitute the predictor set that is used in kernel SVR, with the polynomial or rbf kernels. The motivation for combining PCA and SVR stems from the fact that the original data are high-dimensional, which negatively impacts the performance of SVR. Using a compression stage first (this is often called feature extraction) allows more stable and effective models to be developed.
3.3. Model Comparison Methodology
A double cross-validation scheme was employed in this work to compare the different methods. The root mean squared error, RMSE, was the metric used to assess and compare the predictive performance. Furthermore, multiple runs of Monte Carlo double cross-validation were conducted to characterize the variability of the predicted RMSE for each regression method, resulting in a more robust analysis of their relative performances. In each run of Monte Carlo double cross-validation, the input spectral dataset was split into training and external validation sets, using a stratified scheme based on the percentiles. To perform this step, the reference response measurements (analytical determinations) were grouped into five intervals according to the percentiles (20th, 40th, 60th, and 80th), and 80% of the samples in each group of percentile intervals were used for model training, while the remaining 20% were reserved for the external validation set. During model training, another stratified k-fold cross-validation based on the response percentiles was used to select suitable hyperparameters (Table 2) for each regression method. This was done using seven-fold cross-validation wherein the data were partitioned into seven folds, six used for calibration and one for validation, with the procedure being repeated seven times using a different validation fold each time. Then, each final model was built using the seven folds and the respective best hyperparameter(s) and was applied to predict the external validation set (or independent test set), based on which the corresponding RMSEs were obtained and stored. This process was repeated 30 times and the RMSEs for each run of Monte Carlo double cross-validation were saved for analysis. It is important to note that, in each run, all regression methods made use of the same training dataset for model building, and their performance was assessed in the same validation set (i.e., their RMSEs were correlated). The distribution of RMSEs over all runs of the Monte Carlo double cross-validation characterized the performance of each regression method, and methods with lower RMSE values were preferred.

Table 2.
Hyperparameters settings.
The distribution of the RMSE from the double cross-validation constitutes an informative source to compare the performance of different regression methods. However, visually comparing the RMSE distributions can be cumbersome due to the high number of methods included in this study (16 in total). Thus, besides RMSE, an additional key performance indicator (KPI) was devised to facilitate the ranking of the methods. As an additional advantage, this KPI was based on a rigorous statistical approach of hypothesis testing, allowing for detecting whether differences in performance were statistically significant or not. The KPI included in L&NL-PAC is similar to the one utilized in the PAC framework, and more details can be obtained in the original paper [35]. To compute this KPI, every pair of regression methods was considered, and their average RMSE was compared using a statistical hypothesis test, namely a paired t-test. This allows for the assessment of whether the average RMSE is statistically different across each pair of methods or not. If the differences were found not to be statistically significant, we considered this to be a “tie”, and both methods under comparison received 1 point. When a statistically significant difference was observed, a “win” was attributed to the method with the lowest RMSE and it received 2 points. The method with a higher RMSE was attributed a “loss” and it received 0 points. The KPI for each method was defined as the sum of all the points received from all of the pairwise comparisons. Thus, if a method was statistically superior to all of the others, it obtained the maximum number of points: 2 (nmethods − 1), where nmethods is the number of methods under comparison (the winning method receives 2 points from all the other nmethods − 1 methods). On the other hand, a method that presented statistically inferior results compared with of the all others received no points. This scheme provides an immediate ranking, allowing for the identification of the best regression methods and the best classes. Due to the complementary information provided by the RMSE of Monte Carlo double cross-validation and the KPI, both will be presented in the results section to provide a thorough analysis of the methods’ performance.
All computations were conducted in the MATLAB R2019b environment (MathWorks, Inc., Natick, MA, USA).
4. Results and Discussion
This section presents the results obtained for the prediction of sugar, pH, and anthocyanin contents. Both classes of linear and non-linear regression methods (described in Section 4.2) were considered for each property, and a detailed discussion of the top models is presented for each parameter, highlighting important spectral regions that most contribute to predicting the response variable. However, the spectral preprocessing was first considered in order to select a suitable preprocessing approach. As additional information, a summary of the descriptive statistics for the sugar, pH, and anthocyanin parameters determined by the conventional physic and chemical techniques is presented in Table A1 in Appendix A. These enological measurements were used as reference values to develop and test the proposed models in the following subsections.
4.1. Preprocessing Evaluation
As a significant number of regression methods were developed for L&NL-PAC (16 regression methods in total), testing all the combinations of preprocessing alternatives and regression methods would be a time-consuming task. Therefore, an alternative strategy was devised to select a suitable preprocessing technique, in which representative regression methods from each class were considered and their prediction performance was assessed under different preprocessing techniques. The preprocessing technique that more often led to better and more stable predictions (i.e., lower prediction errors) was selected as the most effective.
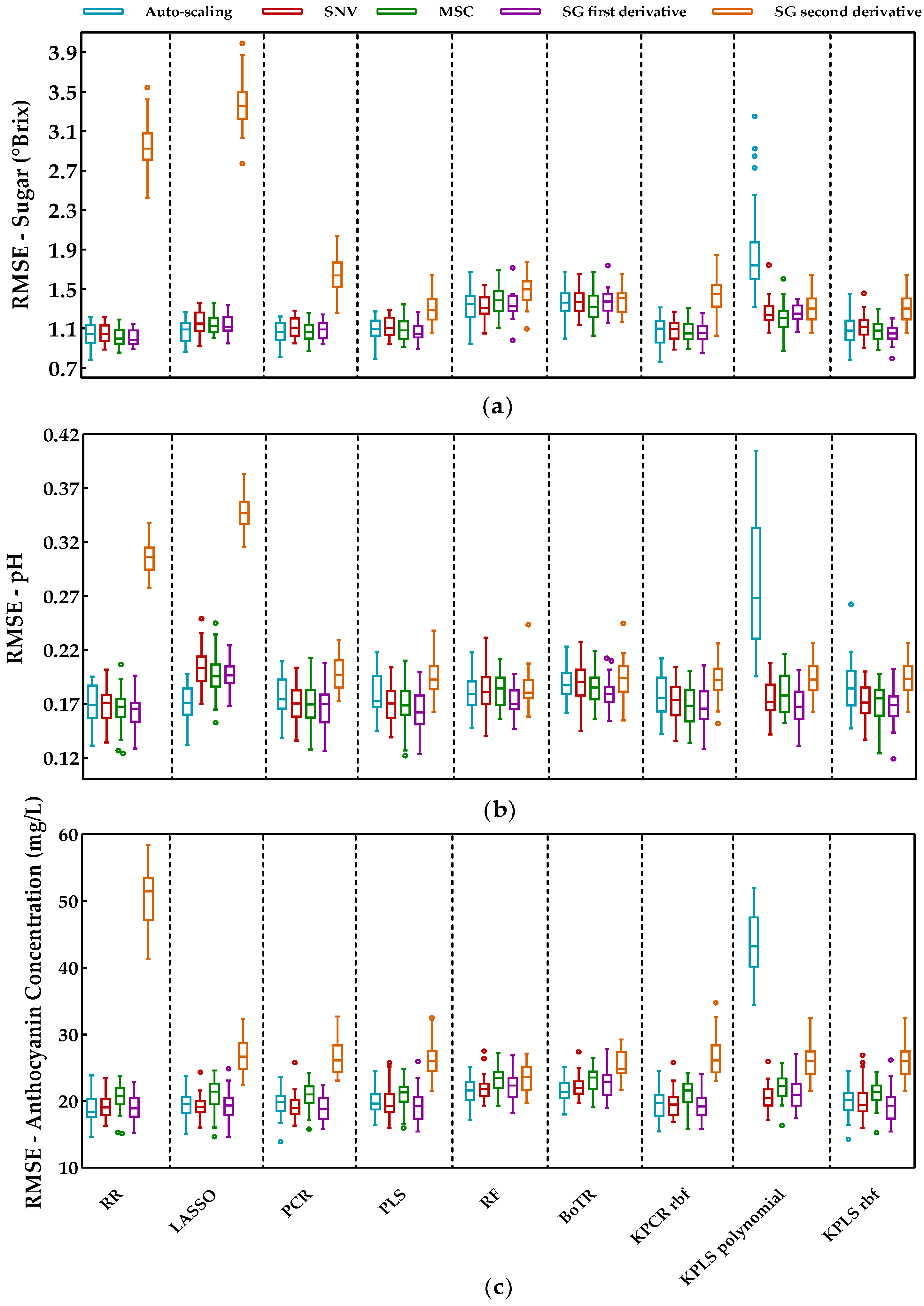
The prediction performances of the selected regression methods (RR and LASSO in the penalized regression class, PCR and PLS from the latent variable class; RF and BoRT from the tree-based ensembles, and kernel PCR and kernel PLS from the kernel-based methods), regarding the three enological parameters (sugar content, pH, and anthocyanin concentration), are presented in Figure 4. The prediction errors, given by the RMSE, indicate which combination of regression methods and preprocessing techniques had the better performance.
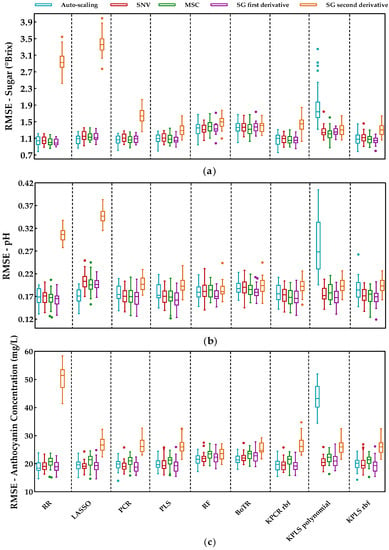
Figure 4.
Tukey boxplot showing the RMSE values of the external validation sets obtained by each preprocessing technique over the selected regression methods: (a) results for sugar contents, (b) results for pH, and (c) results for anthocyanin concentration. The bottom and top of the box represent the first and third quartiles (25th and 75th percentiles), and the internal horizontal lines represent the median (50th percentile). The whiskers represent the maximum and minimum data within the 1.5 interquartile range (IQR) and the circles represent outliers outside 3 IQR.
One can notice from Figure 4 that the SG second derivative was the worst preprocessing technique for this dataset as it often led to very high prediction errors. This was more clearly observed for the penalized regression methods. Excluding the second derivative, the alternative preprocessing techniques showed fewer differences and there was typically a significant overlap between different preprocessing methods for the same regression method. This suggests that the effect of the different preprocessing technique is not the most critical step and the SNV, MSC, or SG first derivative are expected to perform similarly; summary statistics (mean and standard deviation) for the distribution of the RMSE obtained for each preprocessing technique are presented in Table A1 (Appendix A). Moreover, Figure 4 displays a few outliers corresponding to instances where some combinations of models and preprocessing methods showed an abnormally high/low performance. This was expected as some peculiar training and test data splits can occur, leading to an abnormal model performance that is not representative. By observing the Tuckey’s boxplot, the distribution of the RMSE can be better assessed in terms of percentiles, and the few outliers can be ignored.
Concerning the literature mentioned in the introductory section (Table 1), the authors of [9] employed SNV, MSC, SG, and the second derivative to predict the anthocyanin concentration, denoting a better performance with SG and SNV techniques, and without differences between them. However, there are some works where preprocessing does not lead to an improvement in the results [8,11]. In addition, the authors of [10] compared MSC with SNV preprocessing, concluding that the performance is case dependent. In this work, we selected the SG first derivative preprocessing as a suitable approach, as it often came up with smaller median prediction errors (this was observed for RR, PCR, PLS, kernel PCR, and kernel PLS) and presented an error variability that was similar to the other alternatives. Thus, the SG first derivative was used in the remainder of this paper to preprocess the spectra before the development of the regression models.
4.2. Sugar Content Analysis
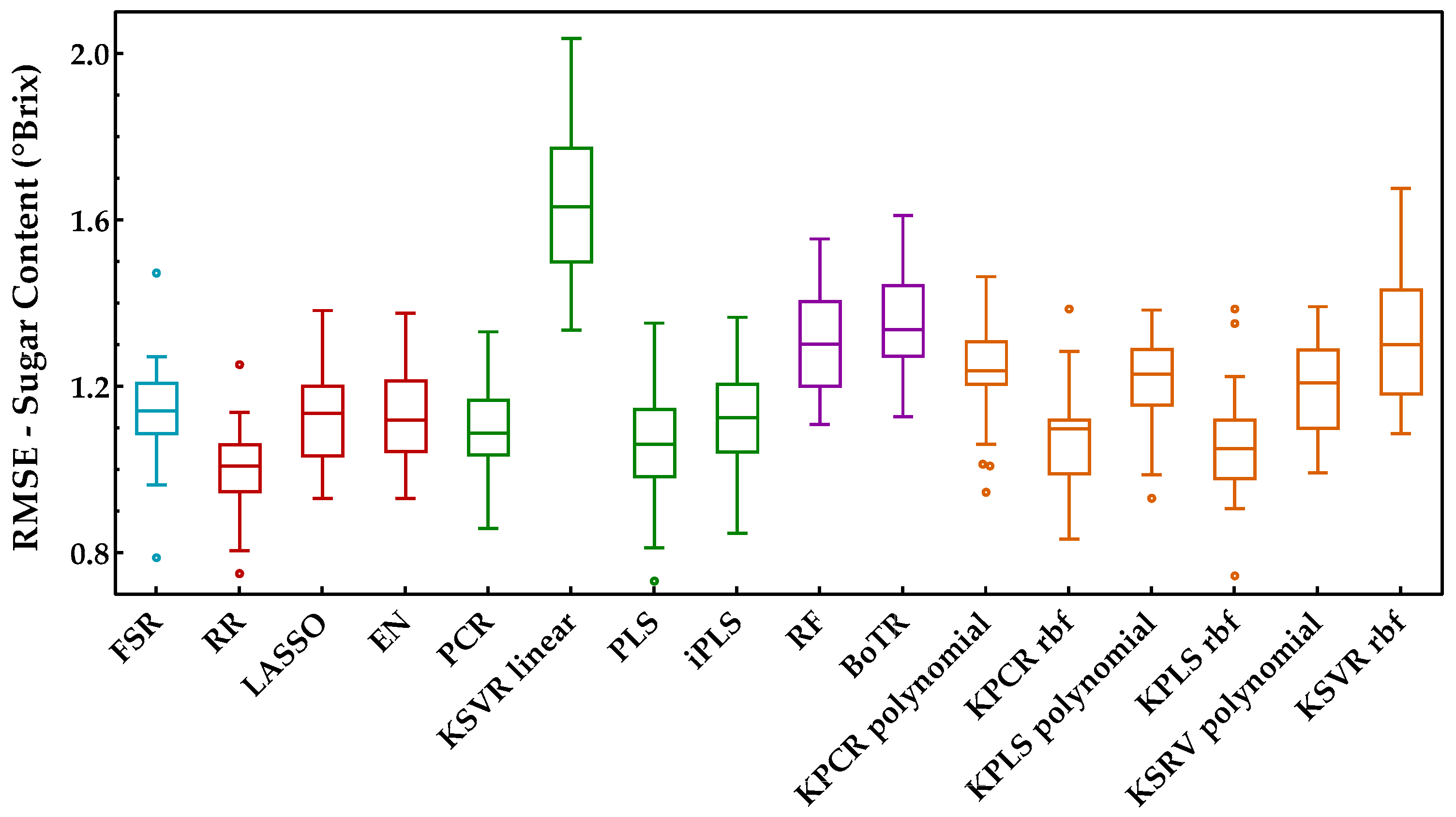
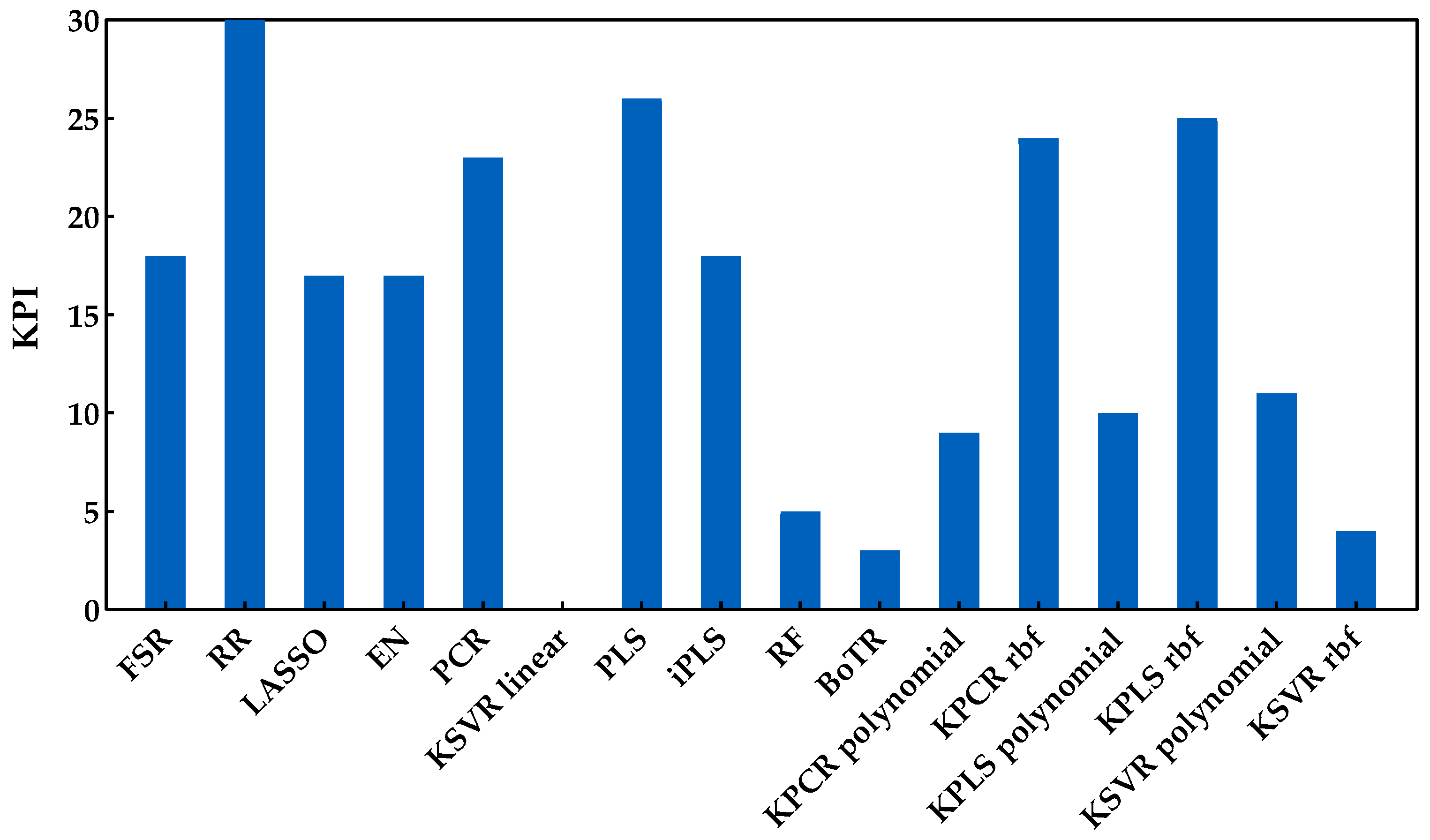
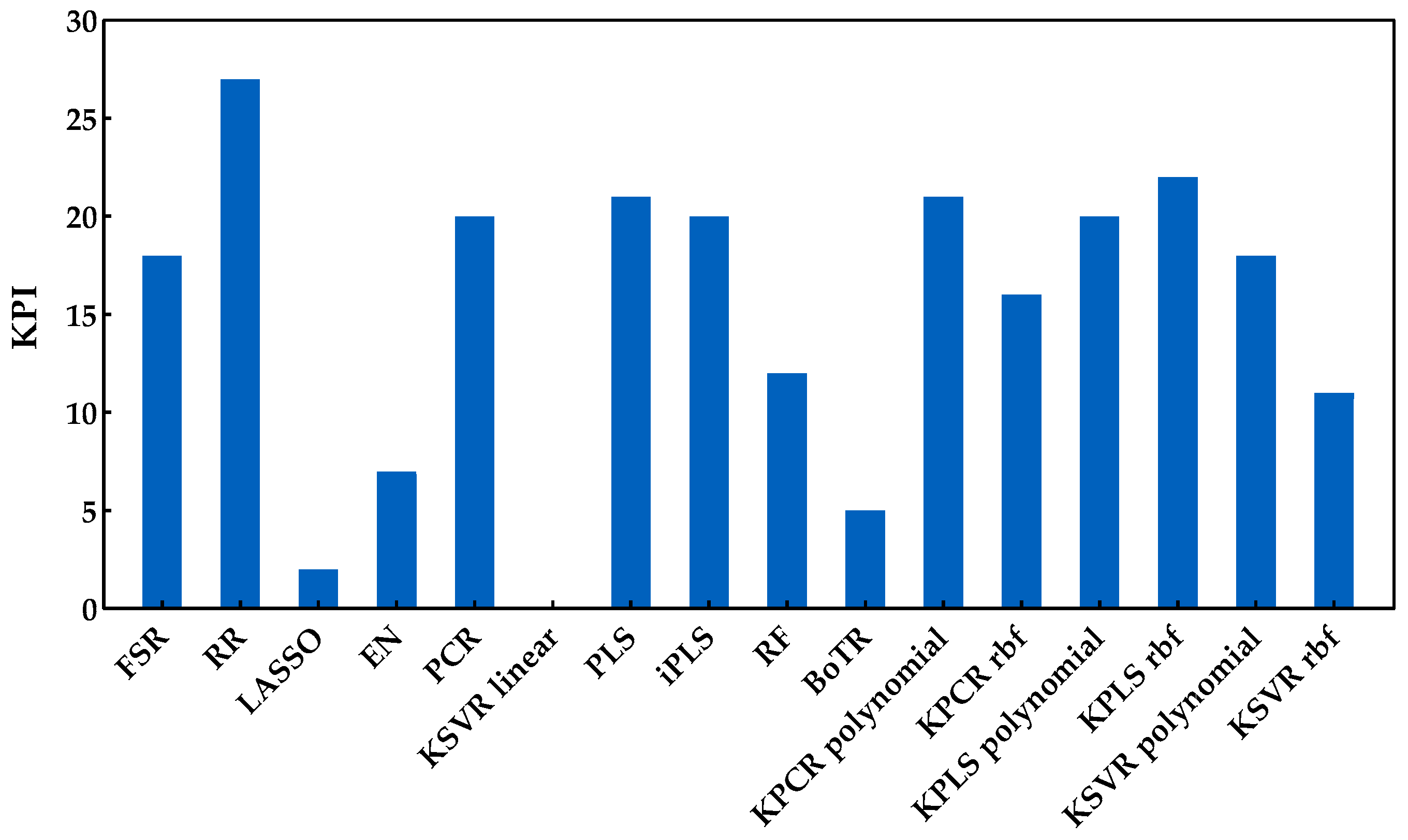
The sugar content is one of the main quality characteristics to evaluate the maturity of grapes and was the first property considered for developing the regression models. The Savitzky−Golay first derivative was applied to the spectra as a preprocessing technique and the RMSEs obtained from the Monte Carlo double cross-validation are presented in Figure 5 for all the regression methods included in this study. This figure clearly demonstrates the need to test a wide range of regression methods from different classes, as the performance of certain methods can be significantly different, even within the same class of methods. Only by testing and comparing their performance can one choose the most suitable regression model for each case. In addition, in order to characterize the overall method’s performance, Figure 6 summarizes the KPI (described in Section 3.3) obtained by each regression method.
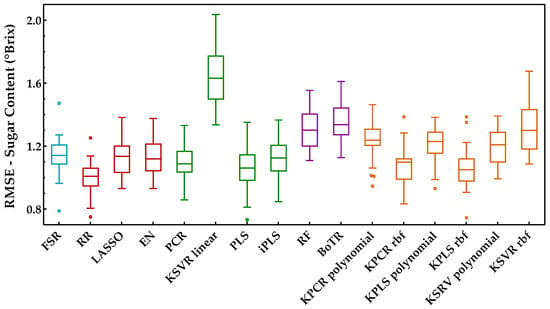
Figure 5.
Tukey boxplot showing the sugar content results to the external validation sets obtained for the classes of regression methods included: (blue) variable selection, (red) penalized regression, (green) latent variables, (purple) tree-based ensemble, and (orange) Kernel methods.
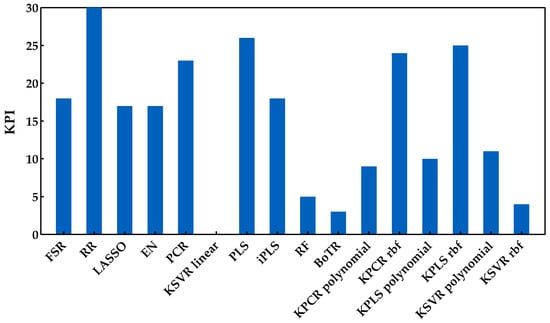
Figure 6.
KPI obtained by each regression method when applied to predict the sugar content.
From the analysis of Figure 5 and Figure 6, it is possible to identify that RR was the best regression method for predicting the sugar content, obtaining the lowest RMSE (Figure 5) and the best overall performance with the highest KPI (Figure 6). More precisely, the difference between RR and the other regression methods was statistically significant (two points were always attributed to RR method, see Section 3.3). One can also notice that the class of tree-based ensembles is not suitable for predicting the sugar content, which can probably be attributed to the regression tree’s stepwise approximation. The underlying relationship between the spectral signals and contents often follows the Beer−Lambert law, which is a continuous function instead of a stepwise relationship. The class of latent variable methods contains the worst method, which is the combination of PCA and a linear kernel SVR. Nevertheless, the other methods in this class presented a good and consistent performance (Figure 5). The newly tested alternatives based on non-linear kernels had rather different performances, namely kernel PLS and PCR (both rbf), which achieved the lowest RMSE (Figure 5), reaching a good position (third and fourth, respectively), among the top performances (Figure 6). Despite this result, the performance of the class of kernel methods suggests that the use of non-linear methods is not particularly advantageous for predicting the sugar content. Nevertheless, other types of non-linear approaches may be more effective, e.g., using neural networks, as in our previously published works [7,13], which presented RMSE values between 0.95 and 0.96 °Brix for the same set of data. Moreover, in [7], a comparison between PLSR and NN was done, where the results showed a similar performance between the two methods. Comparing the results reported in [7,13] to those obtained in this study (Figure 5), one can notice that the results here are more conservative because a double cross-validation procedure was employed in multiple runs, which allowed for a more comprehensive incorporation of all variation and uncertainty sources present in the raw data as well as during model development. Thus, there were testing scenarios where the RMSE was below 0.95 °Brix, but there were others where the observed performance was worse, depending on the particular random data split in training and validation sets. This demonstrates that the data split plays an important role when determining the methods’ performance, and therefore the reported results should have an increased focus on assessing all sources of variability and uncertainty. Regarding the KSVR method, the authors of [12] reported RMSE values of 0.80 °Brix, using SVR with a radial basis function approach for the same dataset acquisition. However, the authors used a genetic algorithm followed by random search to determine the hyperparameters range and the best combination that led to the lowest RMSE. Furthermore, they employed a different splitting strategy where three samples were selected from each grape’s harvest day and reserved for validation (by contrast, we randomly select 20% in each percentile). This might justify the difference in the obtained results and indicates that the data splitting step may have a significant impact on the results. Naturally, the splitting strategy employed should take into account the goals of the research being conducted. Nevertheless, in this work, the focus was on understanding how regression models perform under more strict conditions, where samples in the validation set do not require a counterpart sample in the training set collected on the same day. Furthermore, and as complementary information to the strategy designed for the proposed comparison framework (RMSE and KPI), the ratio of performance deviation (RPD) and range error rate (RER) values are provided in Table 3, in terms of the mean and respective 95% confidence intervals. Both RPD and RER normalize the RMSE values obtained by each model for the external validation sets against the standard deviation and range of their reference data, respectively. The best RPD and RER values were achieved for the RR method, with mean values of 3.32 and 13.83, respectively, indicating a good overall prediction ability of the RR model (following the guideline scale suggested by [39]).

Table 3.
RPD and RER results in the sugar content for the external validation sets—Mean and associated 95% confidence intervals obtained for each regression method.
Taking into account the RR and PLSR approaches, with PLSR being the most employed in the literature for spectroscopic measurements in reflectance mode (Table 1), the RMSE results obtained forthe sugar content in the present work were better than those from [7,8,10,15,16]. On the other hand, the authors of [11] revealed results better than ours, but these authors used a significantly larger number of berries per sample compared with the six berries per sample used in this work. The use of a large number of berries reduces the sample variability, which have an impact on the RMSE. In terms of RPD and/or RER, the authors of [7,8] did not show the values; however, in [15], the authors reported RPD values of 1.88 and 1.54 for Chardonnay and Viura varieties, respectively. Furthermore, the authors of [10] reported RPD and RER values of 4.06 and 13.89, respectively, for a Syrah variety and of 5.83 and 19.68 for a Cabernet Sauvignon variety, respectively; in [16], the RPD was 4.12, and the authors of [11] only mentioned that the RPD was acceptable (the authors did not disclose the value). Nevertheless, the authors of [8,10,16] also used a larger number of berries per sample in their studies, which might justify the better results obtained in terms of RPD and RER. In this work, the decision to use a small number of berries per sample (six) was due to the desire of some wineries to select the best berries from each bunch to produce specific high-quality wines, as stated in [7,12,13].
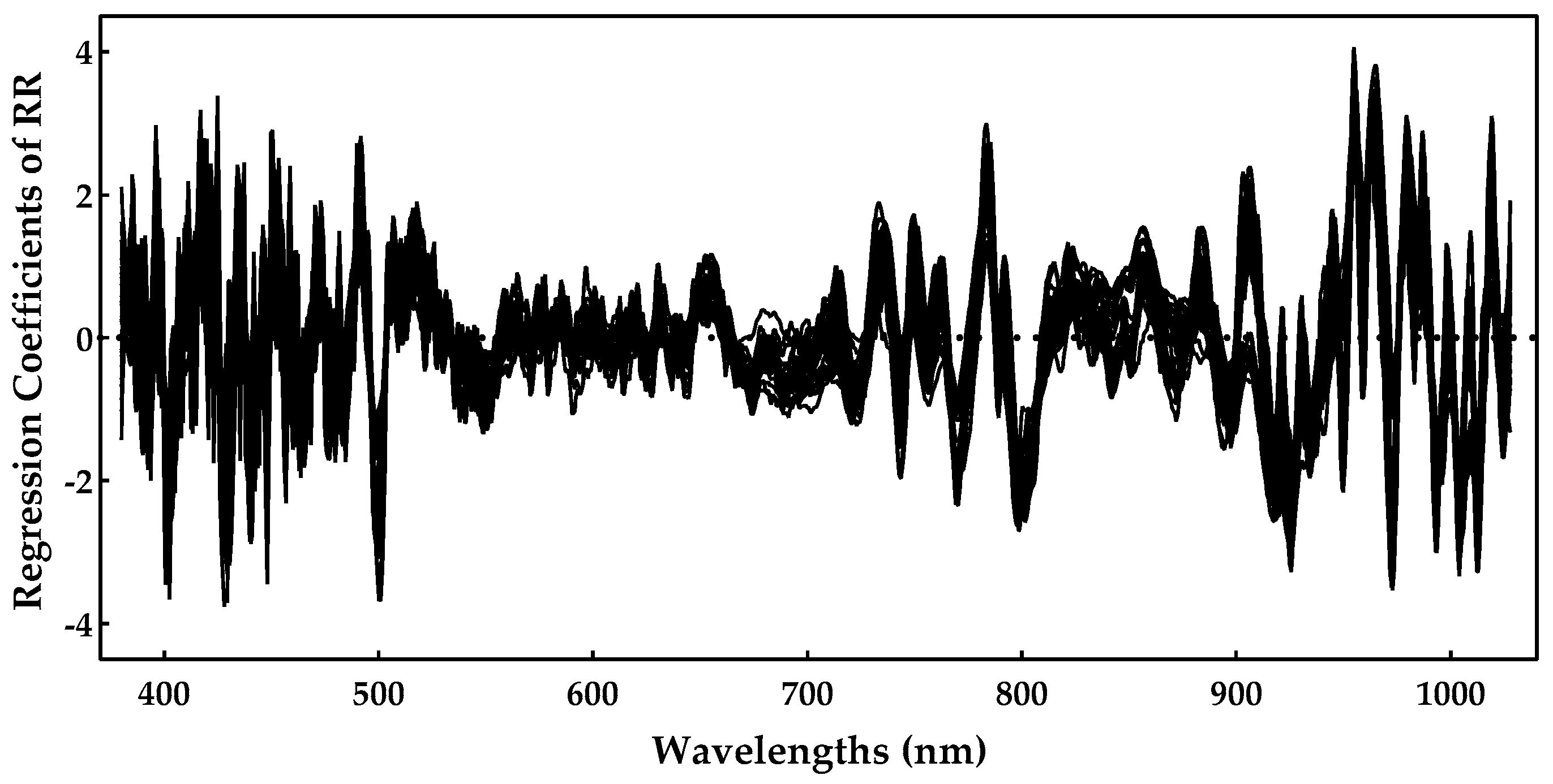
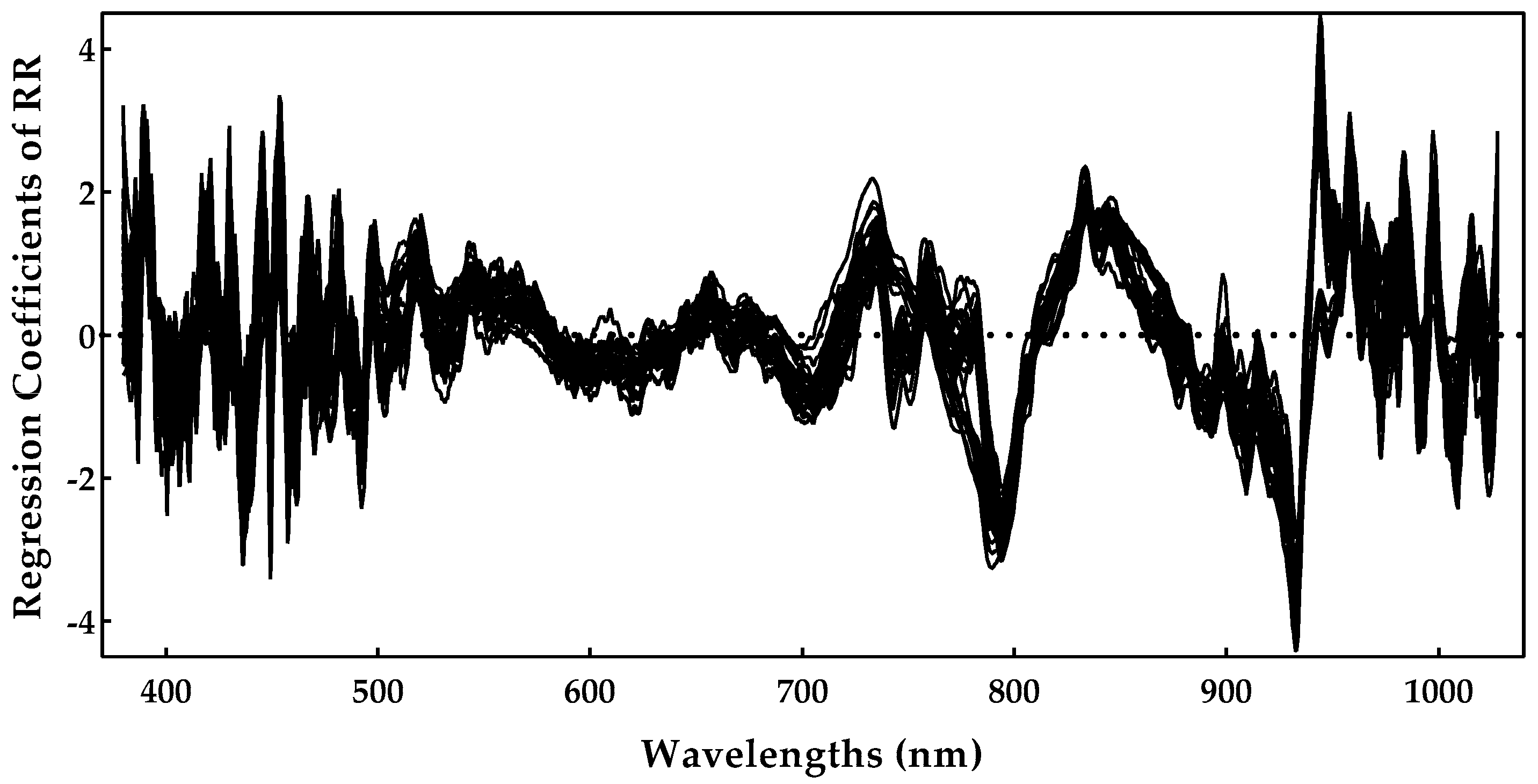
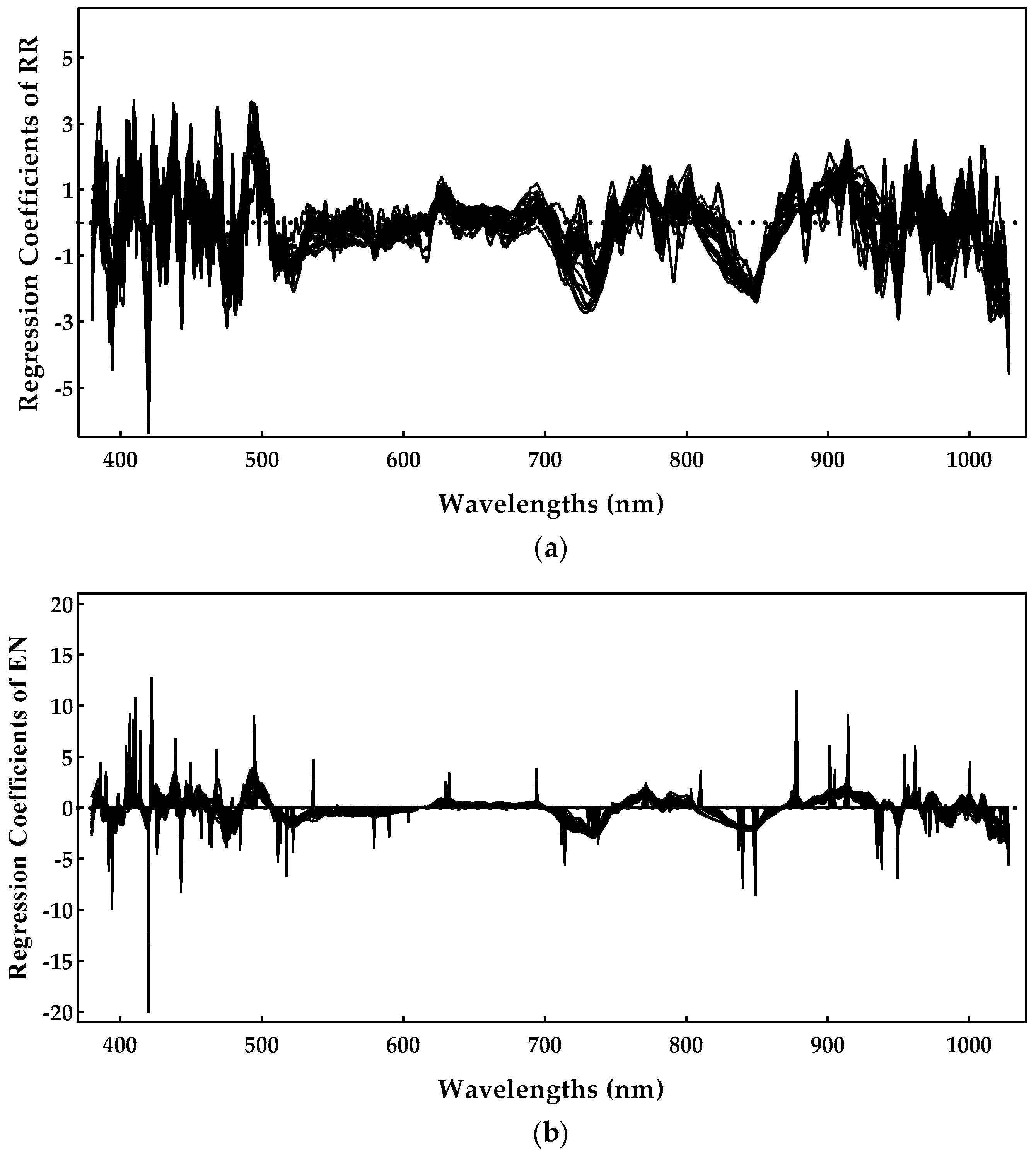
As RR was shown to be the best method, we analyzed the most important spectral regions for this model. Figure 7 presents the regression coefficients for the RR model, and important regions are characterized by higher magnitudes of the regression coefficients.
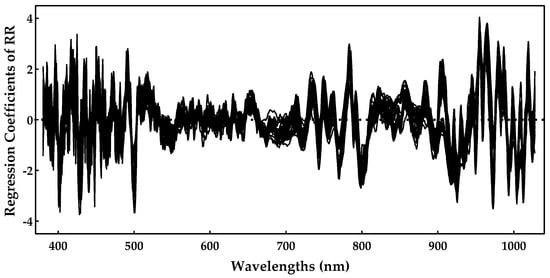
Figure 7.
Sugar regression coefficients obtained during the ridge regression model training.
One can notice by Figure 7 that the region near 900–1000 nm is rather important, as is the region near 750–800 nm. The spectral regions (400–500 nm) also had some important predictors, but they tended to be noisier. The 770, 920, 960, and 980 nm peaks might be related to the sugar absorption, but we should point out that the regions above 960 nm were closer to the end of the sensing spectral range, and, for that reason, they also tended to be noisier. Nevertheless, these results are in line with those previously reported in [7,10], further confirming the effectiveness of these models.
4.3. pH Analysis
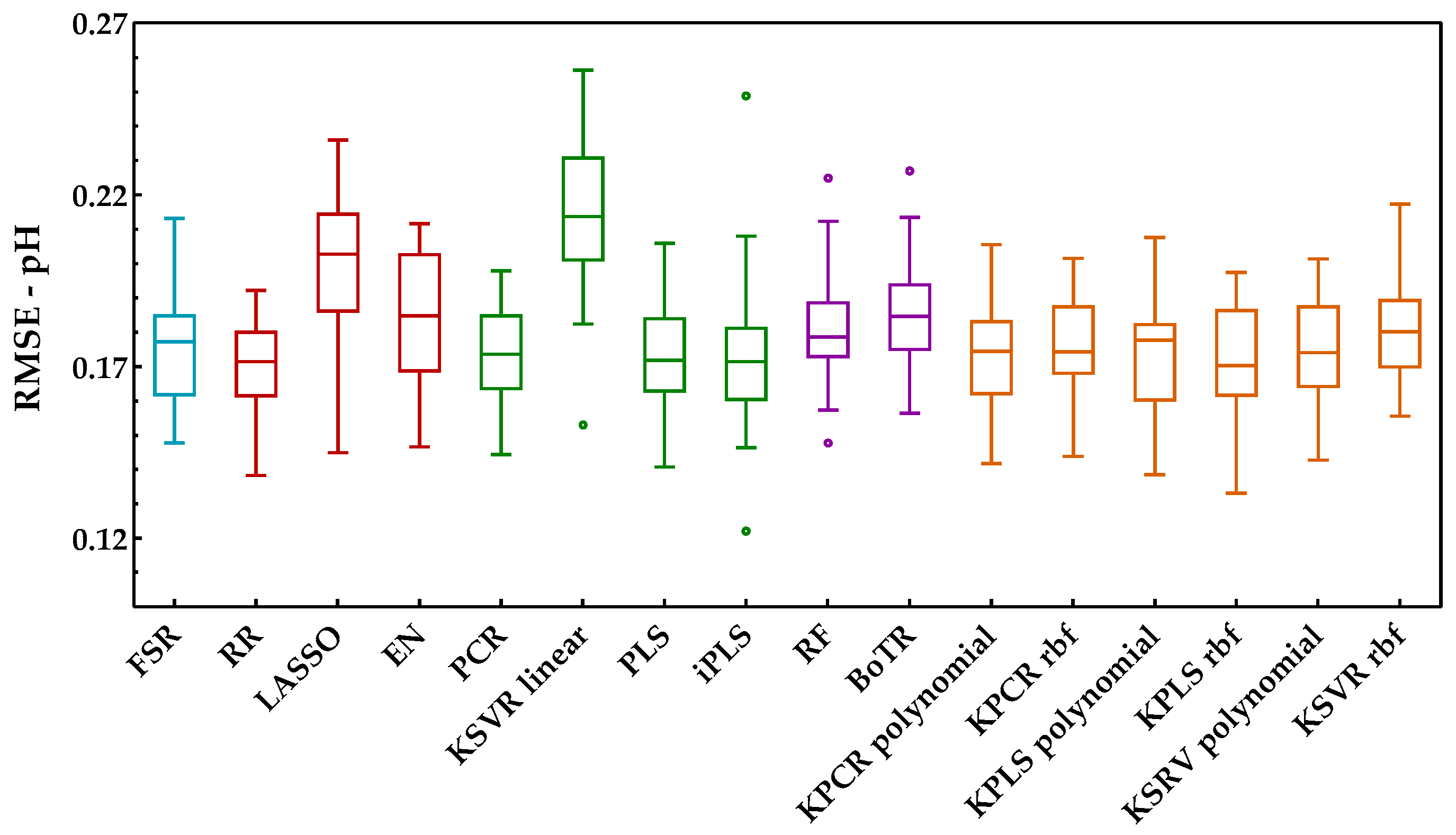
The analysis of this second enological parameter followed the same methodology as the previous one (Savitzky−Golay preprocessing plus development of models). Figure 8 shows the RMSE obtained with the external validation dataset for each run and regression method. Through the analysis of this figure, one may see no major practical differences between the classes included. In fact, except for the combination of PCA and a linear kernel SVR, which presented the worst performance, all other methods seemed to have a similar behavior. In contrast with the sugar content, here, it was not possible to clearly understand which method might be the most suitable to create the predictions. However, through the analysis of Figure 9, which presents the KPI obtained by each regression method, one can observe that RR obtained the best performance (reaching 27 points in a total of 30). Further analysis of the KPI results showed that the difference in the RMSE distribution between the RR and KPCR polynomial function was not statistically significant, and that the difference between RR and KPLS radial basis function was statistically significant, with two points attributed to KPLS rbf.
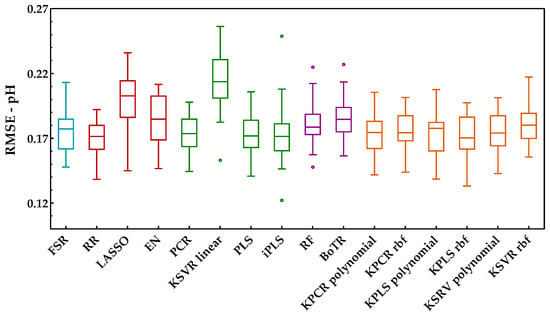
Figure 8.
Tukey boxplot showing the pH results for the external validation set obtained for the class of regression methods included: (blue) variable selection, (red) penalized regression, (green) latent variables, (purple) tree-based ensemble, and (orange) kernel methods.
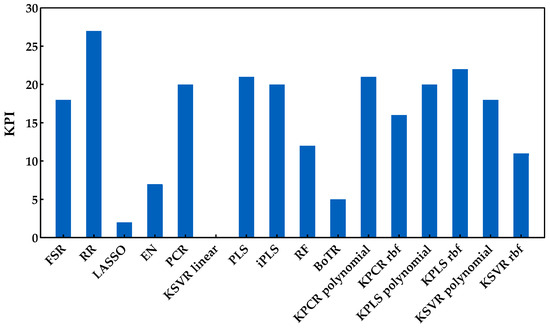
Figure 9.
KPI obtained by each regression method when applied to predict the pH contents.
In addition to the RMSE and KPI metrics, Table 4 present the results obtained for RPD and RER. Here, the RPD values showed low levels for the predictions, but RER was above 5.0, which indicates acceptable predictions [39]. Concerning the existing scientific literature on the same subjects (see Table 1), it was possible to verify that the RMSE values obtained here for the pH were in accordance with the results of [8,12,13,16], but they were worse than those reported in [11], for the same reason already justified in Section 4.2. For RPD and RER, the authors of [8,12,13,16] did not report values and [11] only stated that the models were moderately successful, with average R2 but with low RPD values. In fact, the lower results obtained for the pH might be related to the small distribution in the range of the reference measurements (Table A1, Appendix A), which may increase the difficulty of the models to provide suitable prediction performances.

Table 4.
RPD and RER results in pH for the external validation sets—mean and associated 95% confidence intervals obtained for each regression method.
Following the same procedure as for the sugar content, the ridge regression model was selected to identify the most important spectral regions (Figure 10). Analyzing Figure 10, it is possible to detect some noise between the 400 and 500 nm regions. Additionally, the region near 750–950 nm, with much less noise, seemed to be the most important, presenting relevant peaks at 790, 840, and 930 nm, which could be related with the pH. Regarding the scientific literature, the authors of [40] identified, for table grapes, peaks of 695, 870, and 950 nm sing the highest regression coefficients of PLS, while the authors of [41] implemented a genetic algorithm with the least-squares support vector machine approach to identify the effective wavelengths at 446, 489, 504, and 561 nm. As acidity seems to be a sensitive case, we believe that the most spectral regions depend on the regression method and on the characteristics of the grapes under study.
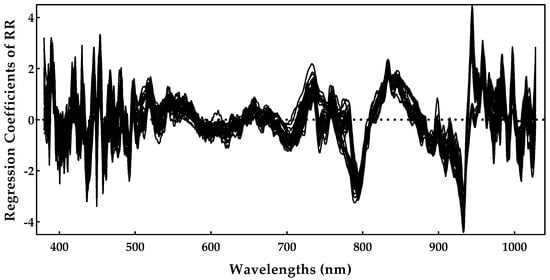
Figure 10.
pH regression coefficients obtained during the ridge regression model training.
4.4. Anthocyanin Concentration Analysis
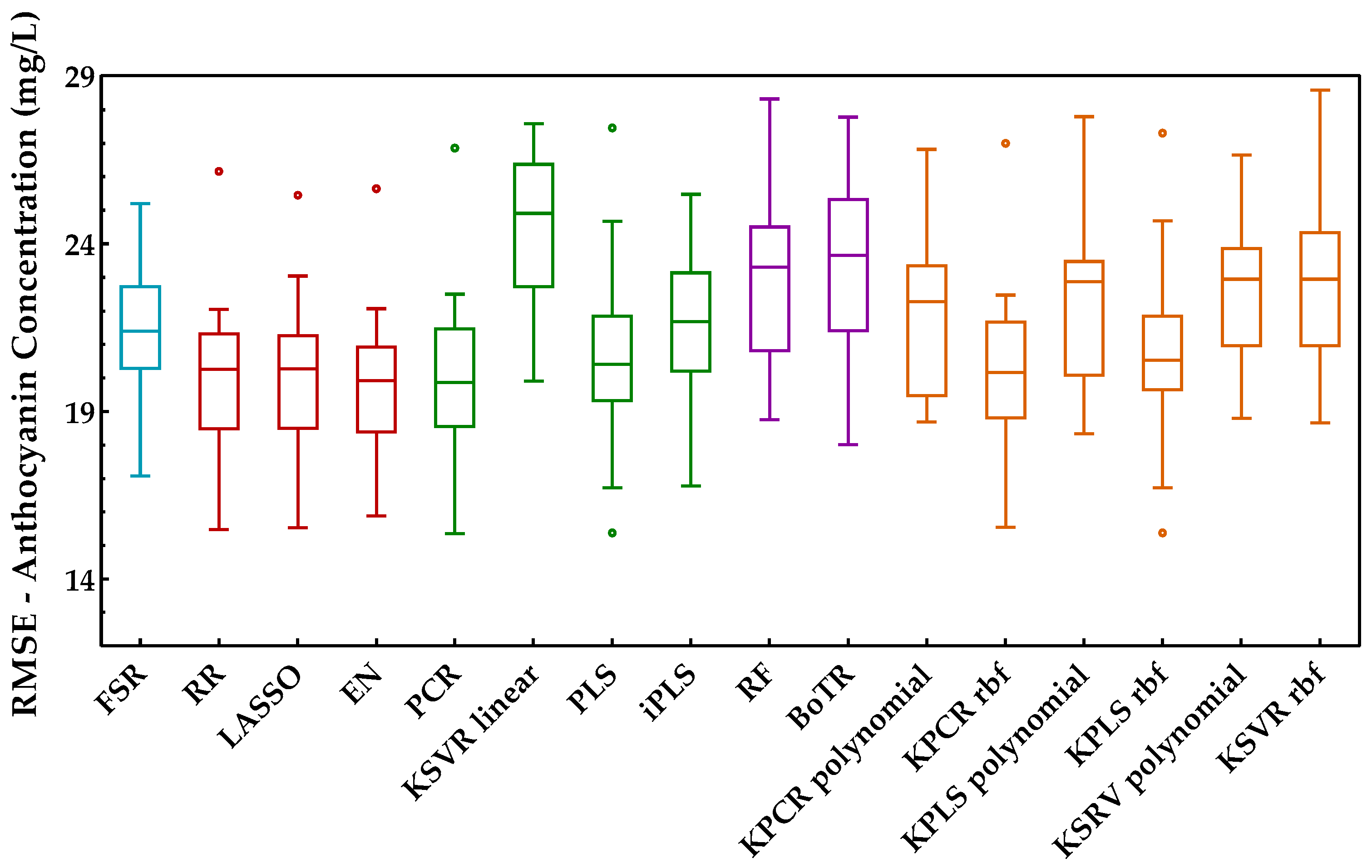
Anthocyanins are the pigments responsible for red wine color and were the last enological parameter considered for the proposed comparison. The procedure adopted was identical to the previous enological parameters, and the results achieved in terms of root mean square error for each class of methods are presented in Figure 11.
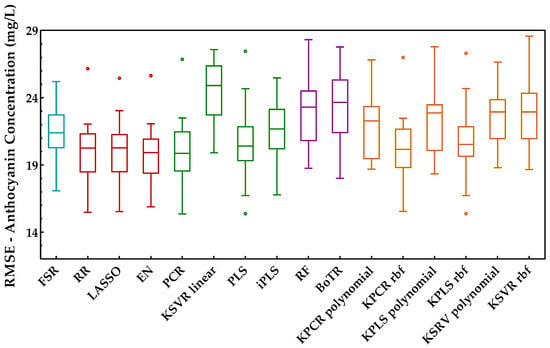
Figure 11.
Tukey boxplot showing the anthocyanin concentration results in the external validation set obtained for the class of regression methods included: (blue) variable selection, (red) penalized regression, (green) latent variables, (purple) tree-based ensemble, and (orange) kernel methods.
From the analysis of this figure, one can identify the class of penalized regression as the most suitable and promising class to predict the anthocyanins concentration. The same can also be confirmed by Figure 12, wherein this class reached the best scores. Within the latent variable’s class, PCR was the best method, with a similar overall performance to the penalized regression class, namely to RR. However, the other regression methods of the latent variables class presented quite different performances. Such differences can also be observed for the kernel methods class. The class of tree-based ensemble showed the worst overall performance for estimating the anthocyanin concentration. Overall, EN presented the best performance (27 points in 30), losing 2 points for PCR (the difference was statistically significant) and 1 point for the LASSO method (not statistically significant). RR, PCR, and LASSO were the next methods with the best KPI, obtaining values between 24 and 26 (in a maximum of 30 points).
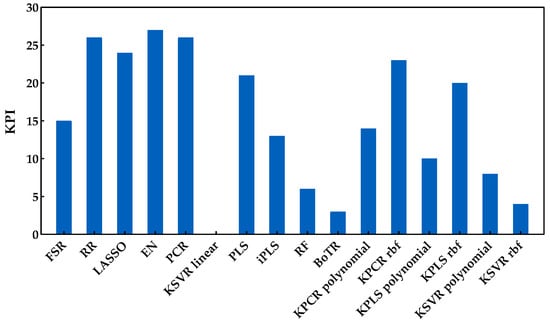
Figure 12.
KPI obtained by each regression method when applied to predict the anthocyanin concentration.
Regarding the results from the literature, some of the works listed in Table 1 are not comparable with ours due to differences in the anthocyanin quantification procedures. The RMSE values presented in our work outperformed those obtained [11]. However, the authors of [13] obtained better RMSEs than those obtained here, indicating that the use of neural networks might be more effective for predicting the anthocyanin concentration. The results of [12] also showed a better performance for the SVR approach, indicating that the use of a genetic algorithm can be a good alternative for predicting the anthocyanin concentration. In addition, Table 5 displays the results for RPD and RER, and from which it is possible to conclude that, overall, moderately successful predictions were achieved, with RDP and RER obtaining values larger than 2.5 and 10, respectively [39]. Comparing these values with those reported in the literature, the authors of [10] reported RPD and RER values of 3.89 and 10.49, respectively, for a Syrah variety and of 5.38 and 12.78, respectively, for a Cabernet Sauvignon variety, but the authors used a large number of berries per sample, while [11] only stated that the models were moderately successful with average R2 but low RPD values. The remaining works from Table 1 did not report values for RPD or RER for the anthocyanin parameter.

Table 5.
RPD and RER results in the anthocyanin parameter for the external validation sets—Mean and associated 95% confidence intervals obtained for each regression method.
As EN was the best method for estimating the anthocyanin concentration and RR appeared on the top of the best methods for the enological parameters considered in this work, both were selected to identify the relevant spectral regions, regarding the anthocyanin property. From the analysis of Figure 13, which presents the regression coefficients obtained for the 30 runs of the Monte Carlo double cross-validation, it is possible to observe an agreement regarding the relevant regions, covering 400–520 nm and 700–900 nm ranges. Nevertheless, spectral regions between 400–480 and 900–1000 nm tend to be noisier for the RR method, but presented much less noise for the EN method. Thus, 420, 450, 490, 520, 730, and 850 nm peaks might be related to the anthocyanins. Additionally, some coefficients were very important in some runs, but were very close to 0 in others (Figure 13b), confirming the instability due to the collinearity that is prevalent in the spectral datasets. In some runs of Monte Carlo double cross-validation, a single wavelength was selected and led to better results, whereas nearby regions were ignored (their coefficients were set to zero, which is a property of the LASSO L1-norm penalty). On other runs, the coefficients converged to a scenario where nearby spectral regions had a similar contribution (a property of the RR L2-norm penalty). Again, this example shows the benefits of Monte Carlo double cross-validation as it allows for a more comprehensive study of the variability that can be expected from different models built under slightly different conditions. Concerning the scientific literature on this subject, most of the works from Table 1 did not identify the important spectral regions for the anthocyanin concentration, nevertheless, the results obtained here are in agreement with those previously reported in [10].
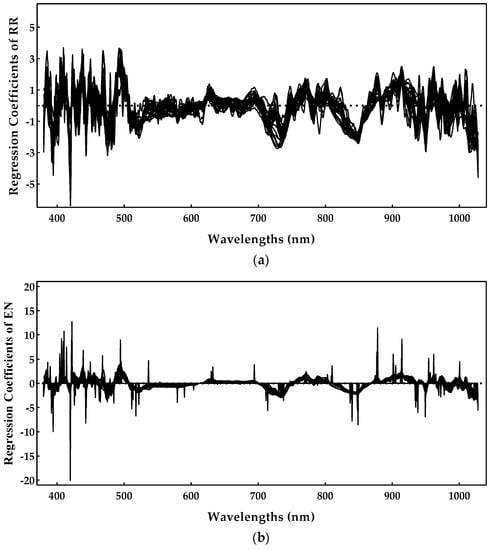
Figure 13.
Anthocyanin concentration regression coefficients obtained during the model training: (a) ridge regression and (b) elastic net.
4.5. Miscellaneous Discussion
The previous sections demonstrate the effectiveness of developing regression methods for estimating the enological parameters considered, namely sugar content, pH, and anthocyanin concentration, expanding the existing approaches on the same subject. The best results obtained for each enological parameter are highlighted in Table A3 (Appendix A) for an easier interpretation and comparison of the finding. The columns contain information’s such as the ranking of the three best methods, the mean and standard deviation of the RMSEs obtained for the external data set, the rather important spectral regions identified by the regression coefficients of the best model, and the relevant peaks that might be related to the enological parameter in the study and identified by the highest regression coefficients of the best model.
Analyzing the overall results, it is possible to verify that sometimes one method was more adequate for one parameter, and another method was more explanatory for another. The reason for this can be justified by the fact that each method has its strengths and weaknesses in terms of modelling the entire spectra vs. the narrow region, linear vs. non-linear relationships, and sparse vs. correlated features. This means that the best method depends on the underlying relationship between the hyperspectral image and a particular enological parameter, and in practice, the best method is never known a priori. This fact supports conducting such extensive studies as the one that we have done here, in order to identify the most suitable regression methods per the observed enological parameters. Interestingly, our study found that the combination of the Savitzky−Golay preprocessing and ridge regression methods led to the best or near best performance across all enological parameters, and was more robust in comparison to other linear approaches, as well as more complex non-linear methods. Therefore, researchers are incentivized to also consider this particular combination when modeling such enological parameters. On the other hand, the present results do not extend to enological parameters that were not considered in this study. Instead, the application of the complete L&NL-PAC is suggested as a robust framework to identify superior methods for modeling additional enological properties of interest.
Another interesting feature of this work concerns the identification of important spectral regions by screening the regression coefficients. In this regard, the complete spectra of the samples (ranging from 380 to 1028 nm; Figure 2) were used to establish and validate (with samples not employed during the training) the models, and the identification of the best method allowed us to understand which spectral regions may be more relevant to further design more specific equipment, reducing the dimensionality of data and providing a faster and more cost-effective methodology (e.g., a smaller number of bands leads to cheaper equipment). Notably, the most important spectral regions seemed to range between 700 and 960 nm for the three enological parameters, with some other important peaks appearing between 400 and 520 nm for the anthocyanin parameter. The spectral regions between 400 and 500 nm also had some important predictors for sugar content and pH, but they tended to be noisier. It is important to denote that, due to differences in terroir, the grape berries were at various ripening stages (even within the same bunch and day), so it was already expected that for the 30 runs of Monte Carlo double cross-validation the highest regression coefficients would present some scenarios of variation (as it is possible to observe in Figure 7, Figure 10 and Figure 13). Furthermore, the measurements were done for samples composed of a small number of berries, which increased the sample variability when compared with a large number of berries per sample. Thus, we recognize the need to expand this study with more varieties and even with more vintages in order to try to capture most of the variations presented in the samples.
5. Conclusions
In this work, a robust framework was developed in order to compare different preprocessing techniques and a wide variety of regression methods for predicting three important enological parameters (related to grape’s maturity) through hyperspectral imaging. In addition, the framework employed a Monte Carlo double cross-validation scheme to assess the prediction performance of each regression method, using its RMSE distribution and a preset key performance indicator (KPI) as comparison metrics. In terms of preprocessing, we have shown that the effect of SNV, MSC, and Savitzky−Golay first derivative on the regression performance was similar, whereas Savitzky−Golay second derivative provided poor results. Thus, Savitzky−Golay first derivative was the select preprocessing approach to make further comparisons between the regression methods. From all 16 regression methods tested, the best results were obtained with ridge regression (that belongs to the class of penalized regression methods), as it was the method that most often appeared with the highest performance. This indicates that the combination Savitzky−Golay first derivative and ridge regression can be a good choice to deal with the prediction of enological parameters based on the use of hyperspectral imaging technology. The wavelengths most contributing to explaining the variability of each enological parameter were also investigated, providing important information for the development of precision viticulture technology. However, future work should assess the performance of RR using a larger dataset (composed by different varieties and vintages) and compare it with the most often used methods from literature (e.g., PLS and ANN), as well as with recent deep learning approaches. The use of ensemble preprocessing methods should also be the subject of future research.
Author Contributions
Conceptualization, V.G., R.R., M.S.R. and P.M.-P.; methodology, V.G., R.R., M.S.R. and P.M.-P.; conventional laboratory analysis, A.M.-F.; software, V.G. and R.R.; validation, V.G. and R.R.; formal analysis, V.G. and R.R.; investigation, V.G., R.R. and M.S.R.; resources, M.S.R., A.M.-F. and P.M.-P.; writing—original draft preparation, V.G. and R.R.; writing—review and editing, V.G., R.R., M.S.R., A.M.-F. and P.M.-P.; supervision, M.S.R. and P.M.-P.; project administration, M.S.R. and P.M.-P.; funding acquisition, M.S.R. and P.M.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Funds from FCT, the Portuguese Foundation for Science and Technology, under the project UIDB/04033/2020 (CITAB); and by Portuguese-FCT (PD/BD/128272/2017), under the Agrichains Doctoral Program (PD/00122/2012).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors acknowledge the support provided by Biosystems and Integrative Sciences Institute by FCT (BioISI; FCT/UIDB/04046/2020); and the support from the Chemical Process Engineering and Forest Products Research Centre (CIEPQPF), which is financed by national funds from FCT/MCTES (reference UID/EQU/00102/2019). The authors also gratefully acknowledge all resources and support provided by Symington Family Estates.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Descriptive statistics for the reference measurements of sugar, pH, and anthocyanin contents obtained by conventional analysis.
Table A1.
Descriptive statistics for the reference measurements of sugar, pH, and anthocyanin contents obtained by conventional analysis.
| Enological Parameters | Mean | SD a | Min b | Max c | Median |
|---|---|---|---|---|---|
| Sugar content (°Brix) | 16.925 | 3.342 | 9.060 | 24.720 | 17.060 |
| pH | 3.552 | 0.346 | 2.850 | 4.230 | 3.580 |
| Anthocyanin concentration (mg·L−1) | 160.278 | 56.860 | 3.894 | 257.819 | 173.841 |
a,b,c Standard deviation, and minimum and maximum values of each enological parameter, respectively.

Table A2.
RMSE of the external validation set obtained by each preprocessing technique over the selected regression methods: mean ± standard deviation for the 30 runs of Monte Carlo double cross-validation.
Table A2.
RMSE of the external validation set obtained by each preprocessing technique over the selected regression methods: mean ± standard deviation for the 30 runs of Monte Carlo double cross-validation.
| Enological Parameter | Methods | Auto-Scaling | SNV | MSC | SG 1D | SG 2D |
|---|---|---|---|---|---|---|
| Sugar | RR | 1.039 ± 0.109 | 1.047 ± 0.096 | 1.008 ± 0.092 | 1.004 ± 0.077 | 2.964 ± 0.254 |
| LASSO | 1.072 ± 0.117 | 1.146 ± 0.119 | 1.145 ± 0.105 | 1.138 ± 0.093 | 3.371 ± 0.270 | |
| PCR | 1.058 ± 0.117 | 1.111 ± 0.096 | 1.071 ± 0.099 | 1.089 ± 0.093 | 1.655 ± 0.189 | |
| PLS | 1.098 ± 0.117 | 1.117 ± 0.104 | 1.085 ± 0.106 | 1.061 ± 0.084 | 1.292 ± 0.135 | |
| RF | 1.330 ± 0.161 | 1.315 ± 0.121 | 1.375 ± 0.149 | 1.338 ± 0.128 | 1.486 ± 0.154 | |
| BoTR | 1.358 ± 0.149 | 1.366 ± 0.127 | 1.315 ± 0.148 | 1.370 ± 0.121 | 1.394 ± 0.144 | |
| KPCR rbf | 1.072 ± 0.136 | 1.081 ± 0.098 | 1.058 ± 0.100 | 1.060 ± 0.098 | 1.431 ± 0.184 | |
| KPLS polynomial | 1.897 ± 0.478 | 1.263 ± 0.131 | 1.201 ± 0.152 | 1.259 ± 0.083 | 1.302 ± 0.140 | |
| KPLS rbf | 1.088 ± 0.137 | 1.114 ± 0.120 | 1.070 ± 0.106 | 1.040 ± 0.088 | 13.09 ± 0.148 | |
| pH | RR | 0.170 ± 0.017 | 0.168 ± 0.019 | 0.165 ± 0.018 | 0.163 ± 0.018 | 0.306 ± 0.015 |
| LASSO | 0.172 ± 0.016 | 0.205 ± 0.018 | 0.197 ± 0.019 | 0.196 ± 0.014 | 0.346 ± 0.016 | |
| PCR | 0.176 ± 0.019 | 0.171 ± 0.018 | 0.168 ± 0.019 | 0.168 ± 0.018 | 0.198 ± 0.017 | |
| PLS | 0.178 ± 0.019 | 0.170 ± 0.018 | 0.168 ± 0.020 | 0.165 ± 0.019 | 0.195 ± 0.016 | |
| RF | 0.180 ± 0.016 | 0.182 ± 0.019 | 0.182 ± 0.016 | 0.174 ± 0.013 | 0.184 ± 0.017 | |
| BoTR | 0.189 ± 0.015 | 0.190 ± 0.020 | 0.186 ± 0.017 | 0.180 ± 0.014 | 0.194 ± 0.017 | |
| KPCR rbf | 0.178 ± 0.018 | 0.172 ± 0.017 | 0.168 ± 0.018 | 0.168 ± 0.018 | 0.192 ± 0.016 | |
| KPLS polynomial | 0.283 ± 0.071 | 0.174 ± 0.016 | 0.181 ± 0.020 | 0.168 ± 0.016 | 0.194 ± 0.015 | |
| KPLS rbf | 0.186 ± 0.023 | 0.173 ± 0.017 | 0.170 ± 0.019 | 0.169 ± 0.017 | 0.194 ± 0.016 | |
| Anthocyanins | RR | 18.835 ± 1.925 | 19.090 ± 1.651 | 20.558 ± 2.016 | 18.952 ± 1.965 | 50.597 ± 3.967 |
| LASSO | 19.618 ± 1.807 | 19.249 ± 1.618 | 20.994 ± 2.296 | 19.394 ± 2.190 | 26.770 ± 2.401 | |
| PCR | 19.871 ± 1.885 | 19.170 ± 1.893 | 20.745 ± 2.097 | 18.949 ± 1.673 | 26.528 ± 2.646 | |
| PLS | 19.894 ± 1.804 | 19.759 ± 2.151 | 21.081 ± 1.960 | 19.232 ± 2.285 | 26.121 ± 2.686 | |
| RF | 21.456 ± 1.951 | 22.010 ± 1.819 | 23.262 ± 1.911 | 22.078 ± 2.265 | 23.487 ± 2.151 | |
| BoTR | 21.479 ± 1.837 | 22.010 ± 1.650 | 23.237 ± 1.873 | 22.444 ± 2.142 | 25.500 ± 2.165 | |
| KPCR rbf | 19.514 ± 2.009 | 19.568 ± 1.961 | 21.245 ± 2.013 | 19.459 ± 2.086 | 26.637 ± 2.865 | |
| KPLS polynomial | 49.425 ± 18.671 | 20.722 ± 1.820 | 22.061 ± 2.004 | 21.047 ± 2.260 | 26.048 ± 2.699 | |
| KPLS rbf | 20.047 ± 0.116 | 20.065 ± 0.468 | 21.192 ± 1.920 | 19.445 ± 2.375 | 26.036 ± 2.677 |
SG 1D = Savitzky−Golay first derivative; SG 2D = Savitzky−Golay second derivative.

Table A3.
Summary of the best results obtained in this work for each enological parameter.
Table A3.
Summary of the best results obtained in this work for each enological parameter.
| Enological Parameter | Ranking Three Best Methods (1st, 2nd and 3rd Positions) | RMSEP (Mean ± Sd Values) | Rather Important Spectral Regions for the Best Model | Relevant Peaks for the Best Model |
|---|---|---|---|---|
| Sugar content | RR | 0.998 ± 0.102 | 750–800 nm and 900–1000 nm | 770, 920, 960, and 980 nm |
| PLS | 1.055 ± 0.127 | |||
| KPLS rbf | 1.060 ± 0.128 | |||
| pH | RR | 0.168 ± 0.014 | 750–950 nm | 790, 840 and 930 nm |
| KPLS rbf | 0.172 ± 0.015 | |||
| PLS/ KPCR polynomial | 0.172 ± 0.015/ 0.175 ± 0.016 | |||
| Anthocyanin concentration | EN | 19.773 ± 2.019 | 400–520 nm and 700–900 nm | 420, 450, 490, 520, 730 and 850 nm |
| PCR | 19.864 ± 2.231 | |||
| RR | 19.961 ± 2.061 |
References
- Chen, Q.; Zhang, C.; Zhao, J.; Ouyang, Q. Recent advances in emerging imaging techniques for non-destructive detection of food quality and safety. TrAC—Trends Anal. Chem. 2013, 52, 261–274. [Google Scholar] [CrossRef]
- Maldonado, A.I.L.; Rodriguez-Fuentes, H.; Contreras, J.A.V. Hyperspectral Imaging in Agriculture, Food and Environment; IntechOpen: London, UK, 2018; ISBN 9781789232905. [Google Scholar]
- Rady, A.M.; Guyer, D.E.; Kirk, W.; Donis-González, I.R. The potential use of visible/near infrared spectroscopy and hyperspectral imaging to predict processing-related constituents of potatoes. J. Food Eng. 2014, 135, 11–25. [Google Scholar] [CrossRef]
- Sun, D.W. Hyperspectral Imaging for Food Quality Analysis and Control; Elsevier: Amsterdam, The Netherlands, 2010; ISBN 9780123747532. [Google Scholar]
- Huang, H.; Liu, L.; Ngadi, M.O. Recent developments in hyperspectral imaging for assessment of food quality and safety. Sensors 2014, 14, 7248–7276. [Google Scholar] [CrossRef]
- Fernandes, A.; Gomes, V.; Melo-Pinto, P. A Review of the Application to Emergent Subfields in Viticulture of Local Reflectance and Interactance Spectroscopy Combined with Soft Computing and Multivariate Analysis BT. In Soft Computing for Sustainability Science; Cruz Corona, C., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 87–115. ISBN 978-3-319-62359-7. [Google Scholar]
- Gomes, V.M.; Fernandes, A.M.; Faia, A.; Melo-Pinto, P. Comparison of different approaches for the prediction of sugar content in new vintages of whole Port wine grape berries using hyperspectral imaging. Comput. Electron. Agric. 2017, 140, 244–254. [Google Scholar] [CrossRef]
- Nogales-Bueno, J.; Hernández-Hierro, J.M.; Rodríguez-Pulido, F.J.; Heredia, F.J. Determination of technological maturity of grapes and total phenolic compounds of grape skins in red and white cultivars during ripening by near infrared hyperspectral image: A preliminary approach. Food Chem. 2014, 152, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, F.; Ning, J.; Liu, X.; Zhang, Z.; Yang, S. Predicting the anthocyanin content of wine grapes by NIR hyperspectral imaging. Food Chem. 2015, 172, 788–793. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Costa, D.; Oliveros Mesa, N.F.; Santos Freire, M.; Pereira Ramos, R.; Teruel Mederos, B.J. Development of predictive models for quality and maturation stage attributes of wine grapes using vis-nir reflectance spectroscopy. Postharvest Biol. Technol. 2019, 150, 166–178. [Google Scholar] [CrossRef]
- Fadock, M.; Brown, R.B.; Reynolds, A.G. Visible-Near Infrared Reflectance Spectroscopy for Nondestructive Analysis of Red Wine Grapes. Am. J. Enol. Vitic. 2016, 67, 38–46. [Google Scholar] [CrossRef]
- Silva, R.; Gomes, V.; Mendes-Faia, A.; Melo-Pinto, P. Using support vector regression and hyperspectral imaging for the prediction of oenological parameters on different vintages and varieties ofwine grape berries. Remote Sens. 2018, 10, 312. [Google Scholar] [CrossRef]
- Fernandes, A.M.; Franco, C.; Mendes-Ferreira, A.; Mendes-Faia, A.; da Costa, P.L.; Melo-Pinto, P. Brix, pH and anthocyanin content determination in whole Port wine grape berries by hyperspectral imaging and neural networks. Comput. Electron. Agric. 2015, 115, 88–96. [Google Scholar] [CrossRef]
- Gomes, V.; Fernandes, A.; Martins-Lopes, P.; Pereira, L.; Mendes Faia, A.; Melo-Pinto, P. Characterization of neural network generalization in the determination of pH and anthocyanin content of wine grape in new vintages and varieties. Food Chem. 2017, 218, 40–46. [Google Scholar] [CrossRef]
- Arana, I.; Jarén, C.; Arazuri, S. Maturity, variety and origin determination in white grapes (Vitis vinifera L.) using near infrared reflectance technology. J. Near Infrared Spectrosc. 2005, 13, 349–357. [Google Scholar] [CrossRef]
- González-Caballero, V.; Pérez-Marín, D.; López, M.-I.; Sánchez, M.-T. Optimization of NIR Spectral Data Management for Quality Control of Grape Bunches during On-Vine Ripening. Sensors 2011, 11, 6109–6124. [Google Scholar] [CrossRef]
- Janik, L.J.; Cozzolino, D.; Dambergs, R.; Cynkar, W.; Gishen, M. The prediction of total anthocyanin concentration in red-grape homogenates using visible-near-infrared spectroscopy and artificial neural networks. Anal. Chim. Acta 2007, 594, 107–118. [Google Scholar] [CrossRef]
- Le Moigne, M.; Dufour, E.; Bertrand, D.; Maury, C.; Seraphin, D.; Jourjon, F. Front face fluorescence spectroscopy and visible spectroscopy coupled with chemometrics have the potential to characterise ripening of Cabernet Franc grapes. Anal. Chim. Acta 2008, 621, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Organisation International de la Vigne e du Vin. Recueil des Méthodes Internationales D’analyse des vins et des Mouts. OIV 2009. Available online: https://www.franceagrimer.fr/content/download/29260/259660/file/recueil_methodes_d_analyse_2009_vol1_fr.pdf (accessed on 21 September 2021).
- Riberéau-Gayon, P.; Stonestreet, E. La dosage des anthocyanes dans les vins rouge. Bull. Société Chim. 1965, 9, 2649. [Google Scholar]
- Purwanto, Y.A.; Sari, H.P.; Budiastra, I.W. Effects of preprocessing techniques in developing a calibration model for soluble solid and acidity in “Gedong Gincu” mango using NIR spectroscopy. Int. J. Eng. Technol. 2015, 7, 1921–1927. [Google Scholar]
- Sarkar, S.; Basak, J.K.; Moon, B.E.; Kim, H.T. A comparative study of PLSR and SVM-R with various preprocessing techniques for the quantitative determination of soluble solids content of hardy kiwi fruit by a portable Vis/NIR spectrometer. Foods 2020, 9, 1078. [Google Scholar] [CrossRef] [PubMed]
- Pahlawan, M.F.R.; Wati, R.K.; Masithoh, R.E. Development of a low-cost modular VIS/NIR spectroscopy for predicting soluble solid content of banana. In Proceedings of the IOP Conference Series: Earth and Environmental Science; IOP Publishing Ltd.: Bristol, UK, 2021; Volume 644, p. 012047. [Google Scholar]
- Zhang, Y.; Guo, W. Moisture content detection of maize seed based on visible/near-infrared and near-infrared hyperspectral imaging technology. Int. J. Food Sci. Technol. 2020, 55, 631–640. [Google Scholar] [CrossRef]
- Yu, H.; Guo, L.; Kharbach, M.; Han, W. Multi-way analysis coupled with near-infrared spectroscopy in food industry: Models and applications. Foods 2021, 10, 802. [Google Scholar] [CrossRef]
- Sun, D.W. Infrared Spectroscopy for Food Quality Analysis and Control; Academic Press, Elsevier: San Diego, CA, USA, 2009; ISBN 9780123741363. [Google Scholar]
- Rinnan, Å.; van den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC—Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Tech. Instrum. 2015, 2, 8. [Google Scholar] [CrossRef]
- Zeaiter, M.; Rutledge, D. Preprocessing Methods. In Comprehensive Chemometrics: Chemical and Biochemical Data Analysis; Brown, S.D., Tauler, R., Walczak, B., Eds.; Elsevier: Amsterdam, The Netherlands, 2009; pp. 121–231. ISBN 978-0-444-52701-1. [Google Scholar]
- Sun, T.; Xu, W.; Wang, X.; Liu, M. Improvement of Soluble Solids Content Prediction in Navel Oranges by Vis/NIR Semi-Transmission Spectra and UVE-GA-LSSVM. In Knowledge Engineering and Management; Springer: Berlin/Heidelberg, Germany, 2014; pp. 363–372. [Google Scholar]
- Jiao, Y.; Li, Z.; Chen, X.; Fei, S. Preprocessing methods for near-infrared spectrum calibration. J. Chemom. 2020, 34, e3306. [Google Scholar] [CrossRef]
- Bi, Y.; Yuan, K.; Xiao, W.; Wu, J.; Shi, C.; Xia, J.; Chu, G.; Zhang, G.; Zhou, G. A local pre-processing method for near-infrared spectra, combined with spectral segmentation and standard normal variate transformation. Anal. Chim. Acta 2016, 909, 30–40. [Google Scholar] [CrossRef]
- Sadeghi, M.; Behnia, F.; Amiri, R. Window Selection of the Savitzky-Golay Filters for Signal Recovery from Noisy Measurements. IEEE Trans. Instrum. Meas. 2020, 69, 5418–5427. [Google Scholar] [CrossRef]
- Rendall, R.; Reis, M.S. Which regression method to use? Making informed decisions in “data-rich/knowledge poor” scenarios—The Predictive Analytics Comparison framework (PAC). Chemom. Intell. Lab. Syst. 2018, 181, 52–63. [Google Scholar] [CrossRef]
- Cao, D.S.; Liang, Y.Z.; Xu, Q.S.; Hu, Q.N.; Zhang, L.X.; Fu, G.H. Exploring nonlinear relationships in chemical data using kernel-based methods. Chemom. Intell. Lab. Syst. 2011, 107, 106–115. [Google Scholar] [CrossRef]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Müller, K.R.; Mika, S.; Rätsch, G.; Tsuda, K.; Schölkopf, B. An introduction to kernel-based learning algorithms. IEEE Trans. Neural Netw. 2001, 12, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.C. Implementation of near-infrared technology. In Near-Infrared Technology in the Agricultural and Food Industries; American Association of Cereal Chemistry: St. Paul, MN, USA, 2001. [Google Scholar]
- Piazzolla, F.; Amodio, M.L.; Colelli, G. Spectra evolution over on-vine holding of italia table grapes: Prediction of maturity and discrimination for harvest times using a Vis-NIR hyperspectral device. J. Agric. Eng. 2017, 48, 109–116. [Google Scholar] [CrossRef][Green Version]
- Cao, F.; Wu, D.; He, Y. Soluble solids content and pH prediction and varieties discrimination of grapes based on visible–near infrared spectroscopy. Comput. Electron. Agric. 2010, 71, S15–S18. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).