Ecological Response in the Integrated Process of Biostimulation and Bioaugmentation of Diesel-Contaminated Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Sampling
2.2. Soil Treatment and Experimental Design
2.3. Determination of Total Diesel
2.4. Determination of Soil Enzyme Activity
2.5. Analysis of Degradation Products
2.6. Extraction of Total Soil DNA
2.7. Real-Time Q-PCR (Quantitative Polymerase Chain Reaction) Analysis
2.8. Microbial Ecology Analysis
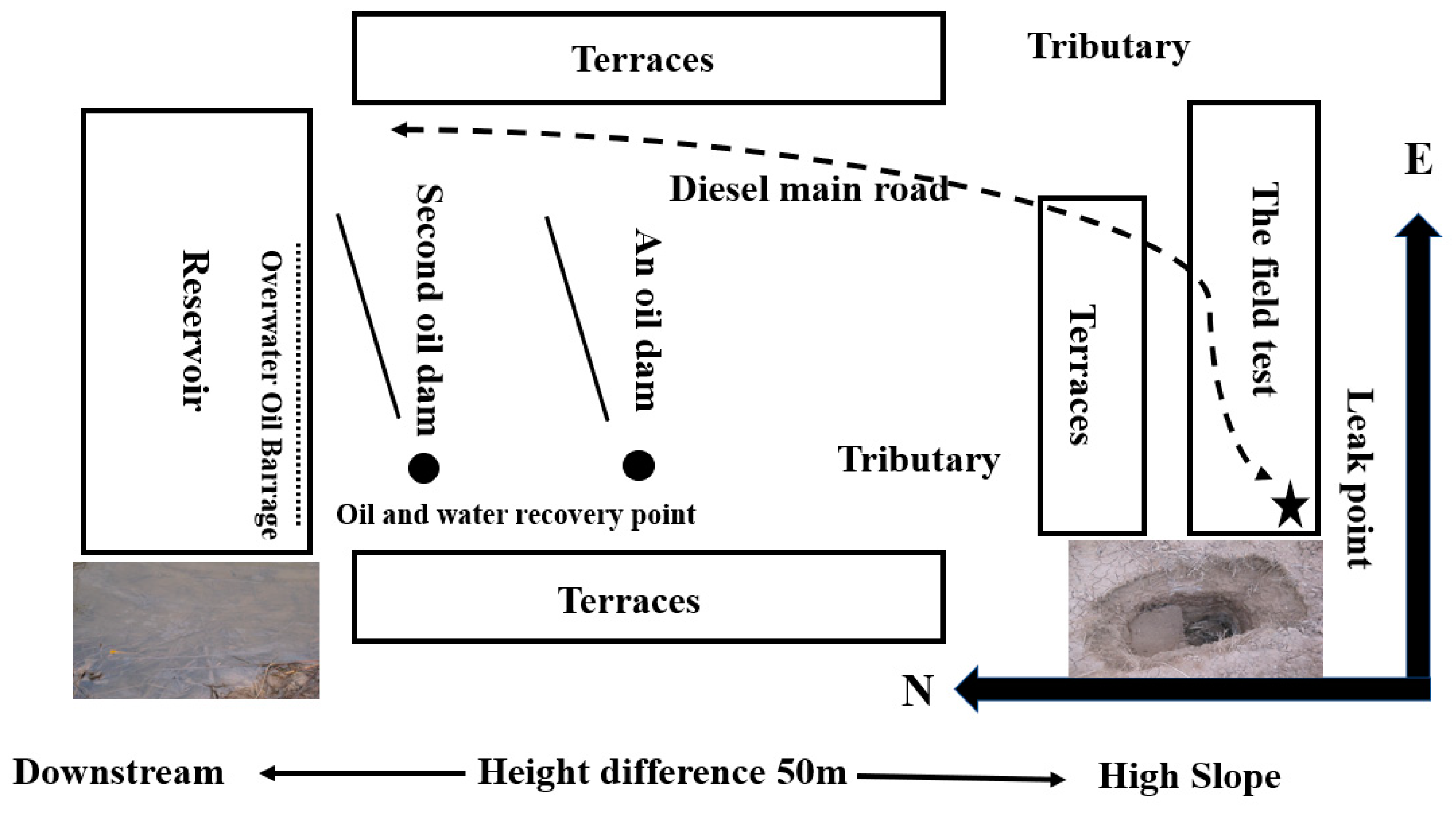
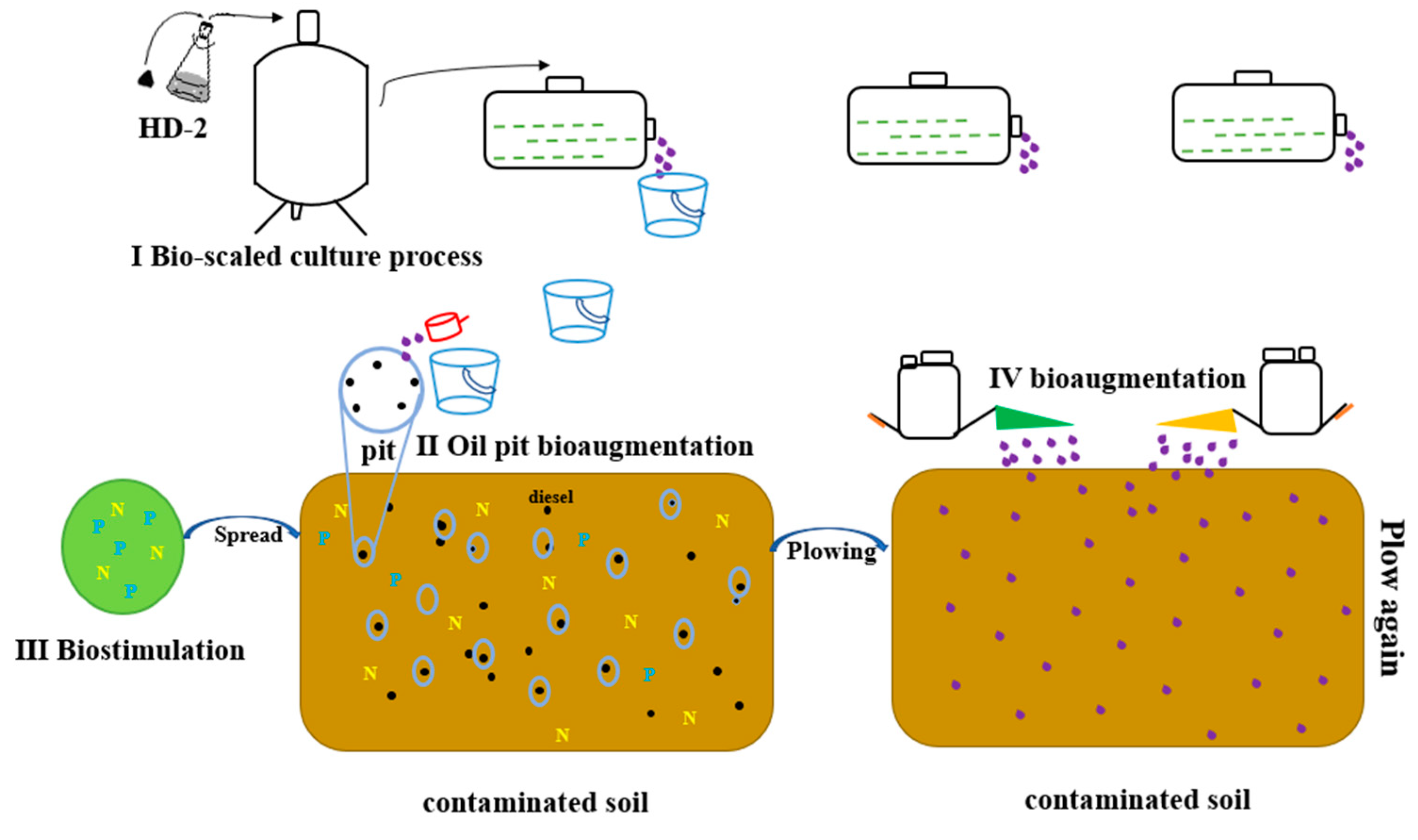
2.9. On-Site Implementation of the Bioremediation Approach
2.10. Statistical Analysis
3. Results and Discussions
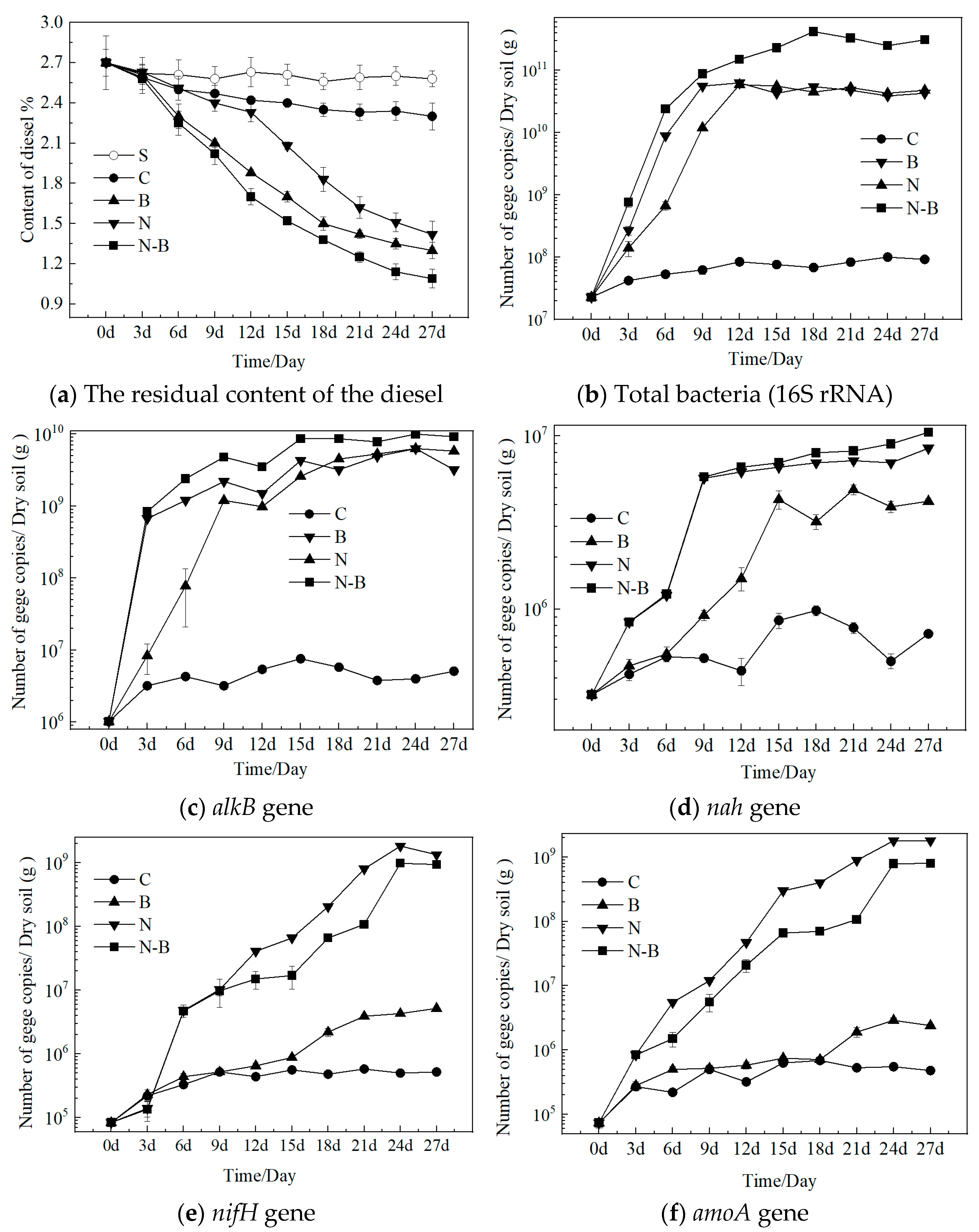
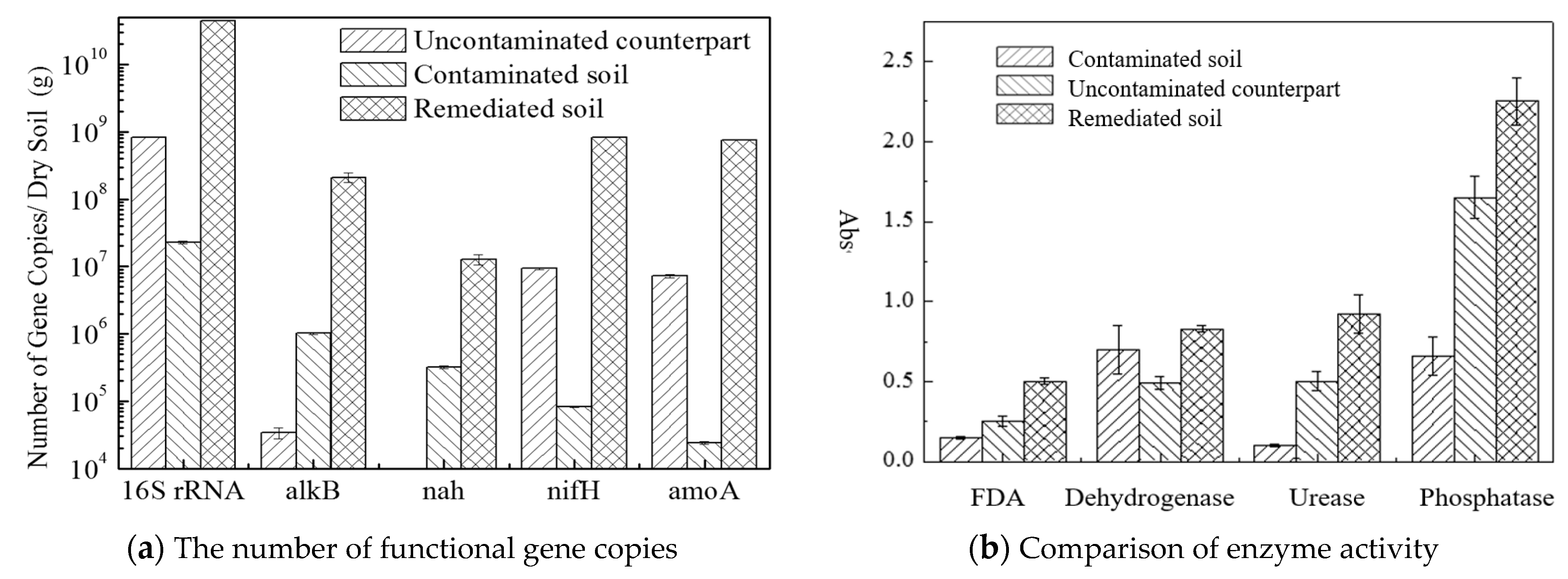
3.1. The Microbial and Gene Kinetics of Different Degradation Approaches
3.2. Soil Enzyme Activities
3.3. Analysis of Diesel Oil Components
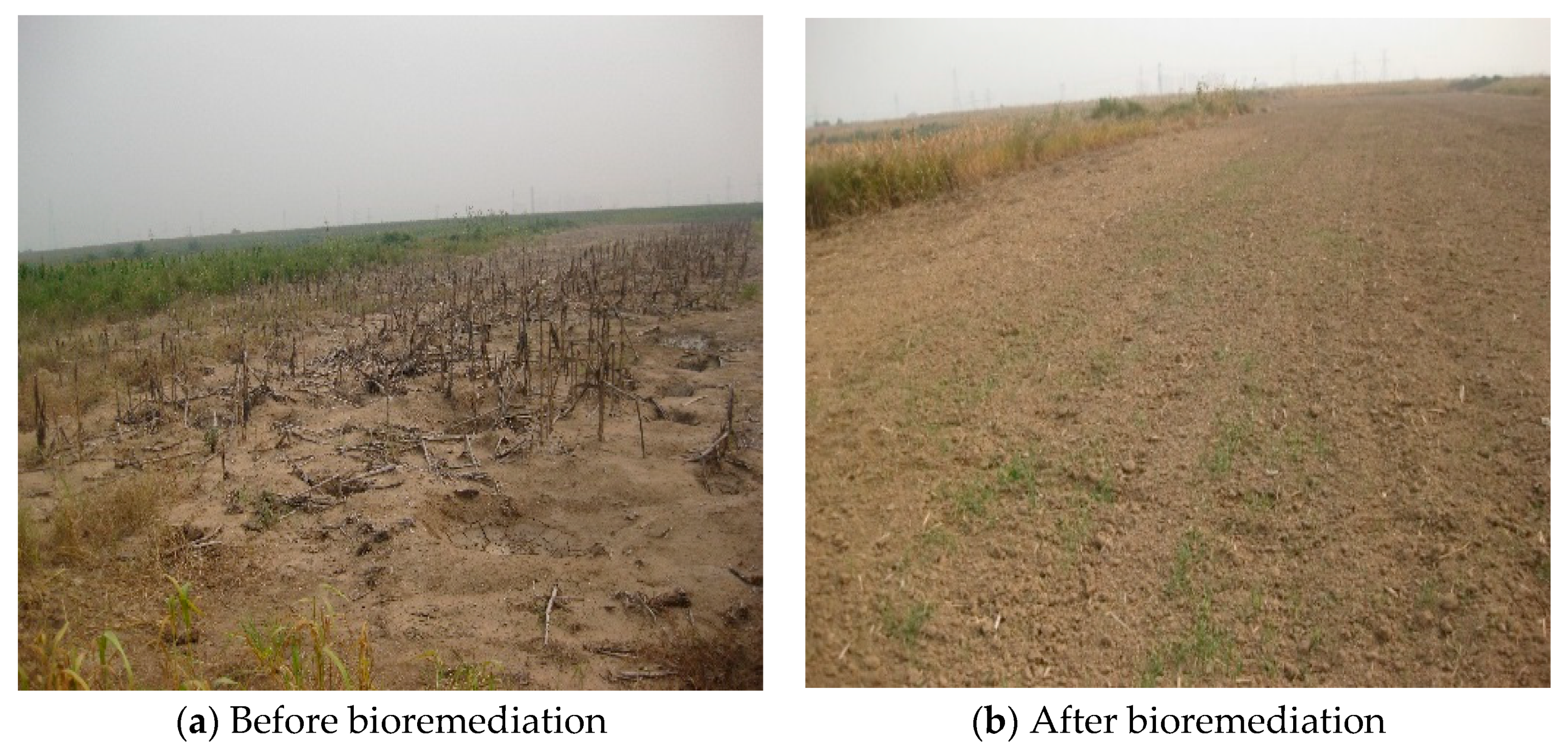
3.4. The On-Site Test of the Biosimulation-Bioaugmentation Joint Remediation
3.5. Profiling of Functional Genes and Enzyme Activity during Biodegradation
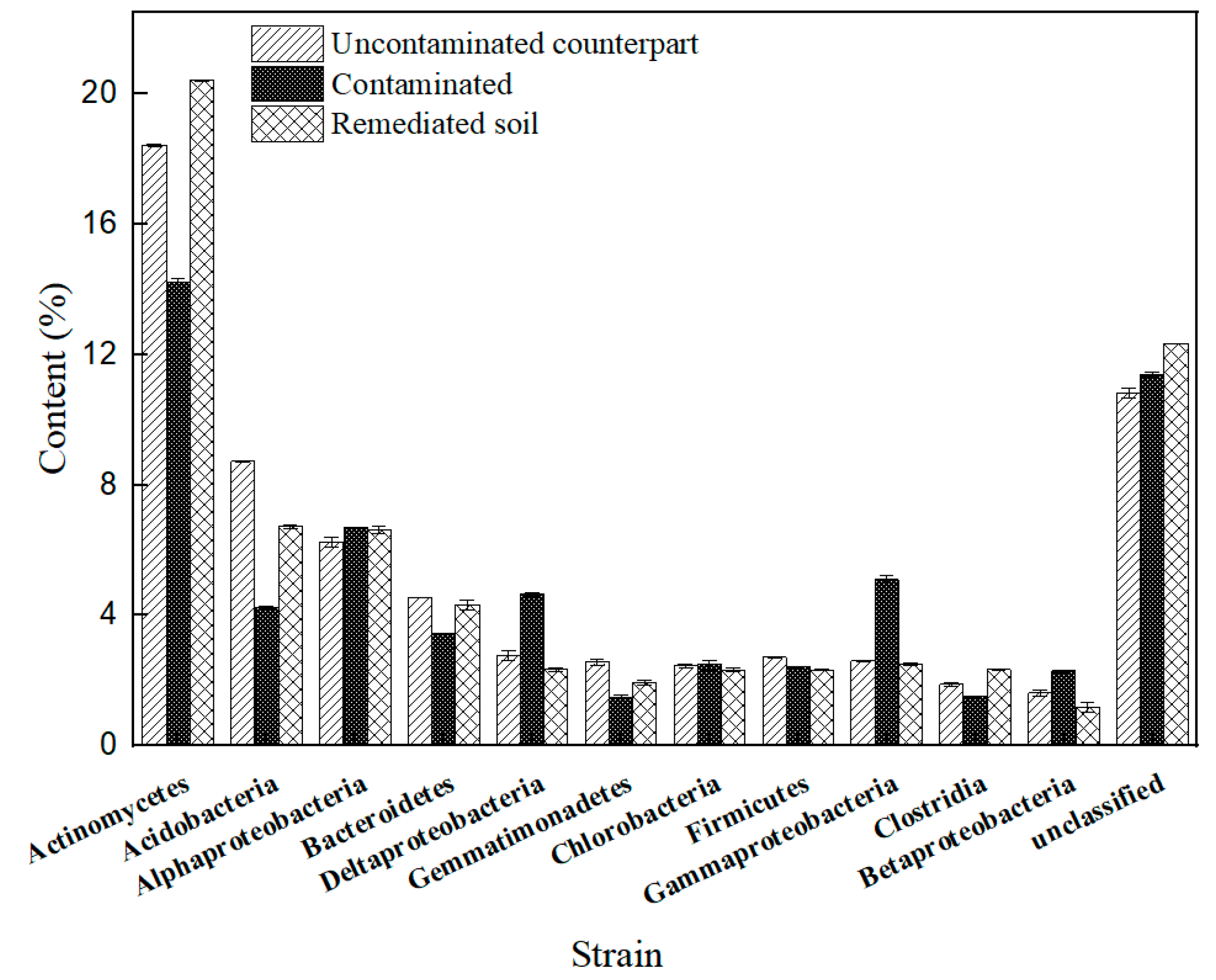
3.6. Profiling of Microbial Community during the Biostimulation-Bioaugmentation Joint Remediation Process
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Abbreviation | |
| OD | Optical Density |
| qPCR | Quantitative Polymerase Chain Reaction |
| GC-MS | Gas chromatography-Mass spectrometry |
| TTC | 2,3,5-triphenyltetrazolium chloride |
| TPF | Triphenylformanzan |
| nifH | nitrogen fixation |
| amoA | Ammoxidation |
| alkB | alkane degradation |
| nah | naphthalene degradation |
| UB | universal bacteria |
| NFB | nitrogen-fixing bacteria |
| AOB | ammonia-oxidizing bacteria |
| ADB | alkane degradation bacteria |
| NDB | naphthalene degradation bacteria |
| PAHs | Polycyclic Aromatic Hydrocarbons |
| TPH | Total Petroleum Hydrocarbon |
| FDA hydrolase | Fluorescein Diacetate Hydrolase |
Appendix B. Physicochemical Properties of Soil Samples
| Samples | Results | |||||||
| Organic Matter | Ammonium Nitrogen | Available Phosphorus | Soil pH | EC | Soil Texture (%) | |||
| g/kg | mg/kg | mg/kg | mS/m | Sand | Silt | Clay | ||
| Uncontaminated counterpart | 20.8 | 7.23 | 144 | 8.19 | 10.1 | 2.44 | 13.68 | 24 |
| Diesel contaminated soil | 29.7 | 7.59 | 114 | 8.2 | 8.83 | 3.98 | 12.14 | 26 |
| Remediated soil | 17.8 | 10.3 | 150 | 7.99 | 24.7 | 2.82 | 13.24 | 25 |
| Referenced value | >15 | >20 | >100 | 7.5–8.5 | <50 | |||
Appendix C. The Isolation of the Strain from the Contaminated Soil
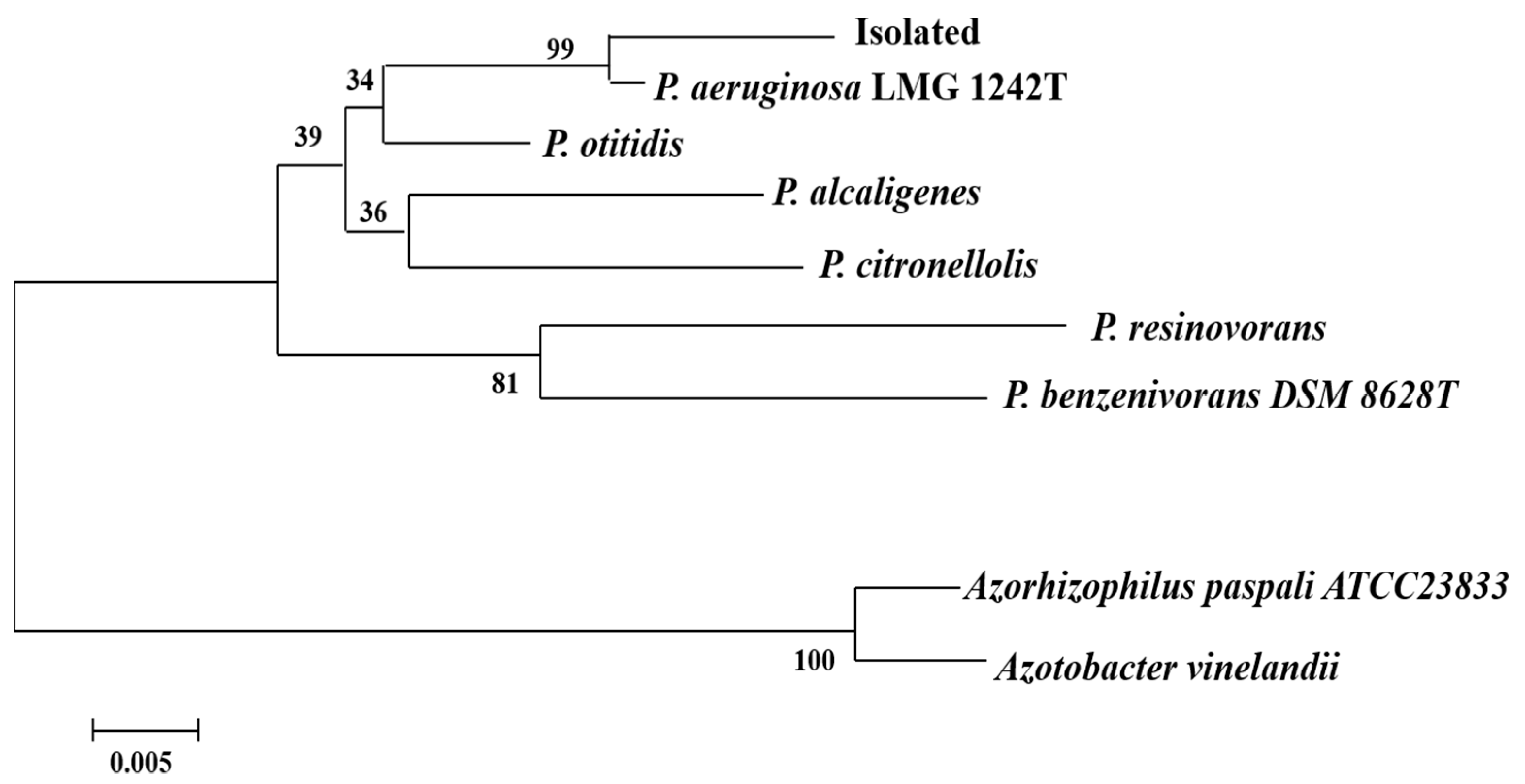
Appendix D. The Primer Sequences and Normal PCR Procedure
| Target Gene | Primer (5′-3′) | PCR Program |
| 16S rRNA Universal | AAACTCAAAKGAATTGACGG | 10 min at 95 °C; followed by 40 cycles of 15 s at 95 °C, 30 s at 50 °C, 50 s at 72 °C |
| CTCACRRCACGAGCTGCTGAC | ||
| alkB Partial alkane hydroxylase gene | AACTACMTCGARCAYTACGG | 5 min at 94 °C; followed by 40 cycles of 15 s at 95 °C, 30 s at 50 °C, 45 s at 72 °C |
| TGAMGATGTGGTYRCTGTTCC | ||
| nah Partial naphthalene dioxygenase gene | CAAAARCACCTGATTYATGG | 10 min at 95 °C; followed by 40 cycles of 1 min at 95 °C, 1 min at 49 °C, 2 min at 72 °C |
| AYRCGRGSGACTTCTTTCAA | ||
| nifH Partial nitrogen fixation gene | AAAGGYGGWATCGGYAARTCCACCAC | 5 min at 95 °C; followed by 40 cycles of 1 min at 94 °C, 1 min at 55 °C, 2 min at 72 °C |
| TTGTTSGCSGCRTACATSGCCATCAT | ||
| amoA Partial nitrification gene | GGGGTTTCTACTGGTGGT | 5 min at 94 °C; followed by 40 cycles of 1 min at 94 °C; 90 s at 60 °C, 90 s at 72 °C |
Appendix E. Standard Curves for Quantitative PCR (qPCR)
Appendix F. Total Diesel Analysis
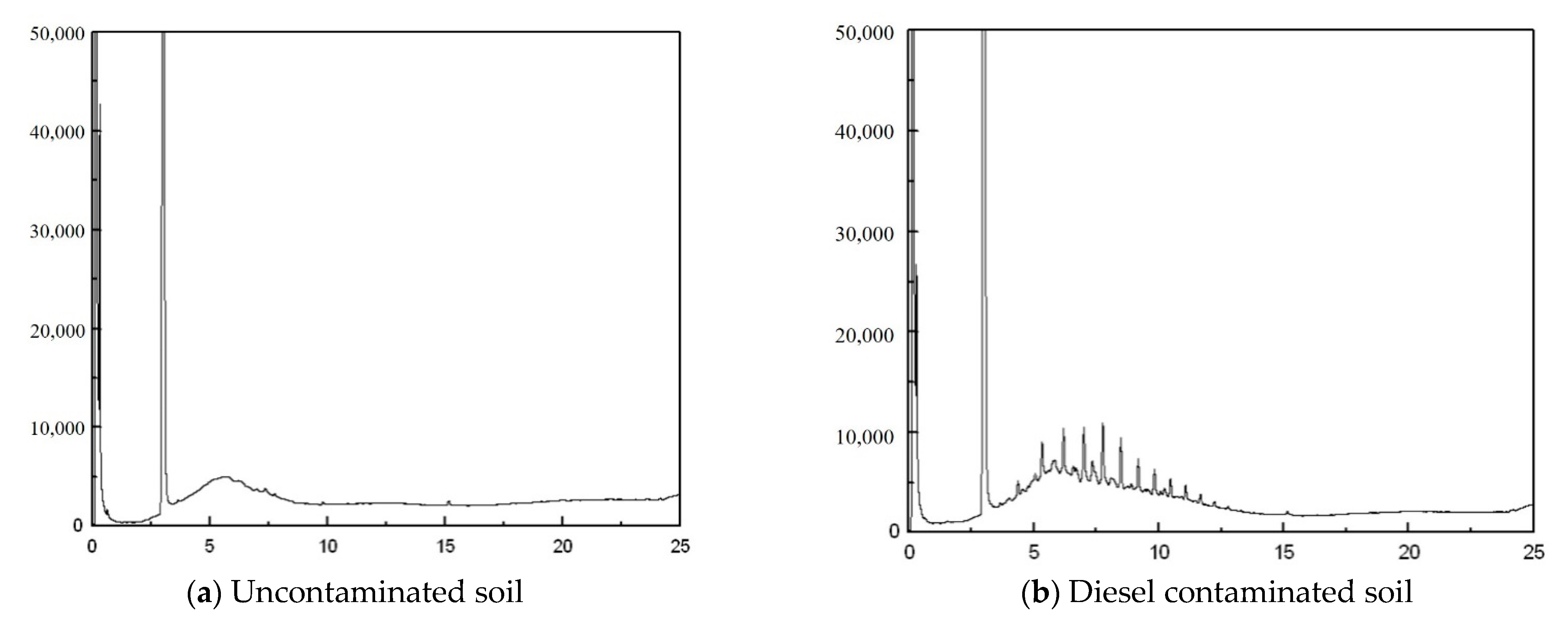
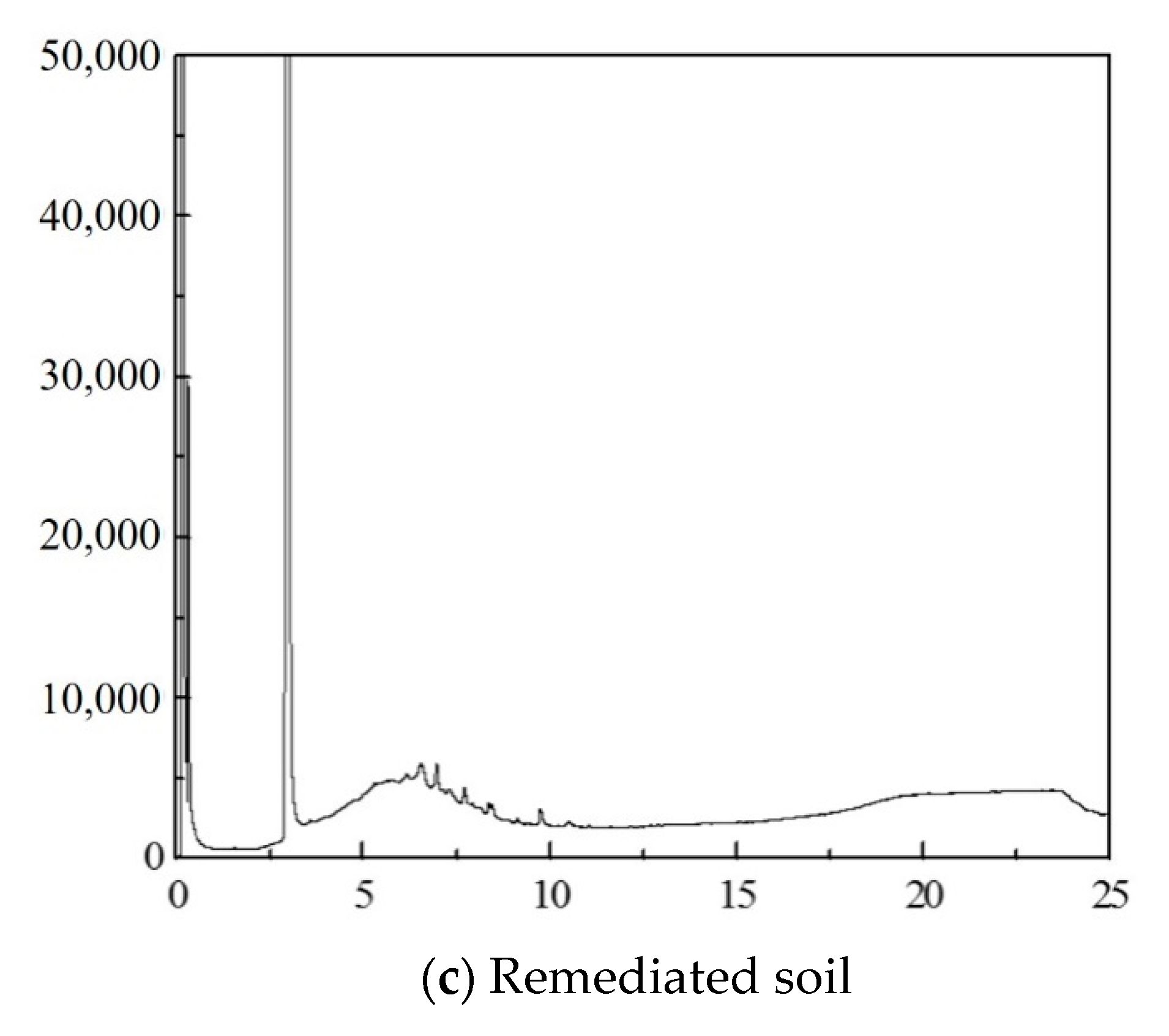
Appendix G
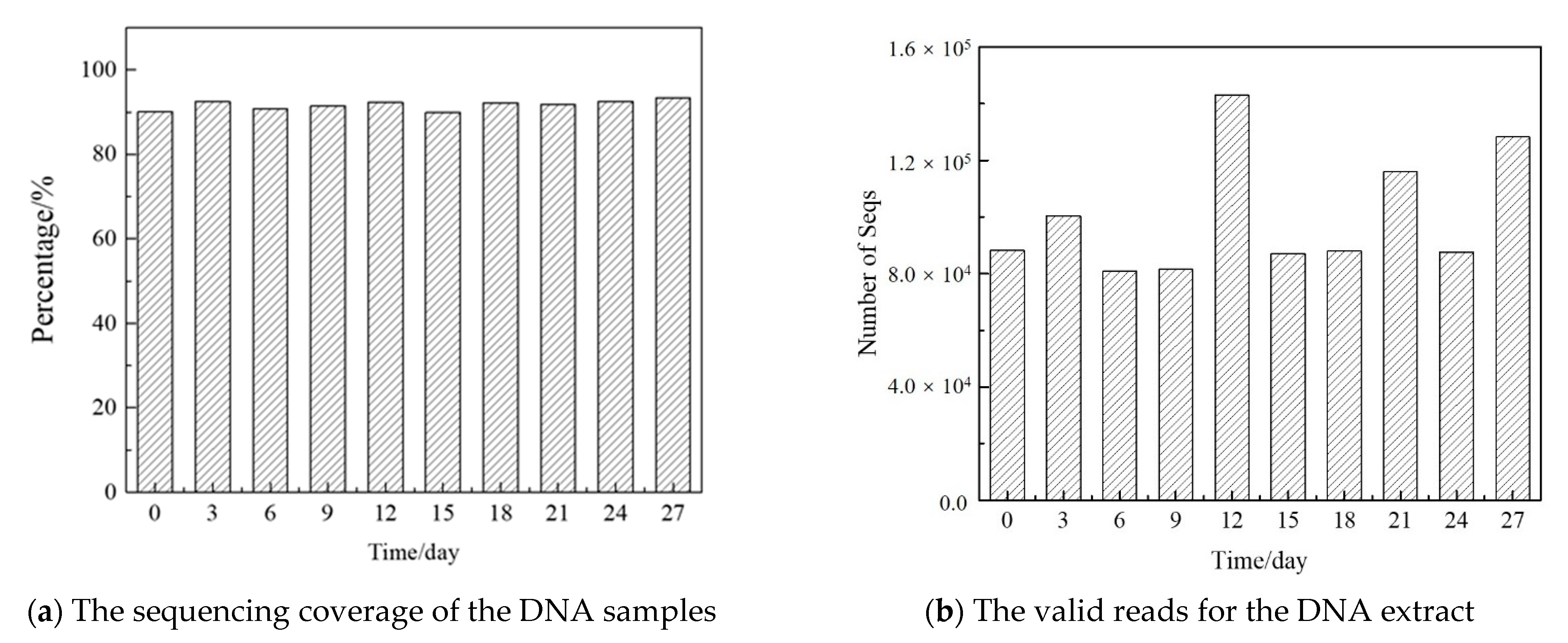
| Start | End | NBases | Ambigs | Polymer | NumSeqs | |
| Minimum: | 1 | 387 | 150 | 0 | 3 | 1 |
| 2.5%-tile: | 1 | 391 | 168 | 0 | 2 | 835,020 |
| 25%-tile: | 1 | 391 | 170 | 0 | 2 | 835,020 |
| Median: | 1 | 391 | 188 | 0 | 4 | 670,039 |
| 75%-tile: | 1 | 391 | 193 | 0 | 5 | 1,005,058 |
| 97.5%-tile: | 1 | 391 | 194 | 0 | 5 | 1,306,575 |
| Maximum: | 2 | 391 | 216 | 0 | 6 | 1,431,033 |
| Mean: | 1.00028 | 391 | 182.774 | 0 | 3.8571 | 775,285 |
| unique | seqs: | 174,834 | ||||
| total | seqs: | 1,431,033 |
References
- Guan, Z.; Tang, X.Y.; Nishimura, T.; Katou, H.; Liu, H.Y.; Qing, J. Surfactant-enhanced flushing enhances colloid transport and alters macroporosity in diesel-contaminated soil. J. Environ. Sci. 2018, 64, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Pasha, A.Y.; Hu, L.M.; Meegoda, J.A.; Aflaki, E.; Du, J. Centrifuge modeling of in situ surfactant enhanced flushing of diesel contaminated soil. Geotech. Test. J. 2011, 34, 1–11. [Google Scholar] [CrossRef]
- Nazri, M.K.H.M.; Sapawe, N. A Short Review on Photocatalytic Reaction in Diesel Degradation. Mater. Today Proc. 2020, 31, A33–A37. [Google Scholar] [CrossRef]
- Al-Baldawi, I.A.; Abdullah, S.R.S.; Anuar, N. Phytodegradation of total petroleum hydrocarbon (TPH) in diesel-contaminated water using Scirpus grossus. Ecol. Eng. 2015, 74, 463–473. [Google Scholar] [CrossRef]
- Fahid, M.; Arslan, M.; Shabir, G.; Younus, S.; Yasmeen, T.; Rizwan, M.; Siddique, K.; Ahmad, S.R.; Tahseen, R.; Iqbal, S.; et al. Phragmites australis in combination with hydrocarbons degrading bacteria is a suitable option for remediation of diesel-contaminated water in floating wetlands. Chemosphere 2020, 240, 124890. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.J.; Duca, D.; Dan, T.; Knopper, L.D. Petroleum hydrocarbon (PHC) uptake in plants: A literature review. Environ. Pollut. 2019, 245, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Moreira, I.T.A.; Oliveira, O.M.C.; Triguis, J.A.; Santos, A.M.P.D.; Queiroz, A.F.S.; Martins, C.M.S.; Silva, C.S.; Jesus, R.S. Phytoremediation using Rhizophora mangle L. in mangrove sediments contaminated by persistent total petroleum hydrocarbons (TPH′s). Microchem. J. 2011, 99, 376–382. [Google Scholar] [CrossRef]
- Vasenev, V.I.; Ananeva, N.D.; Ivashchenko, K.V. The effect of pollutants (heavy metals and diesel fuel) on the respiratory activity of constructozems (artificial soils). Russ. J. Ecol. 2013, 44, 475–483. [Google Scholar] [CrossRef]
- Al-Turki, A.I. Microbial Polycyclic Aromatic Hydrocarbons Degradation in Soil. Res. J. Environ. Toxicol. 2009, 3, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Borowik, A.; Wyszkowska, J.; Mirosław, M.; Kucharski, J. Implications of Soil Pollution with Diesel Oil and BP Petroleum with ACTIVE Technology for Soil Health. Int. J. Environ. Res. Public Health 2019, 16, 2474. [Google Scholar] [CrossRef] [Green Version]
- Gimžauskaitė, D.; Tamošiūnas, A.; Tučkutė, S.; Snapkauskienė, V.; Aikas, M.; Uscila, R. Treatment of diesel-contaminated soil using thermal water vapor arc plasma. Environ. Sci. Pollut. Res. 2020, 27, 43–54. [Google Scholar] [CrossRef]
- Labianca, C.; Gisi, S.D.; Picardi, F.; Todaro, F.; Notarnicola, M. Remediation of a Petroleum Hydrocarbon-Contaminated Site by Soil Vapor Extraction: A Full-Scale Case Study. Appl. Sci. 2020, 10, 4261. [Google Scholar] [CrossRef]
- Bortone, I.; Labianca, C.; Todaro, F.; De Gisi, S.; Coulon, F.; Notarnicola, M. Experimental investigations and numerical modeling of in-situ reactive caps for PAH contaminated marine sediments—ScienceDirect. J. Hazard. Mater. 2020, 387, 121724. [Google Scholar] [CrossRef]
- Li, X.; Wu, B.; Zhang, Q.; Liu, Y.; Gu, Q. Effects of soil properties on the remediation of diesel-contaminated soil by Triton X-100-aided washing. Environ. Sci. Pollut. Res. 2020, 27, 23323–23330. [Google Scholar] [CrossRef]
- Asadollahfardi, G.; Rezaee, M. Electrokinetic remediation of diesel-contaminated silty sand under continuous and periodic voltage application. Environ. Eng. Res. 2018, 24, 456–462. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Song, X.; Ding, D. Sustainable remediation of diesel-contaminated soil by low-temperature thermal treatment: Improved energy efficiency and soil reusability. Chemosphere 2019, 241, 124952. [Google Scholar] [CrossRef]
- Uyizeye, O.C.; Thiet, R.K.; Knorr, M.A. Effects of community-accessible biochar and compost on diesel-contaminated soil. Bioremediation J. 2019, 23, 107–117. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Seo, Y.; Ha, M.; Lee, J.; Yang, H.; Cho, K.S. Evaluation of rhizoremediation and methane emission in diesel-contaminated soil cultivated with tall fescue (Festuca arundinacea). Environ. Res. 2021, 194, 110606. [Google Scholar] [CrossRef]
- Lofrano, G.; Libralato, G.; Minetto, D.; De Gisi, S.; Todaro, F.; Conte, B.; Calabro, D.; Quatraro, L.; Notarnicola, M. In situ remediation of contaminated marine sediments: An overview. Environ. Sci. Pollut. Res. 2017, 24, 5189–5206. [Google Scholar] [CrossRef] [Green Version]
- Eman, K.; Esmaeil, S.; Arturo, A.-M.; Mohamed, T.; Nagalakshmi, H.; Tanvih, M.; Paul, D.M.; Andrew, S.B. Bioremediation potential of diesel-contaminated Libyan soil. Ecotoxicol. Environ. Saf. 2016, 133, 297–305. [Google Scholar] [CrossRef]
- Safdari, M.S.; Kariminia, H.R.; Ghobadi Nejad, Z.; Fletcher, T.H. Study Potential of Indigenous Pseudomonas aeruginosa and Bacillus subtilis in Bioremediation of Diesel-Contaminated Water. Water Air Soil Pollut. 2017, 228, 37. [Google Scholar] [CrossRef]
- Yasushi, M.; Atsushi, S.; Yuko, M.; Atsushi, F.; Kosuke, S. Enhancing bioremediation of oil-contaminated soils by controlling nutrient dispersion using dual characteristics of soil pore structure. Ecol. Eng. 2013, 51, 237–243. [Google Scholar] [CrossRef]
- Łebkowska, M.; Zborowska, E.; Karwowska, E.; Miaśkiewicz-Pęska, E.; Muszyński, A.; Tabernacka, A.; Naumczyk, J.; Jęczalik, M. Bioremediation of soil polluted with fuels by sequential multiple injection of native microorganisms: Field-scale processes in Poland. Ecol. Eng. 2011, 37, 1895–1900. [Google Scholar] [CrossRef]
- Lei, H.; Xu, G.; Ming, L.; Gang, D.; Guo, J.S.; Theoneste, N. Correlation among soil microorganisms, soil enzyme activities, and removal rates of pollutants in three constructed wetlands purifying micro-polluted river water. Ecol. Eng. 2012, 46, 98–106. [Google Scholar] [CrossRef]
- Ortega, G.; Katherine, O.G.; Zaragoza, D. Degradation of benzene, toluene, and xylene isomers by a bacterial consortium obtained from rhizosphere soil of Cyperus sp grown in a petroleum-contaminated area. Folia Microbiol. 2013, 58, 569–577. [Google Scholar] [CrossRef]
- Coenye, T. Classification of Ralstonia pickettii-like isolates from the environment and clinical samples as Ralstonia insidiosa sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Funke, G.; Ramos, C.P.; Collins, M.D. Identification of some clinical strains of CDC coryneform group A-3 and A-4 bacteria as Cellulomonas species and proposal of Cellulomonas hominis sp. nov. for some group A-3 strains. J. Clin. Microbiol. 1995, 33, 2091–2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hanada, S.; Shigematsu, T.; Shibuya, K.; Kamagata, Y.; Kanagawa, T.; Kurane, R. Burkholderia kururiensis sp. nov. a trichloroethylene (TCE)-degrading bacterium isolated from an aquifer polluted with TCE. Int. J. Syst. Evol. Microbiol. 2000, 50, 743–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimont, F.; Grimont, P.A.D. Serratia marcescens. Bizio 1823, 288. [Google Scholar] [CrossRef]
- Reena, D.; Tiwary, B.N. Isolation of a novel strain of Planomicrobium chinense from diesel contaminated soil of tropical environment. J. Basic Microbiol. 2013, 53, 723–732. [Google Scholar] [CrossRef]
- Wannarak, N.; Parichat, N.; Onruthai, P. Diesel oil removal by immobilized Pseudoxanthomonas sp. RN402. Biodegradation 2013, 24, 387–397. [Google Scholar] [CrossRef]
- Daghio, M.; Vaiopoulou, E.; Patil, S.A.; Suárez-Suárez, A.; Head, I.M.; Franzetti, A.; Rabaey, K. Anodes stimulate anaerobic toluene degradation via sulfur cycling in marine sediments. Appl. Environ. Microbiol. 2015, 82, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Heider, J.; Schühle, K. Anaerobic Biodegradation of Hydrocarbons Including Methane. Prokaryotes 2013, 605–634. [Google Scholar] [CrossRef]
- Sutton, N.B.; Maphosa, F.; Morillo, J.A.; Al-Soud, W.A.; Langenhoff, A.A.M.; Grotenhuis, T.; Rijnaarts, H.H.M. Impact of long-term diesel contamination on soil microbial community structure. Appl. Environ. Microbiol. 2013, 79, 619–630. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Lan, T.; Lu, D.; Liu, Z. Ecological and enzymatic responses to petroleum contamination. Environ. Sci. Process. Impacts 2014, 16, 1501–1509. [Google Scholar] [CrossRef]
- Dong, Y.; Lang, Z.; Kong, X.; Lu, D.; Liu, Z. Kinetic and multidimensional profiling of accelerated degradation of oil sludge by biostimulation. Environ. Sci. Process. Impacts 2015, 17, 763–774. [Google Scholar] [CrossRef]
- Wu, Z.; Dong, H.; Zou, L.; Lu, D.; Liu, Z. Enriched Microbial Community in Bioaugmentation of Petroleum-Contaminated Soil in the Presence of Wheat Straw. Appl. Biochem. Biotechnol. 2011, 164, 1071–1082. [Google Scholar] [CrossRef]
- Bahrampour, T.; Moghanlo, V.S. Evaluation of soil biological activity after soil contaminating by crude oil. Int. J. Agric. Res. Rev. 2012, 2, 671–679. [Google Scholar]
- Gupta, S.; Pathak, B.; Fulekar, M.H. Molecular approaches for biodegradation of polycyclic aromatic hydrocarbon compounds: A review. Rev. Environ. Sci. Bio Technol. 2015, 14, 241–269. [Google Scholar] [CrossRef]
- Zhou, L.S.; Li, H.; Zhang, Y.; Han, S.Q.; Xu, H. Sphingomonas from petroleum-contaminated soils in Shenfu, China and their PAHs degradation abilities. Braz. J. Microbiol. 2016, 47, 271–278. [Google Scholar] [CrossRef] [Green Version]
- Pratiwi, E.; Satwika, T.D.; Agus, F. Analysis of peat bacterial diversity in oil palm plantations and a logged forest in Jambi, Indonesia, using PCR-DGGE technique. IOP Conf. Ser. Earth Environ. Sci. 2021, 648, 012200. [Google Scholar] [CrossRef]
- Wolff, D.C.; Cryder, Z.; Gan, J. Soil bacterial community dynamics following surfactant addition and bioaugmentation in pyrene-contaminated soils. Chemosphere 2019, 231, 93–102. [Google Scholar] [CrossRef]
- Sutton, N.B.; Kalisz, M.; Krupanek, J.; Marek, J.; Grotenhuis, T.; Smidt, H.; de Weert, J.; Rijnaarts, H.H.M.; van Gaans, P.; Keijzer, T. Geochemical and Microbiological Characteristics during in Situ Chemical Oxidation and in Situ Bioremediation at a Diesel Contaminated Site. Environ. Sci. Technol. 2014, 48, 2352. [Google Scholar] [CrossRef]
- Bücker, F.; de Moura, T.M.; da Cunha, M.E.; de Quadros, P.D.; Beker, S.A.; Cazarolli, J.C.; Caramao, E.B.; Frazzon, A.P.G.; Bento, F.M. Evaluation of the deteriogenic microbial community using qPCR, n-alkanes and FAMEs biodegradation in diesel, biodiesel and blends (B5, B10, and B50) during storage. Fuel 2018, 233, 911–917. [Google Scholar] [CrossRef]
- Peel, M.C.; Finlayson, B.L.; Mcmahon, T.A. Updated world map of the Köppen-Geiger climate classification. Hydrol. Earth Syst. Sci. 2007, 11, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Trindade, P.V.O.; Sobral, L.G.; Rizzo, A.C.L. Evaluation of the biostimulation and bioaugmentation techniques in the bioremediation process of petroleum hydrocarbons contaminated soil. In Proceedings of the Contribuição Técnica a 9th International Petroleum Environmental Conference, Albuquerque, NM, USA, 21–25 October 2002. [Google Scholar]
- USEPA. Hazardous Waste Test Methods, Method 3546: Microwave Extraction; USEPA: Washington, DC, USA, 2007.
- Dai, R.; Wu, J.; Xu, J.; Yu, M.; Shi, J. Improvement of determination dehydrogenase activity in wheat rhizospheric soil. Jiangsu J. Agric. Sci. 2013, 29, 772–776. [Google Scholar]
- Guo, H.; Yao, J.; Cai, M.; Qian, Y.; Guo, Y.; Richnow, H.H.; Blake, R.E.; Doin, S.; Ceccanti, B. Effects of petroleum contamination on soil microbial numbers, metabolic activity and urease activity. Chemosphere 2012, 87, 1273–1280. [Google Scholar] [CrossRef]
- Alvarez, H.M. Relationship between β-oxidation pathway and the hydrocarbon-degrading profile in actinomycetes bacteria. Int. Biodeterior. Biodegrad. 2003, 52, 35–42. [Google Scholar] [CrossRef]
- Calvo, C.; Silva, C.G.A.; Uad, I.; Manzanera, M.; Perucha, C.; Laguna, J.; Gónzalez, L.J. Biostimulation combined treatments for remediation of diesel contaminated soil. WIT Trans. Ecol. Environ. 2010, 132, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Abraham, J.; Gea, T.; Vicent, T. Impact of Biostimulation and Bioaugmentation as Bioremediation Systems on Diesel Contaminated Soil. Environ. Eng. Manag. J. 2016, 15, 1743–1753. [Google Scholar] [CrossRef]
- Bosco, F.; Casale, A.; Mazzarino, I.; Godio, A.; Ruffino, B.; Mollea, C.; Chiampo, F. Microcosm evaluation of bioaugmentation and biostimulation efficacy on diesel-contaminated soil. J. Chem. Technol. Biotechnol. 2020, 95, 904–912. [Google Scholar] [CrossRef]
- Suja, F.; Rahim, F.; Taha, M.R.; Hambali, N.; Razali, R.; Khalid, A.; Hamzah, A. Effects of local microbial bioaugmentation and biostimulation on the bioremediation of total petroleum hydrocarbons (TPH) in crude oil contaminated soil based on laboratory and field observations. Int. Biodeterior. Biodegrad. 2014, 90, 115–122. [Google Scholar] [CrossRef]
- Knowles, R.; Wishart, C. Nitrogen-fixation in arctic marine-sediments-effect of oil and hydrocarbon fractions. Environ. Pollut. 1977, 13, 133–149. [Google Scholar] [CrossRef]
- Padayachee, D.; Lin, J. The effect of fertilizer amendment on diesel biodegradation in contaminated soils. Afr. J. Microbiol. Res. 2011, 5, 1729–1739. [Google Scholar] [CrossRef]
- Greenwood, P.F.; Wibrow, S.; George, S.J.; Tibbett, M. Hydrocarbon biodegradation and soil microbial community response to repeated oil exposure. Org. Geochem. 2009, 40, 293–300. [Google Scholar] [CrossRef]
- Zhang, Q.R.; Zhou, Q.X.; Ren, L.P.; Zhu, Y.G.; Sun, S.L. Ecological effects of crude oil residues on the functional diversity of soil microorganisms in three weed rhizospheres. J. Environ. Sci. 2006, 18, 1101–1110. [Google Scholar] [CrossRef]
- Ramsay, M.A.; Swannell, R.P.J.; Shipton, W.A.; Duke, N.C.; Hill, R.T. Effect of bioremediation on the microbial community in oiled mangrove sediments. Mar. Pollut. Bull. 2000, 41, 413–419. [Google Scholar] [CrossRef]
- Harayama, S.; Kasai, Y.; Hara, A. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 2004, 15, 205–214. [Google Scholar] [CrossRef]

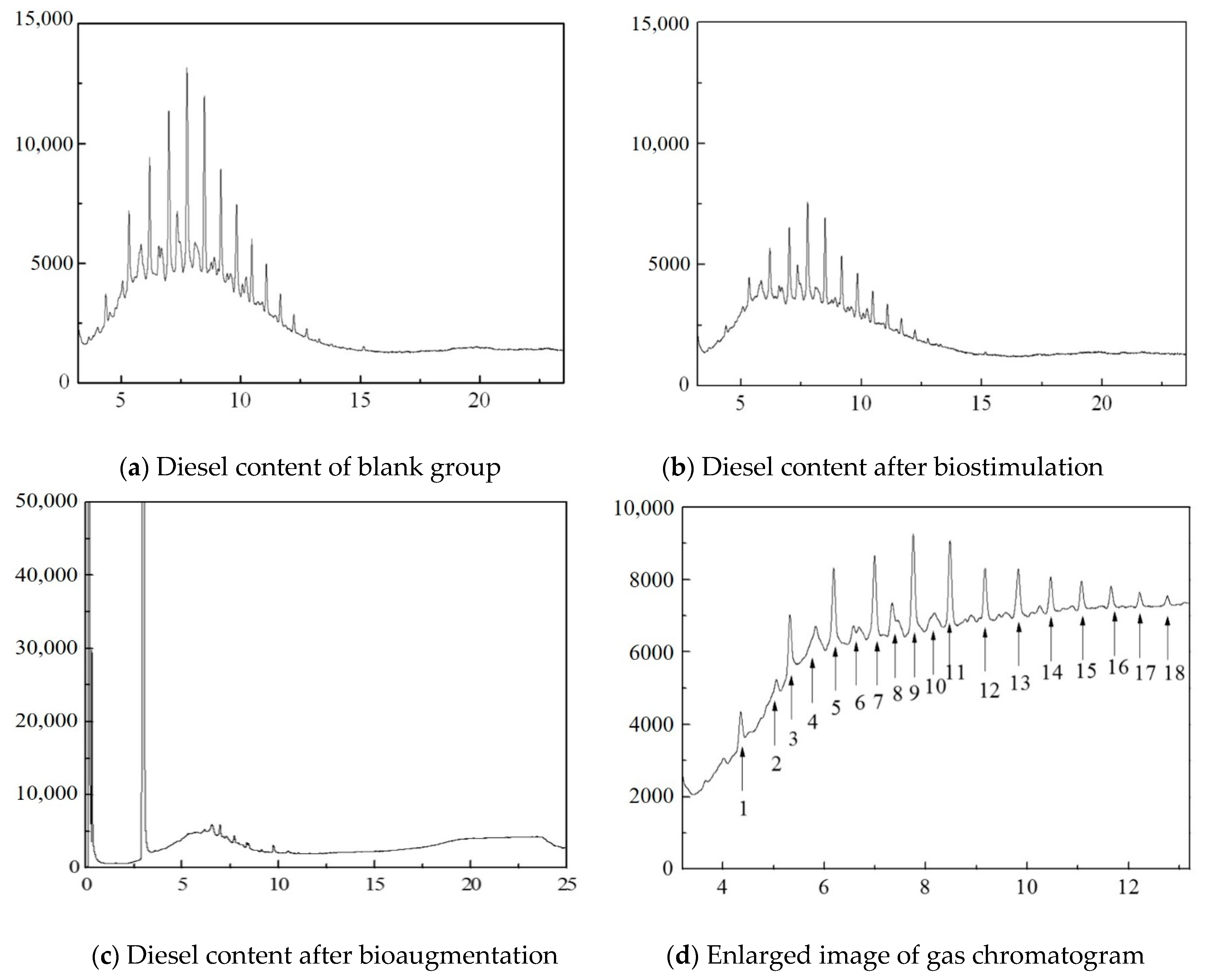
| Number | Component | Number | Component | Number | Component |
|---|---|---|---|---|---|
| 1 | C12H24 | 7 | C15H32 | 13 | C19H40 |
| 2 | C7H5N | 8 | C14H30O | 14 | C20H42 |
| 3 | C13H28 | 9 | C16H34 | 15 | C21H44 |
| 4 | C12H24O | 10 | C15H32O | 16 | C22H46 |
| 5 | C14H30 | 11 | C17H36 | 17 | C23H48 |
| 6 | C7H12O2 | 12 | C18H38 | 18 | C24H50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Chen, Y.; Du, X.; Zheng, J.; Lu, D.; Liu, Z. Ecological Response in the Integrated Process of Biostimulation and Bioaugmentation of Diesel-Contaminated Soil. Appl. Sci. 2021, 11, 6305. https://doi.org/10.3390/app11146305
Li X, Chen Y, Du X, Zheng J, Lu D, Liu Z. Ecological Response in the Integrated Process of Biostimulation and Bioaugmentation of Diesel-Contaminated Soil. Applied Sciences. 2021; 11(14):6305. https://doi.org/10.3390/app11146305
Chicago/Turabian StyleLi, Xiaosen, Yakui Chen, Xianyuan Du, Jin Zheng, Diannan Lu, and Zheng Liu. 2021. "Ecological Response in the Integrated Process of Biostimulation and Bioaugmentation of Diesel-Contaminated Soil" Applied Sciences 11, no. 14: 6305. https://doi.org/10.3390/app11146305
APA StyleLi, X., Chen, Y., Du, X., Zheng, J., Lu, D., & Liu, Z. (2021). Ecological Response in the Integrated Process of Biostimulation and Bioaugmentation of Diesel-Contaminated Soil. Applied Sciences, 11(14), 6305. https://doi.org/10.3390/app11146305






