Unveiling Biological Activities of Marine Fungi: The Effect of Sea Salt
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains
2.2. Detection of Extracellular Enzymes
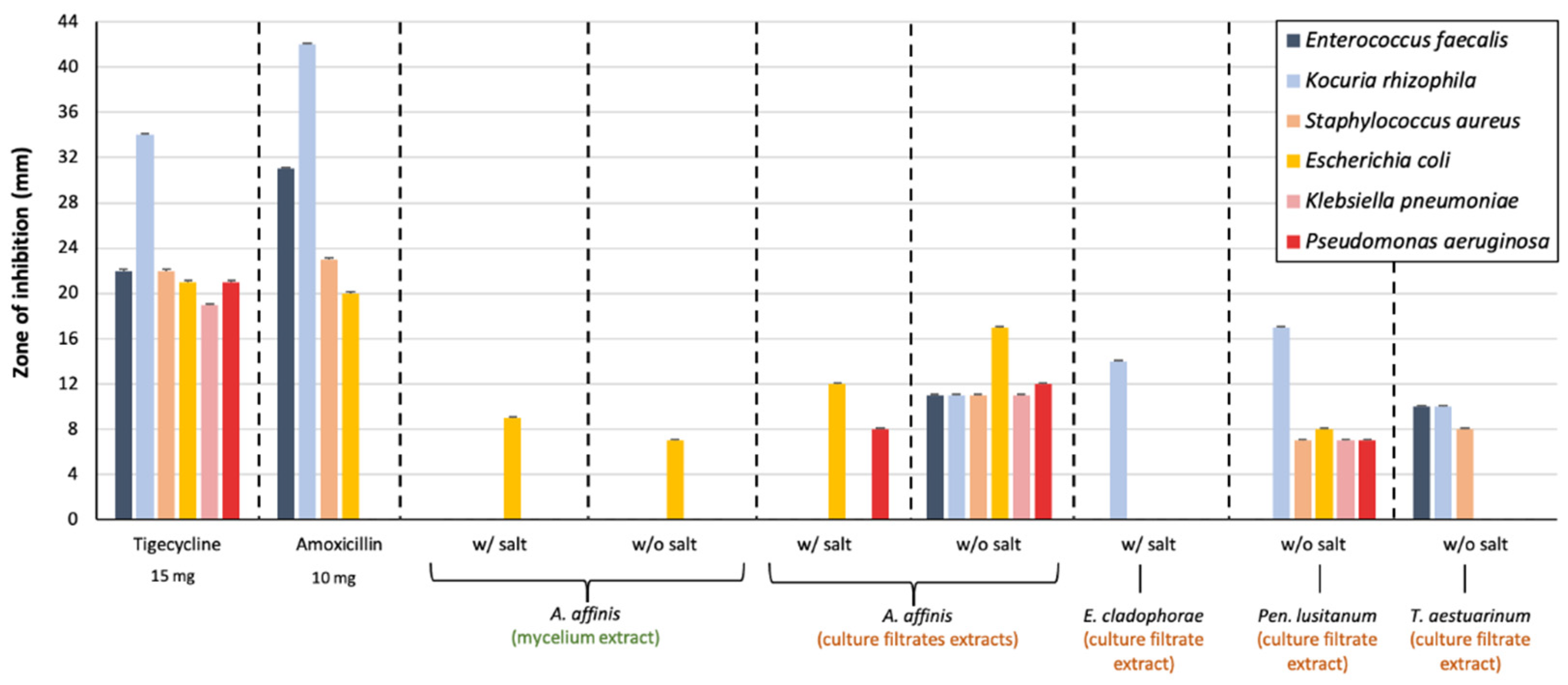
2.3. Detection of Antibacterial Activity Based on Dual Culture Test-Variant
2.4. Production of Crude Extracts and Extraction
2.5. Antibacterial Activity of Fungal Extracts
2.6. Anti-Fungal Activity of Fungal Extracts
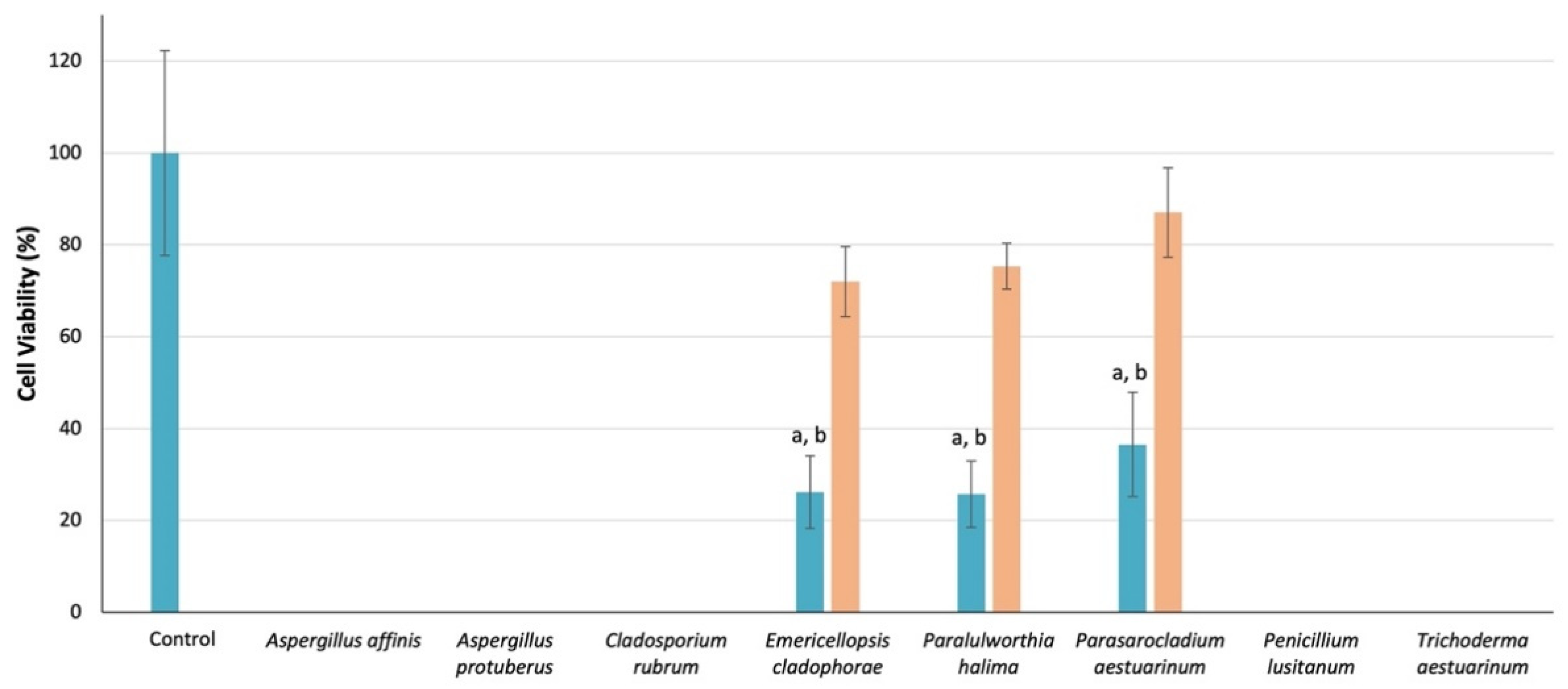
2.7. Cytotoxic Activity of Culture Media Extracts
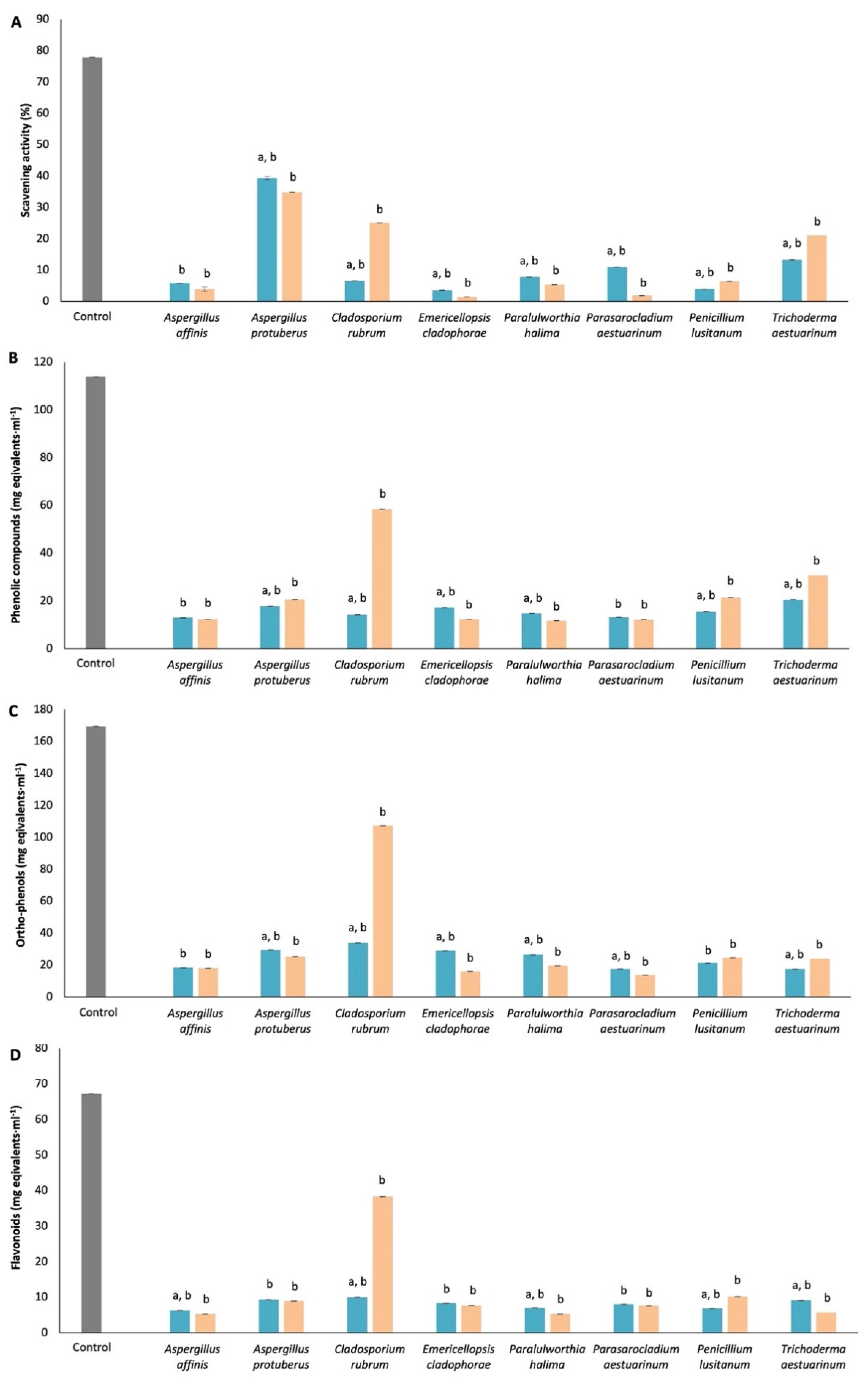
2.8. Antioxidant Activity of Mycelium
2.8.1. DPPH-Free Radical Scavenging Activity
2.8.2. Phenolic Compounds Quantification
2.8.3. Ortho-Phenols Quantification
2.8.4. Flavonoid’s Quantification
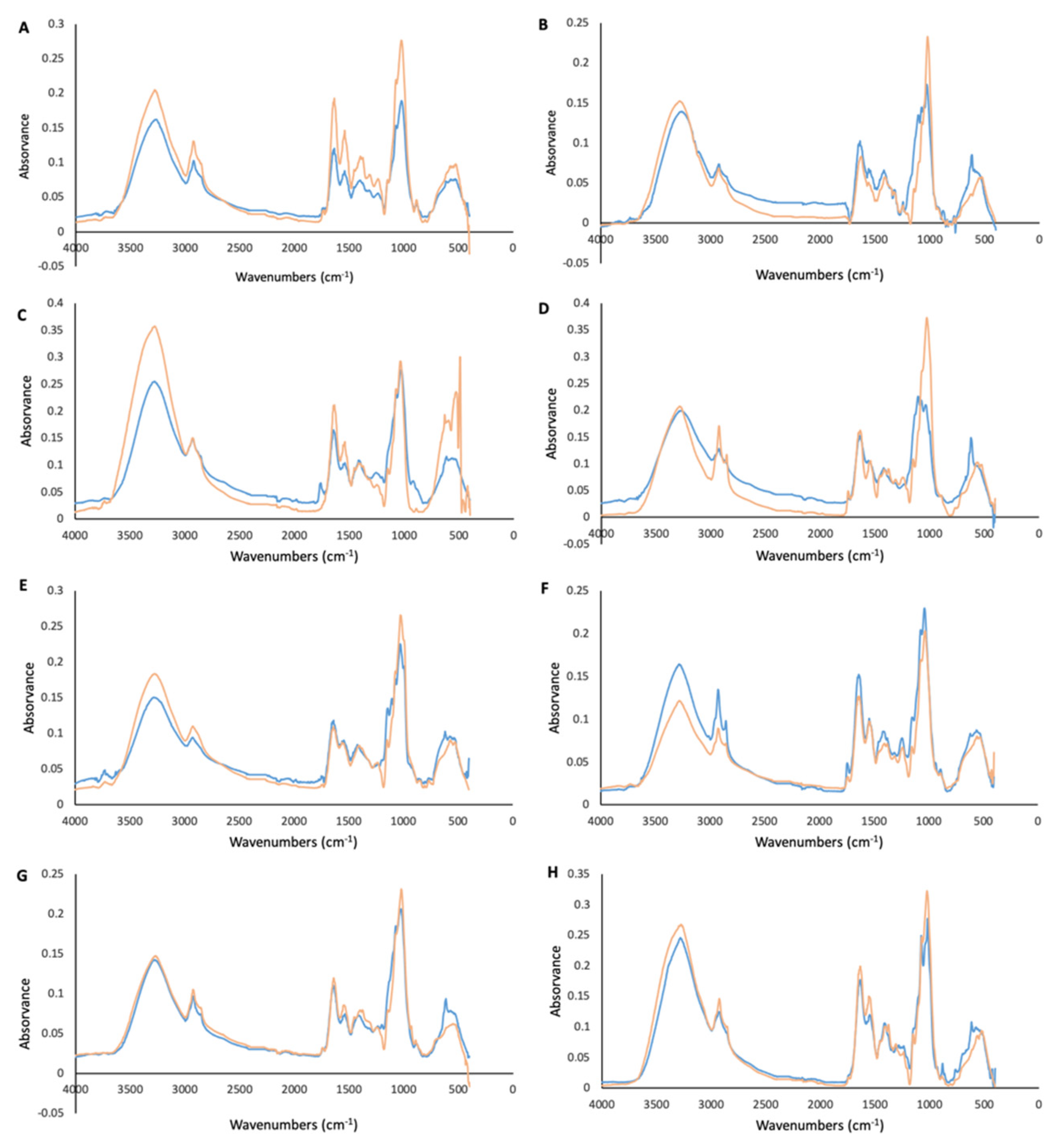
2.9. FTIR-ATR Analysis of Mycelium
3. Results
3.1. Screening of Enzymatic and Antibacterial Activity
3.2. FTIR-ATR Analysis of Mycelium
3.3. Antibacterial Activity of Fungal Extracts
3.4. Anti-Fungal Activity of Fungal Extracts
3.5. Cytotoxic Activity of Culture Media Extracts
3.6. Antioxidant Activity of Mycelium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, E.B.G. Are there more marine fungi to be described? Bot. Mar. 2011, 54, 343–354. [Google Scholar] [CrossRef]
- Richards, T.A.; Jones, M.D.; Leonard, G.; Bass, D. Marine fungi: Their ecology and molecular diversity. Annu. Rev. Mar. Sci. 2012, 4, 495–522. [Google Scholar] [CrossRef] [PubMed]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nad. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Labes, A.; Wiese, J. Bio-mining the microbial treasures of the ocean: New natural products. Biotechnol. Adv. 2011, 29, 468–482. [Google Scholar] [CrossRef]
- Overy, D.P.; Rämä, T.; Oosterhuis, R.; Walker, A.K.; Pang, K.-L. The neglected marine fungi, sensu stricto, and their isolation for natural products’ discovery. Mar. Drugs 2019, 17, 42. [Google Scholar] [CrossRef]
- Gladfelter, A.S.; James, T.Y.; Amend, A.S. Marine fungi. Curr. Biol. 2019, 29, R191–R195. [Google Scholar] [CrossRef]
- Sakayaroj, J.; Pang, K.L.; Jones, E.G. Multi-gene phylogeny of the Halosphaeriaceae: Its ordinal status, relationships between genera and morphological character evolution. Fungal Divers. 2011, 46, 87–109. [Google Scholar] [CrossRef]
- Pang, K.L.; Overy, D.P.; Jones, E.B.G.; Calado, M.; Burgaud, G.; Walker, A.K.; Johnson, J.A.; Kerr, R.G.; Cha, H.-J.; Bills, J.F. ‘Marine fungi’ and ‘marine-derived fungi’ in natural product chemistry research: Toward a new consensual definition. Fungal Biol. Rev. 2016, 30, 163–175. [Google Scholar] [CrossRef]
- Raghukumar, S. Fungi in Coastal and Oceanic Marine Ecosystems; Springer: New York, NY, USA, 2017; 364p. [Google Scholar]
- Mayer, A.M.S.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2009–2011: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2013, 11, 2510–2573. [Google Scholar]
- Li, X.; Li, X.M.; Xu, G.M.; Li, C.S.; Wang, B.G. Antioxidant metabolites from marine alga-derived fungus Aspergillus wentii EN-48. Phytochem. Lett. 2014, 7, 120–123. [Google Scholar] [CrossRef]
- Chen, C.J.; Zhou, Y.Q.; Liu, X.X.; Zhang, W.J.; Hu, S.S.; Lin, L.P.; Huo, G.M.; Jiao, R.H.; Tan, R.X.; Ge, H.M. Antimicrobial and anti-inflammatory compounds from a marine fungus Pleosporales sp. Tetrahedron Lett. 2015, 56, 6183–6189. [Google Scholar] [CrossRef]
- Moghadamtousi, S.Z.; Nikzad, S.; Kadir, H.A.; Abubakar, S.; Zandi, K. Potential antiviral agents from marine fungi: An overview. Mar. Drugs 2015, 13, 4520–4538. [Google Scholar] [CrossRef]
- Bonugli-Santos, R.C.; dos Santos Vasconcelos, M.R.; Passarini, M.R.; Vieira, G.A.; Lopes, V.C.; Mainardi, P.H.; dos Santos, J.A.; Duarte, L.A.; Otero, I.V.R.; Yoshida, A.M.S.; et al. Marine-derived fungi: Diversity of enzymes and biotechnological applications. Front. Microbiol. 2015, 6, 269. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Meng, W.; Cao, C.; Wang, J.; Shan, W.; Wang, Q. Antibacterial and antifungal compounds from marine fungi. Mar. Drugs 2015, 13, 3479–3513. [Google Scholar] [CrossRef]
- Han, J.; Liu, M.; Jenkins, I.D.; Liu, X.; Zhang, L.; Quinn, R.J.; Feng, Y. Genome-inspired Chemical exploration of marine fungus Aspergillus fumigatus MF071. Mar. Drugs 2020, 18, 352. [Google Scholar] [CrossRef]
- Yasuhara-Bell, J.; Lu, Y. Marine compounds and their antiviral activities. Antivir. Res. 2010, 86, 231–240. [Google Scholar] [CrossRef]
- Kumar, A.; Henrissat, B.; Arvas, M.; Syed, M.F.; Thieme, N.; Benz, J.P.; Sørensen, J.L.; Record, E.; Pöggeler, S.; Kempken, F. De novo assembly and genome analyses of the marine-derived Scopulariopsis brevicaulis strain LF580 unravels life-style traits and anticancerous scopularide biosynthetic gene cluster. PLoS ONE 2015, 10, e0140398. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Prakash, V.; Ranjan, N. Marine fungi: A source of potential anticancer compounds. Front. Microbiol. 2018, 8, 2536. [Google Scholar] [CrossRef] [PubMed]
- Jans, P.E.; Mfuh, A.M.; Arman, H.D.; Shaffer, C.V.; Larionov, O.V.; Mooberry, S.L. Cytotoxicity and mechanism of action of the marine-derived fungal metabolite trichodermamide B and synthetic analogues. J. Nat. Prod. 2017, 80, 676–683. [Google Scholar] [CrossRef]
- Sun, H.H.; Mao, W.J.; Chen, Y.; Guo, S.D.; Li, H.Y.; Qi, X.H.; Chen, Y.-L.; Xu, J. Isolation, chemical characteristics and antioxidant properties of the polysaccharides from marine fungus Penicillium sp. F23-2. Carbohydr. Polym. 2009, 78, 117–124. [Google Scholar] [CrossRef]
- Russell, J.R.; Huang, J.; Anand, P.; Kucera, K.; Sandoval, A.G.; Dantzler, K.W.; Hickman, D.; Jee, J.; Kimovec, F.M.; Koppstein, D.; et al. Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 2011, 77, 6076–6084. [Google Scholar] [CrossRef] [PubMed]
- Paço, A.; Duarte, K.; da Costa, J.P.; Santos, P.S.; Pereira, R.; Pereira, M.E.; Freitas, A.C.; Duarte, A.C.; Rocha-Santos, T.A.P. Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Sci. Total Environ. 2017, 586, 10–15. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Lombard, L.; Roets, F.; Swart, W.J.; Alvarado, P.; Carnegie, A.J.; Moreno, G.; Luangsa-Ard, J.; Thangavel, R.; et al. Fungal Planet description sheets: 951–1041. Persoonia 2019, 43, 223–425. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.F.M.; Aleixo, A.; Vicente, T.F.L.; Esteves, A.C.; Alves, A. Three new species of Neocamarosporium isolated from saline environments: N. aestuarinum sp. nov., N. endophyticum sp. nov. and N. halimiones sp. nov. Mycosphere 2019, 10, 608–621. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Vicente, T.F.L.; Esteves, A.C.; Alves, A. Neptunomyces aureus gen. et sp. nov. (Didymosphaeriaceae, Pleosporales) isolated from algae in Ria de Aveiro, Portugal. MycoKeys 2019, 60, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.F.M.; Santos, L.; Silva, B.M.V.; Abreu, A.C.; Vicente, T.F.L.; Esteves, A.C.; Alves, A. Biodiversity of Penicillium species from marine environments in Portugal and description of Penicillium lusitanum sp. nov., a novel species isolated from sea water. Int. J. Syst. Evol. Microbiol. 2019, 69, 3014–3021. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.F.M.; Silva, B.M.V.; Esteves, A.C.; Alves, A. Verrucoconiothyrium ambiguum sp. nov., a novel species isolated from sea water, and affiliation of the genus Verrucoconiothyrium to the family Didymellaceae. Int. J. Syst. Evol. Microbiol. 2019, 69, 3769–3776. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Esteves, A.C.; Alves, A. Revealing the hidden diversity of marine fungi in Portugal with the description of two novel species, Neoascochyta fuci sp. nov. and Paraconiothyrium salinum sp. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 5337–5354. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Vicente, T.F.L.; Esteves, A.C.; Alves, A. Novel halotolerant species of Emericellopsis and Parasarocladium associated with macroalgae in an estuarine environment. Mycologia 2020, 112, 154–171. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Abreu, A.C.; Hilário, S.; Alves, A. Diversity of marine fungi associated with wood baits in the estuary Ria de Aveiro with descriptions of Paralulworthia halima comb. nov., Remispora submersa sp. nov. and Zalerion pseudomaritima sp. nov. Mycologia 2021, 113, 664–683. [Google Scholar] [CrossRef] [PubMed]
- Vicente, T.F.L.; Gonçalves, M.F.M.; Brandão, C.; Fidalgo, C.; Alves, A. Diversity of fungi associated with macroalgae from an estuarine environment and description of Cladosporium rubrum sp. nov. and Hypoxylon aveirense sp. nov. Int. J. Syst. Evol. Microbiol. 2021, 71, 004630. [Google Scholar] [CrossRef] [PubMed]
- Esteves, A.C.; Saraiva, M.; Correia, A.; Alves, A. Botryosphaeriales fungi produce extracellular enzymes with biotechnological potential. Can. J. Microbiol. 2014, 60, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Fenice, M.; Selbmann, L.; Di Giambattista, R.; Federici, F. Chitinolytic activity at low temperature of an Antarctic strain (A3) of Verticillium lecanii. Res. Microbiol. 1998, 149, 289–300. [Google Scholar] [CrossRef]
- Agrawal, T.; Kotasthane, A.S. Chitinolytic assay of indigenous Trichoderma isolates collected from different geographical locations of Chhattisgarh in Central India. SpringerPlus 2012, 1, 73. [Google Scholar] [CrossRef]
- Teixeira, P.; Tacão, M.; Pureza, L.; Gonçalves, J.; Silva, A.; Cruz-Schneider, M.P.; Henriques, I. Occurrence of carbapenemase-producing Enterobacteriaceae in a Portuguese river: blaNDM, blaKPC and blaGES among the detected genes. Environ. Pollut. 2020, 260, 113913. [Google Scholar] [CrossRef]
- Azmir, J.; Zaidul, I.S.M.; Rahman, M.M.; Sharif, K.M.; Mohamed, A.; Sahena, F.; Jahurul, M.H.A.; Ghafoor, K.; Norulaini, N.A.N.; Omar, A.K.M. Techniques for extraction of bioactive compounds from plant materials: A review. J. Food Eng. 2013, 117, 426–436. [Google Scholar] [CrossRef]
- National Committee for Clinical Laboratory Standards (NCCLS). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Testing; Fourteenth Informational Supplement NCCLS documents M100-S14; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2004.
- Duarte, A.S.; Cavaleiro, E.; Pereira, C.; Merino, S.; Esteves, A.C.; Duarte, E.P.; Tomás, J.M.; Correia, A.C. Aeromonas piscicola AH-3 expresses an extracellular collagenase with cytotoxic properties. Lett. Appl. Microbiol. 2015, 60, 288–297. [Google Scholar] [CrossRef]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and maintenance of Vero cell lines. Curr. Protoc. Microbiol. 2009. [Google Scholar] [CrossRef]
- Xu, B.J.; Chang, S.K.C. A comparative study on phenolic profiles and antioxidant activities of legumes as affected by extraction solvents. J. Food Sci. 2007, 72, S159–S166. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar]
- Chang, C.-C.; Yang, M.-H.; Wen, H.-M.; Chern, J.-C. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J. Food Drug Anal. 2002, 10, 178–182. [Google Scholar]
- Chhetri, D.R.; Chhetri, A.; Shahi, N.; Tiwari, S.; Karna, S.K.L.; Lama, D.; Pokharel, Y.R. Isaria tenuipes Peck, an entomopathogenic fungus from Darjeeling Himalaya: Evaluation of in-vitro antiproliferative and antioxidant potential of its mycelium extract. BMC Complement. Med. Ther. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Girometta, C.; Dondi, D.; Baiguera, R.M.; Bracco, F.; Branciforti, D.S.; Buratti, S.; Lazzaroni, S.; Savino, E. Characterization of mycelia from wood-decay species by TGA and IR spectroscopy. Cellulose 2020, 27, 6133–6148. [Google Scholar] [CrossRef]
- Di Mario, F.; Rapana, P.; Tomati, U.; Galli, E. Chitin and chitosan from Basidiomycetes. Int. J. Biol. Macromol. 2008, 43, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Jones, E.G.; Leaño, E.; Pointing, S.B.; Poonyth, A.D.; Vrijmoed, L.L. Role of fungi in marine ecosystems. Biodivers. Conserv. 1998, 7, 1147–1161. [Google Scholar] [CrossRef]
- Liu, X.; Kokare, C. Microbial enzymes of use in industry. In Biotechnology of Microbial Enzymes; Academic Press: Cambridge, MA, USA, 2017; pp. 267–298. [Google Scholar]
- Bajpai, P. Xylanolytic Enzymes; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Bucher, V.V.C.; Hyde, K.D.; Pointing, S.B.; Reddy, C.A. Production of wood decay enzymes, mass loss and lignin solubilization in wood by marine ascomycetes and their anamorphs. Fungal Divers. 2004, 15, 1–14. [Google Scholar]
- Raghukumar, C.; Raghukumar, S.; Chinnaraj, A.; Chandramohan, D.; D’Souza, T.M.; Reddy, C.A. Laccase and other lignocellulose modifying enzymes of marine fungi isolated from the coast of India. Bot. Mar. 1994, 37, 515–523. [Google Scholar] [CrossRef]
- Pointing, S.B.; Hyde, K.D. Lignocellulose-degrading marine fungi. Biofouling 2000, 15, 221–229. [Google Scholar] [CrossRef]
- Sarkar, S.; Pramanik, A.; Mitra, A.; Mukherjee, J. Bioprocessing data for the production of marine enzymes. Mar. Drugs 2010, 8, 1323–1372. [Google Scholar] [CrossRef]
- Homaei, A.; Ghanbarzadeh, M.; Monsef, F. Biochemical features and kinetic properties of α-amylases from marine organisms. Int. J. Biol. Macromol. 2016, 83, 306–314. [Google Scholar] [CrossRef]
- Sundarram, A.; Murthy, T.P.K. α-amylase production and applications: A review. J. Appl. Environ. Microbiol. 2014, 2, 166–175. [Google Scholar]
- Sahoo, K.; Dhal, N.K.; Das, R. Production of amylase enzyme from mangrove fungal isolates. Afr. J. Biotechnol. 2014, 13, 4338–4346. [Google Scholar]
- Sguros, P.L.; Rodrigues, J.; Simms, J. Role of marine fungi in the biochemistry of the oceans. V. Patterns of constitutive nutritional growth responses. Mycologia 1973, 65, 161–174. [Google Scholar] [CrossRef]
- Kumari, D.; Qian, X.Y.; Pan, X.; Achal, V.; Li, Q.; Gadd, G.M. Microbially-induced carbonate precipitation for immobilization of toxic metals. Adv. Appl. Microbiol. 2016, 94, 79–108. [Google Scholar] [PubMed]
- Li, Q.; Gadd, G.M. Biosynthesis of copper carbonate nanoparticles by ureolytic fungi. Appl. Microbiol. Biotechnol. 2017, 101, 7397–7407. [Google Scholar] [CrossRef]
- Berini, F.; Casartelli, M.; Montali, A.; Reguzzoni, M.; Tettamanti, G.; Marinelli, F. Metagenome-sourced microbial chitinases as potential insecticide proteins. Front. Microbiol. 2019, 10, 1358. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, N.; Tewari, R.; Hoondal, G.S. Biotechnological aspects of chitinolytic enzymes: A review. Appl. Microbiol. Biotechnol. 2006, 71, 773–782. [Google Scholar] [CrossRef]
- Pasqualetti, M.; Barghini, P.; Giovannini, V.; Fenice, M. High production of chitinolytic activity in halophilic conditions by a new marine strain of Clonostachys rosea. Molecules 2019, 24, 1880. [Google Scholar] [CrossRef] [PubMed]
- Reiss, R.; Ihssen, J.; Richter, M.; Eichhorn, E.; Schilling, B.; Thöny-Meyer, L. Laccase versus laccase-like multi-copper oxidase: A comparative study of similar enzymes with diverse substrate spectra. PLoS ONE 2013, 8, e65633. [Google Scholar]
- Couto, S.R.; Herrera, J.L.T. Laccases in pollution control. Ter. Aquat. Environ. Tox. 2007, 1, 34–45. [Google Scholar]
- Bonugli-Santos, R.C.; Durrant, L.R.; da Silva, M.; Sette, L.D. Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzyme Microb. Technol. 2010, 46, 32–37. [Google Scholar] [CrossRef]
- Ben Ali, W.; Chaduli, D.; Navarro, D.; Lechat, C.; Turbé-Doan, A.; Bertrand, E.; Faulds, C.B.; Sciara, G.; Lesage-Meessen, L.; Record, E.; et al. Screening of five marine-derived fungal strains for their potential to produce oxidases with laccase activities suitable for biotechnological applications. BMC Biotechnol. 2020, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.S.; Correia, A.; Esteves, A.C. Bacterial collagenases—A review. Crit. Rev. Microbiol. 2016, 42, 106–126. [Google Scholar] [CrossRef]
- Chamekh, R.; Deniel, F.; Donot, C.; Jany, J.L.; Nodet, P.; Belabid, L. Isolation, identification and enzymatic activity of halotolerant and halophilic fungi from the Great Sebkha of Oran in Northwestern of Algeria. Mycobiology 2019, 47, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Arfi, Y.; Chevret, D.; Henrissat, B.; Berrin, J.G.; Levasseur, A.; Record, E. Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Xue, D.-S.; Feng, X.-Y.; Yao, S.-J. Screening and production of ligninolytic enzyme by a marine-derived fungal Pestalotiopsis sp. J63. Appl. Biochem. Biotechnol. 2011, 165, 1754–1769. [Google Scholar] [CrossRef] [PubMed]
- Bonugli-Santos, R.C.; Durrant, L.R.; Sette, L.D. The production of ligninolytic enzymes by marine-derived basidiomycetes and their biotechnological potential in the biodegradation of recalcitrant pollutants and the treatment of textile effluents. Water Air Soil Pollut. 2012, 223, 2333–2345. [Google Scholar] [CrossRef]
- Kong, F.D.; Zhou, L.M.; Ma, Q.Y.; Huang, S.Z.; Wang, P.; Dai, H.F.; Zhao, Y.X. Metabolites with Gram-negative bacteria quorum sensing inhibitory activity from the marine animal endogenic fungus Penicillium sp. SCS-KFD08. Arch. Pharm. Res. 2017, 40, 25–31. [Google Scholar] [CrossRef]
- Inostroza, A.; Lara, L.; Paz, C.; Perez, A.; Galleguillos, F.; Hernandez, V.; Becerra, J.; González-Rocha, G.; Silva, M. Antibiotic activity of Emerimicin IV isolated from Emericellopsis minima from Talcahuano Bay, Chile. Nat. Prod. Res. 2018, 32, 1361–1364. [Google Scholar] [CrossRef]
- De Siqueira, V.M.; Conti, R.; De Araújo, J.M.; Souza-Motta, C.M. Endophytic fungi from the medicinal plant Lippia sidoides Cham. and their antimicrobial activity. Symbiosis 2011, 53, 89–95. [Google Scholar] [CrossRef]
- dos Santos, I.P.; da Silva, L.C.; da Silva, M.V.; de Araújo, J.M.; Cavalcanti, M.D.S.; Lima, V.L. Antibacterial activity of endophytic fungi from leaves of Indigofera suffruticosa Miller (Fabaceae). Front. Microbiol. 2015, 6, 350. [Google Scholar] [CrossRef] [PubMed]
- Marcellano, J.P.; Collanto, A.S.; Fuentes, R.G. Antibacterial activity of endophytic fungi isolated from the bark of Cinnamomum mercadoi. Pharmacogn. J. 2017, 9, 405–409. [Google Scholar] [CrossRef]
- Komai, S.-I.; Hosoe, T.; Itabashi, T.; Nozawa, K.; Yaguchi, T.; Fukushima, K.; Kawai, K. New penicillide derivatives isolated from Penicillium simplicissimum. J. Nat. Med. 2006, 60, 185–190. [Google Scholar] [CrossRef]
- Kaleem, S.; Ge, H.; Yi, W.; Zhang, Z.; Wu, B. Isolation, structural elucidation, and antimicrobial evaluation of the metabolites from a marine-derived fungus Penicillium sp. ZZ1283. Nat. Prod. Res. 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kaleem, S.; Qin, L.; Yi, W.; Lian, X.Y.; Zhang, Z. Bioactive metabolites from the Mariana Trench sediment-derived fungus Penicillium sp. SY2107. Mar. Drugs 2020, 18, 258. [Google Scholar] [CrossRef] [PubMed]
- Pal, M. Candidiasis: An important opportunistic mycosis of global public health concern. Res. Infect. Dis. Trop. Med. 2020, 2, 49–51. [Google Scholar]
- Rosa, N.; Campos, B.; Esteves, A.C.; Duarte, A.S.; Correia, M.J.; Silva, R.M.; Barros, M. Tracking the functional meaning of the human oral-microbiome protein-protein interactions. In Advances in Protein Chemistry and Structural Biology; Academic Press: Cambridge, MA, USA, 2020; Volume 121, pp. 199–235. [Google Scholar]
- Pal, M. Aspergillosis: A highly infectious global mycosis of human and animal. Clin. Biotechnol. Microbiol. 2017, 1, 46–49. [Google Scholar]
- Ben-Yaacov, R.; Knoller, S.; Caldwell, G.A.; Becker, J.M.; Koltin, Y. Candida albicans gene encoding resistance to benomyl and methotrexate is a multidrug resistance gene. Antimicrob. Agents Chemother. 1994, 38, 648–652. [Google Scholar] [CrossRef][Green Version]
- Sasnauskas, K.; Jomantienė, R.; Lebedienė, E.; Lebedys, J.; Januska, A.; Janulaitis, A. Cloning and sequence analysis of a Candida maltosa gene which confers resistance to cycloheximide. Gene 1992, 116, 105–108. [Google Scholar] [CrossRef]
- El-Kashef, D.H.; Youssef, F.S.; Reimche, I.; Teusch, N.; Müller, W.E.; Lin, W.; Frank, M.; Liu, Z.; Proksch, P. Polyketides from the marine-derived fungus Aspergillus falconensis: In silico and in vitro cytotoxicity studies. Bioorg. Med. Chem. 2020, 29, 115883. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Kimishima, A.; Ishida, R.; Setiawan, A.; Arai, M. Selective cytotoxicity of epidithiodiketopiperazine DC1149B, produced by marine-derived Trichoderma lixii on the cancer cells adapted to glucose starvation. J. Nat. Med. 2020, 74, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.N.; Lu, H.M.; Gao, C.H.; Guo, L.; Zhan, Z.Y.; Wang, J.J.; Liu, Y.-H.; Xiang, S.-T.; Wang, J.; Luo, X.W. Cytotoxic benzopyranone and xanthone derivatives from a coral symbiotic fungus Cladosporium halotolerans GXIMD 02502. Nat. Prod. Res. 2020, 1–8. [Google Scholar] [CrossRef]
- Marchese, P.; Garzoli, L.; Gnavi, G.; O’Connell, E.; Bouraoui, A.; Mehiri, M.; Murphy, J.M.; Varese, G.C. Diversity and bioactivity of fungi associated with the marine sea cucumber Holothuria poli: Disclosing the strains potential for biomedical applications. J. Appl. Microbiol. 2020, 129, 612–625. [Google Scholar] [CrossRef]
- Bhanja, T.; Kumari, A.; Banerjee, R. Enrichment of phenolics and free radical scavenging property of wheat koji prepared with two filamentous fungi. Bioresour. Technol. 2009, 100, 2861–2866. [Google Scholar] [CrossRef] [PubMed]
- Tavares, D.G.; Barbosa, B.V.L.; Ferreira, R.L.; Duarte, W.F.; Cardoso, P.G. Antioxidant activity and phenolic compounds of the extract from pigment-producing fungi isolated from Brazilian caves. Biocatal. Agric. Biotechnol. 2018, 16, 148–154. [Google Scholar] [CrossRef]
- Hameed, A.; Hussain, S.A.; Yang, J.; Ijaz, M.U.; Liu, Q.; Suleria, H.A.R.; Song, Y. Antioxidants potential of the filamentous fungi (Mucor circinelloides). Nutrients 2017, 9, 1101. [Google Scholar] [CrossRef]
- Belozerskaya, T.A.; Gessler, N.N. Reactive Oxygen Species and the strategy of antioxidant defense in fungi: A Review. Appl. Biochem. Microbiol. 2007, 43, 506–515. [Google Scholar] [CrossRef]
- Smith, H.; Doyle, S.; Murphy, R. Filamentous fungi as a source of natural antioxidants. Food Chem. 2015, 185, 389–397. [Google Scholar] [CrossRef]
- Danilova, O.A.; Ianutsevich, E.A.; Bondarenko, S.A.; Georgieva, M.L.; Vikchizhanina, D.A.; Groza, N.V.; Bilanenko, E.N.; Tereshina, V.M. Osmolytes and membrane lipids in the adaptation of micromycete Emericellopsis alkalina to ambient pH and sodium chloride. Fungal Biol. 2020, 124, 884–891. [Google Scholar] [CrossRef]

| Species | Strain | Host/Substrate | Reference |
|---|---|---|---|
| Aspergillus affinis | CMG 70 | Sea water | [30] |
| Aspergillus protuberus | CMG 71 | Sea water | [30] |
| Cladosporium rubrum | MUM 19.39/CMG 28 | Enteromorpha sp. | [33] |
| Emericellopsis cladophorae | MUM 19.33/CMG 25 | Cladophora sp. | [31] |
| Emericellopsis enteromorphae | MUM 19.34/CMG 26 | Enteromorpha sp. | [31] |
| Emericellopsis phycophila | MUM 19.32/CMG 15 | Filamentous green alga | [31] |
| Lulworthia cf. purpurea | MUM 20.50/CMG 55 | Submerged wood | [32] |
| Lulworthia cf. purpurea | MUM 20.56/CMG 57 | Submerged wood | [32] |
| Neocamarosporium aestuarinum | MUM 18.55/CMG 4 | Sea water | [26] |
| Neocamarosporium endophyticum | MUM 18.56/CAA 808 | Halimione portulacoides | [26] |
| Neocamarosporium haliniones | MUM 18.54/CAA 807 | Halimione portulacoides | [26] |
| Neodevriesia aestuarina | MUM 19.27/CMG 9 | Saline water | [25] |
| Neptunomyces aureus | MUM 19.38/CMG 10A | Gracilaria gracilis | [27] |
| Paraconiothyrium salinum | MUM 19.91/CMG 49 | Sponge | [30] |
| Paralulworthia halima | MUM 20.56/CMG 69 | Submerged wood | [32] |
| Parasarocladium aestuarinum | MUM 19.35/CMG 30 | Fucus sp. | [31] |
| Parasarocladium alavariense | MUM 19.36/CMG 32 | Rhodophyta | [31] |
| Parasarocladium fusiforme | MUM 19.37/CMG 36 | Ulva sp. | [31] |
| Penicillium lusitanum | MUM 18.49/CMG 8 | Sea water | [28] |
| Remispora submersa | MUM 20.48/CMG 53 | Submerged wood | [32] |
| Sedecimiella taiwanensis | CMG 51 | Submerged wood | [32] |
| Trichoderma aestuarinum | MUM 19.05/CMG 1 | Saline water | [24] |
| Verrucoconiothyrium ambiguum | MUM 18.57/CMG 5 | Sea water | [29] |
| Zalerion maritima | MUM 20.138/CMG 67 | Submerged wood | [32] |
| Zalerion pseudomaritima | MUM 20.49/CMG 65 | Submerged wood | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, M.F.M.; Paço, A.; Escada, L.F.; Albuquerque, M.S.F.; Pinto, C.A.; Saraiva, J.A.; Duarte, A.S.; Rocha-Santos, T.A.P.; Esteves, A.C.; Alves, A. Unveiling Biological Activities of Marine Fungi: The Effect of Sea Salt. Appl. Sci. 2021, 11, 6008. https://doi.org/10.3390/app11136008
Gonçalves MFM, Paço A, Escada LF, Albuquerque MSF, Pinto CA, Saraiva JA, Duarte AS, Rocha-Santos TAP, Esteves AC, Alves A. Unveiling Biological Activities of Marine Fungi: The Effect of Sea Salt. Applied Sciences. 2021; 11(13):6008. https://doi.org/10.3390/app11136008
Chicago/Turabian StyleGonçalves, Micael F. M., Ana Paço, Luís F. Escada, Manuela S. F. Albuquerque, Carlos A. Pinto, Jorge A. Saraiva, Ana Sofia Duarte, Teresa A. P. Rocha-Santos, Ana Cristina Esteves, and Artur Alves. 2021. "Unveiling Biological Activities of Marine Fungi: The Effect of Sea Salt" Applied Sciences 11, no. 13: 6008. https://doi.org/10.3390/app11136008
APA StyleGonçalves, M. F. M., Paço, A., Escada, L. F., Albuquerque, M. S. F., Pinto, C. A., Saraiva, J. A., Duarte, A. S., Rocha-Santos, T. A. P., Esteves, A. C., & Alves, A. (2021). Unveiling Biological Activities of Marine Fungi: The Effect of Sea Salt. Applied Sciences, 11(13), 6008. https://doi.org/10.3390/app11136008










