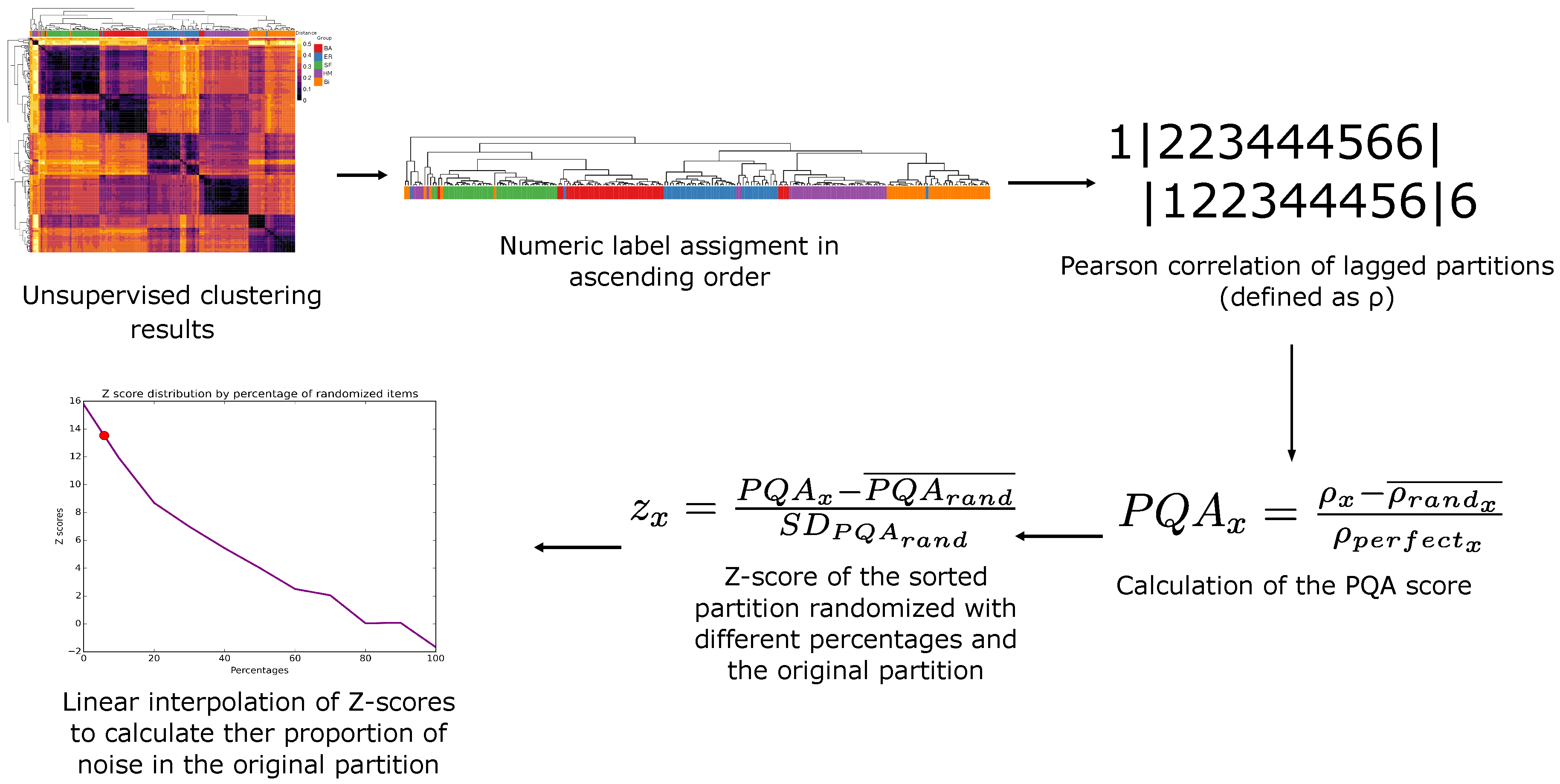
1. Introduction
A common task in today’s research is the identification of specific markers, as predictors of a classification yielded in clustering analysis of the data. For instance, this approach is particularly useful after high-throughput experiments to compare gene expression or methylation profiles among different cell lines [
1]. This task is used in the nascent field of single-cell sequencing, leading to the important step of clustering cells for further classification or as a qualifying metric of the sequencing process [
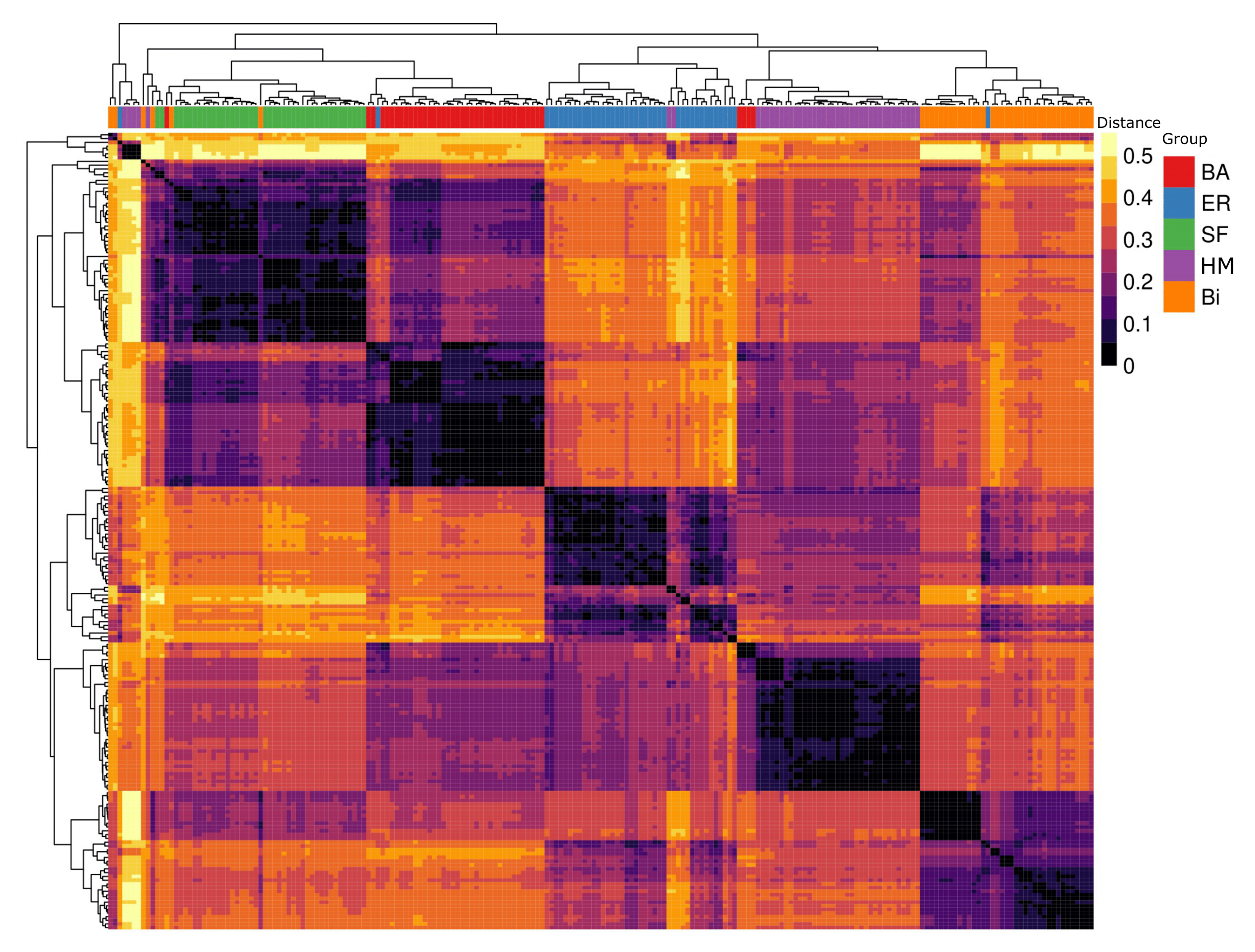
2]. Regarding the vastly used gene expression assays, the vector of profiles for each marker across different cell lines is recorded using hierarchical clustering algorithms. These algorithms yield a dendrogram and a heat map representing the vector of marker profiles, illustrating the arrangement of the clusters. To assess how well the clustering is segregating different cell lines, a class stating the desired partitioning of each cell line is provided a posteriori. Then, a simple visual inspection of the vector of classes is used to estimate whether the clustering is providing a good partition. Such a partition vector is colored according to the classification that each item is associated with, and it is expected that similar items will be contiguous, so the groups formed are assessed qualitatively against the biological background of each item.
This procedure should not be confused with “supervised clustering”, which provides a vector of classes starting the desired partitioning a priori. This is then used to guide the clustering algorithms by allowing the learning of the metric distances that optimizes the partitioning [
3]. Additionally, it may become confused with the metric assessment of the clustering algorithms, especially with the external cluster evaluation. For this, various metrics have been developed to qualify the clustering algorithm itself, such as intrinsic and extrinsic measures. The extrinsic validation compares the clustering to a goal to say whether it is good clustering or not. The internal validation compares the elements within the cluster and their differences [
4]. Partition quantitative assessment (
PQA) involves characteristics of both kinds of validation, through using both the crafted goal standard and the yielded signal itself (clustered vector). However,
PQA gathers these elements not qualifying the clustering algorithm itself but to quantify the noise embedded in the cluster, this noise may be due to the intrinsic metric or marker used to order the data set.
A possible caveat of the qualitative assessment discussed above is that humans tend to perceive meaningful patterns within random data leading to a cognitive bias known as apophenia [
5]. While interpreting the partitions obtained from unsupervised clustering analysis, researchers attempt to visually assess how close the classifications are to each other, finding patterns that are not well supported by the data. Such an effect is created because the adjacency between items may give a notion of the dissimilarity distance in the dendrogram leaves. Unfortunately, as far as we know, there is no method to quantitatively assess the quality of the groups of classifications from the clustering or, at least, there is no attempt to quantify whether certain configuration or order of the items may be due to randomness. This is a serious caveat, since the insertion of noise can lead to a false conclusion or misleading results. Furthermore, the purging of this noise can lead to more efficient descriptions of markers and its phenomena, accelerating the advance in many fields.
In statistics, serial correlation (SC) is a term used to describe the relationship between observations of the same variable over specific periods. It was originally used in engineering to determine how a signal, for instance, a radio wave, varies with itself over time. Later, SC was adapted to econometrics to analyze economic data over time principally to predict stock prices and, in other fields, to model-independent random variables [
6]. We applied the SC to propose a manner to quantify how good the grouping is of a posterior classification just by retrieving the results of unsupervised clustering analysis. Thus, we propose a novel relative score,
PQA, to solve the subjectivity of the visual inspection and to quantify statistically how much noise is embedded in the results of clustering analysis.
2. Methodology
2.1. Assigning Numeric Labels to Classifications
A vector denoting the putative similarities among the variables in a study is usually obtained after a clustering analysis. Each variable is classified to generate a vector of profiles (VP). Such a vector of classifications is usually translated into a colors vector, in which each color represents a classification. It is common to inspect this vector to find groups that make sense according to the analyzed data. To the method presented in this work, the VP may be as simple as a vector of strings or numbers that represent the input.
Whatever representation of the classifications may be, it is necessary to transform the classifications to a vector of numeric labels, in which a number represents a classification, to be able to calculate SC. To accomplish this, we assign the first numeric label (number 1) to the first item in the vector, which usually lays at one of the vector’s extremes. Then, if the classification of the next item is different from the previous one, the next number in the sequence is assigned, and so on. This way of labeling assures that the changes in the SC values are due to the order of numbers, that is to say, the grouping of the classifications resulting from the clustering, and it is not an artifact of the labeling itself (
Figure 1).
2.2. Partition Quantitative Assessment (PQA) Score
Because the order of the VP could be interpreted as the grouping of the classifications, we measure how well the same classifications are held together in the VP through a SC shifted one position. Such a type of correlation is defined as the Pearson-product-moment correlation between the VP discarding the first item, and the VP discarding the last (Equation (1), xi (order vector i-th position), n (length of x), (resulting SC)).
We then define the PQA as the SC of the VP after removing background noise, normalized for the SC of the perfect grouping of the partitions (defined as the sorted vector in ascending order). Thus, the more similar VP is to its sorted vector, the higher the score is yielded (Equation (2), (SC of the VP), (mean of the SC of 1000 randomizations), SC of the sorted vector in ascending order)).
2.3. Background-Noise Correlation Factor in the PQA Score
To compute the background-noise correlation factor in the PQA score definition, we sample the indexes of the VP and the swapping of the corresponding items. This background correction is aimed to remove inherent noise in the data, even though the score may still be subjected to noise from the chosen clustering algorithm or discrepancies in the posterior classification.
2.4. Statistical Significance of the PQA Score
To quantify the statistical significance of the
PQA score, we calculate a Z-score (Equation (3)),
where
is the
PQA score of the VP,
is the mean of
PQA scores of 1000 randomizations of the VP. These randomizations have the purpose of generating a solid random background to compare it to the real signal. The number of randomizations does not depend on the size of the VP. It is worth noting that there are two randomization processes, one generate the input population of random vectors to calculate the
PQA score to further calculate a Z-score and the other represents the noise in Equation (2).
2.5. Defining Noise Proportions
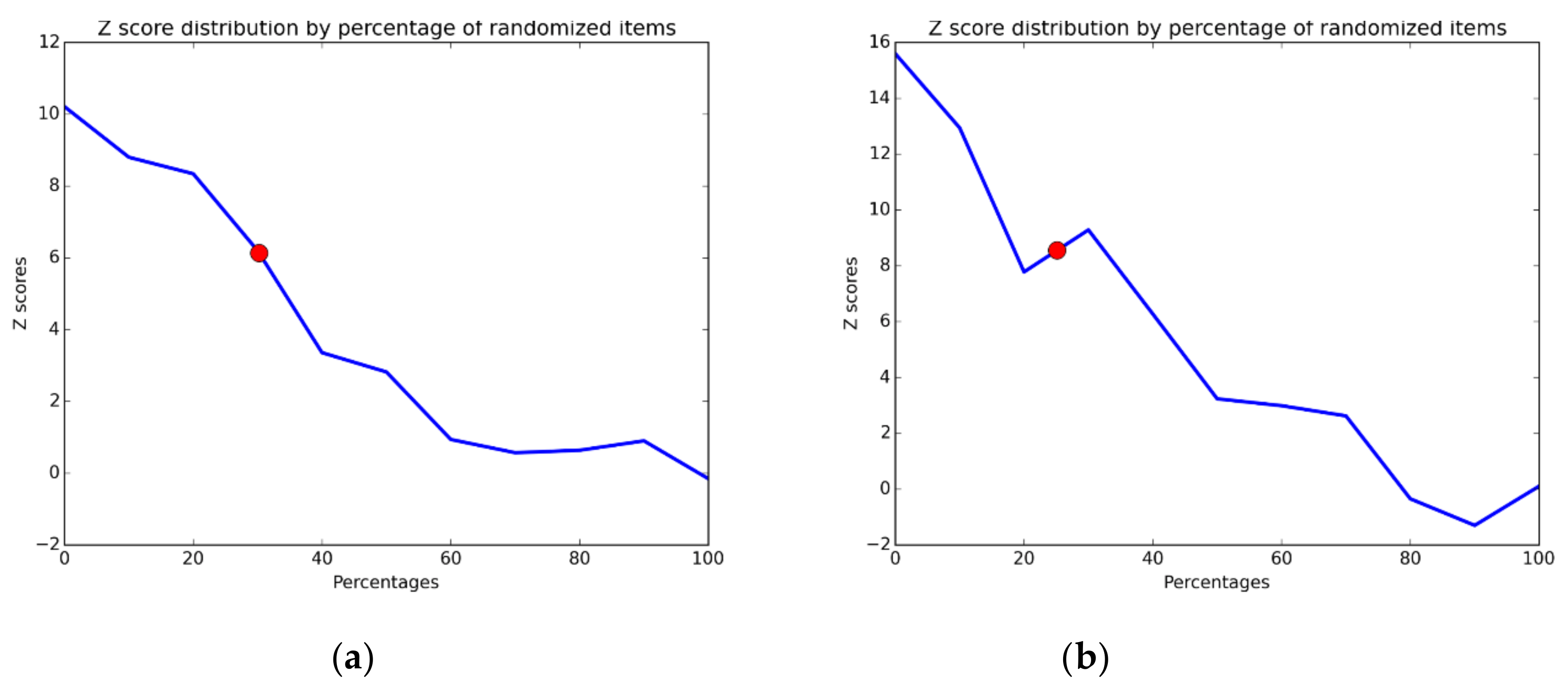
To provide a quantification of the embedded noise in the VP, we calculate the Z-scores from the distribution of PQA values of the randomized vectors. This shuffling is yielded by scrambling the vector. Then this Z-score is interpolated to retrieve the estimated noise in the VP cluster.
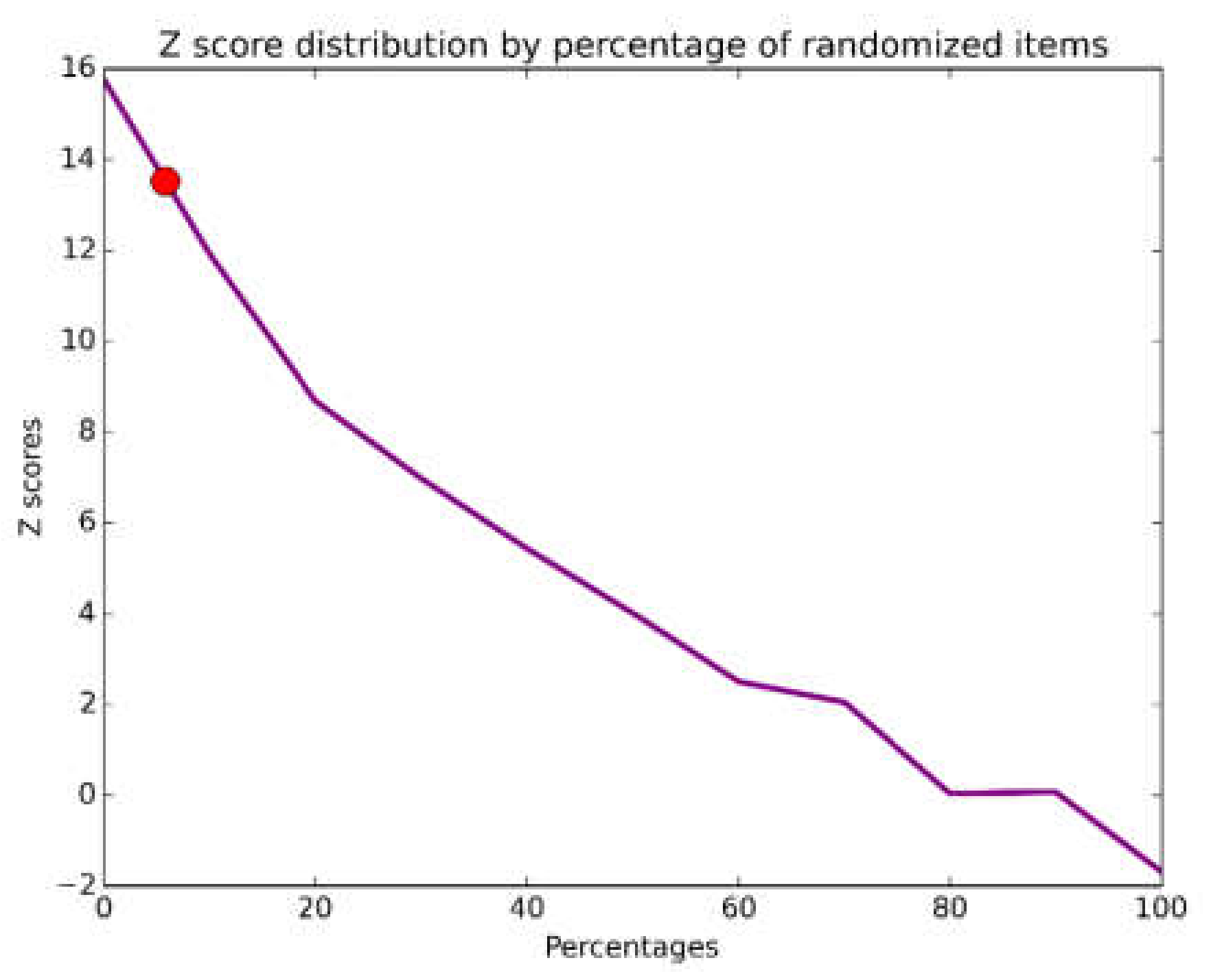
2.6. Effect of the Length and Number of Partitions of the Vector in the Z-Score Distributions
Since we want to compare the PQA with the noise, we randomized 1000 times the VP. We opted to describe the dynamic of the Z-score given the different percentage of noise and the number of partitions. For this, we synthetically crafted a vector of both ranging from 0 to 100 elements and number of classifications. The Z-scores were retrieved from the crafted vectors using the formulas described above.
4. Conclusions
In this work, we presented a novel method to quantify the proportion of noise embedded in the grouping of associated classes of the elements in hierarchical clustering. We proposed a relative score derived from a SC of the VP from the dendrogram of any clustering analysis and calculated Z-statistics as well as an interpolation to deliver an estimation of noise in the VP. We explain how the method is formulated and show the tests we made to systematically refine it.
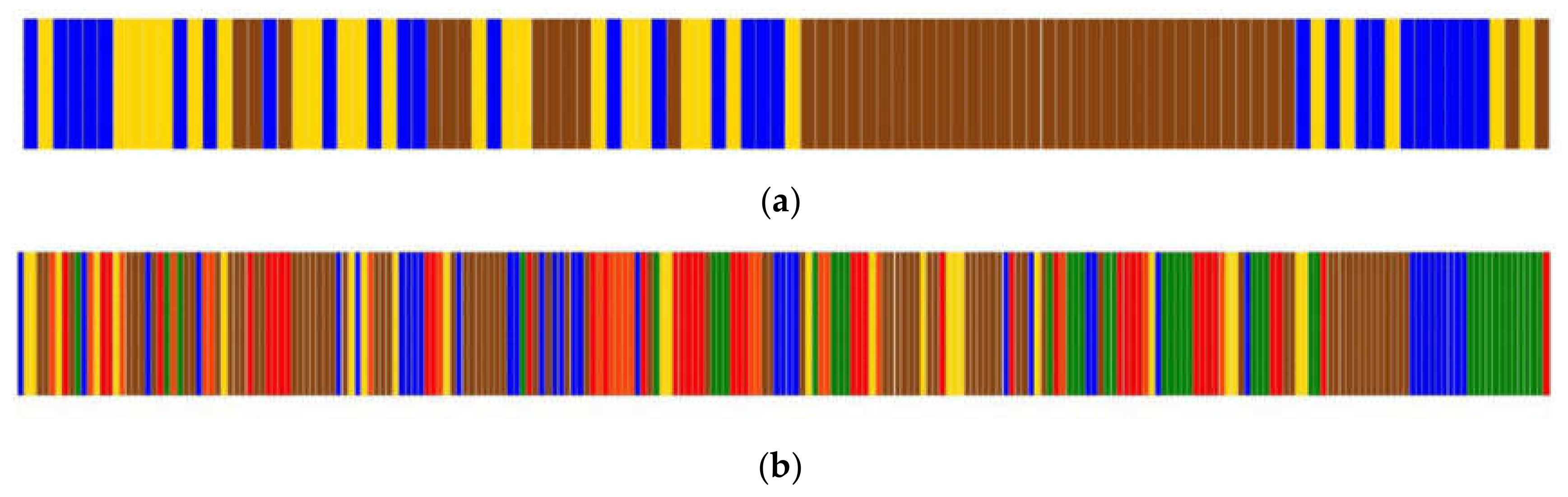
Additionally, we made a proof of concept by using clustering data from two works that we think perfectly represent overfitting by apophenia. Also, we added an example from network biology where clustered networks are separated by intrinsic characteristics. Although in this work we focused on examples where hierarchical clustering is performed, this framework can apply to any partition algorithm in which the elements are identified and a VP can be acquired.
We concluded that the clustered sets of biological data have a high measure of noise, despite looking well grouped. We proved what a minimum number of classifications should be considered in this sort of clustering analysis to have a significant reduction of noise. On the other hand, we permuted the labels of the associated classes and concluded that the effect is negligible. We proved that randomness still plays an important role by biasing the results, although it may not be evident through visual inspection.
The PQA could be used as a benchmark to test which clustering algorithm should be appropriate for the analyzed dataset by minimizing the noise proportion and to guide omics experimental designs. Nevertheless, a word of caution, the PQA score alone can be subject to subjectivity if not used properly since it depended on the characteristics of the analyzed data. Thus, the PQA score is thought to be considered a quantification of noise in clustered data and should be used with discretion.