In Vitro Assessment of Kurdish Rice Genotypes in Response to PEG-Induced Drought Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Genetic Diversity
2.2. Callus Induction
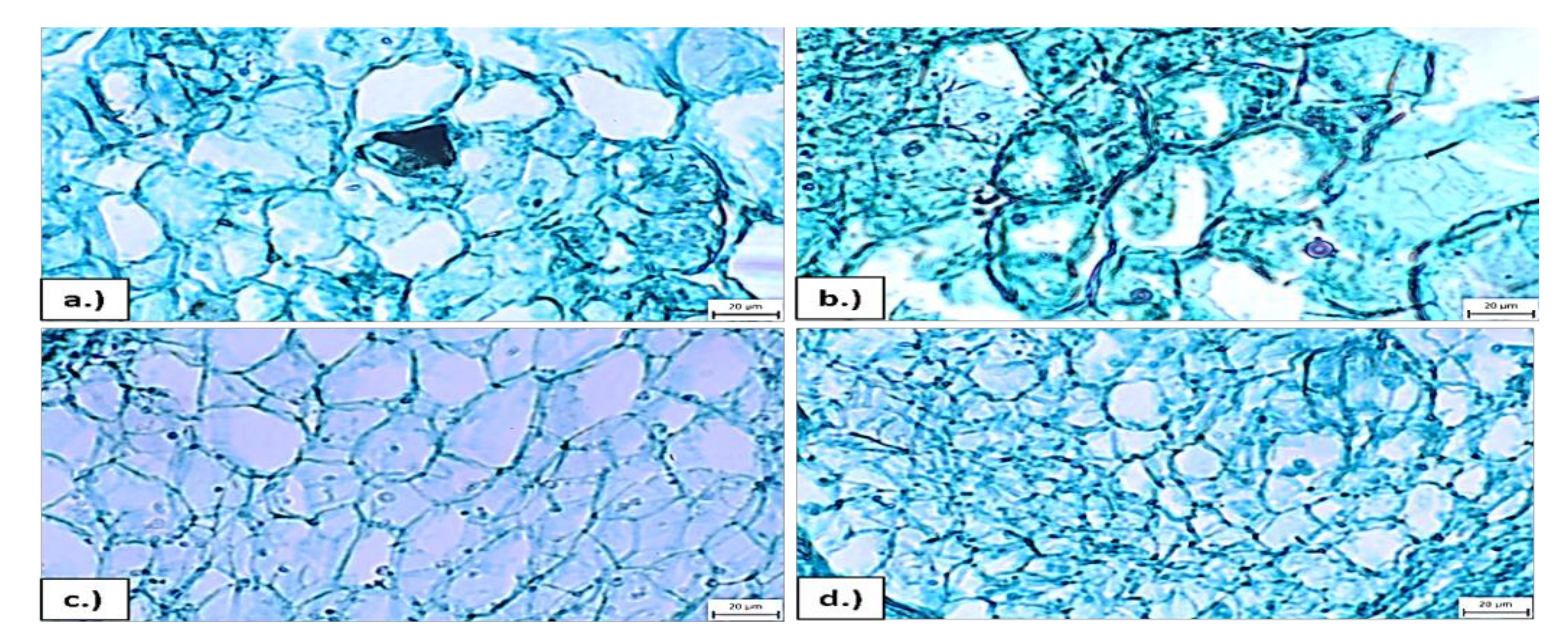
2.3. Microscopic Analyses of Cell Dimension
2.4. Antioxidant Enzyme Activity
2.4.1. Measurement of Ascorbate Peroxidase Activity (APX)
2.4.2. Measurement of Catalase Activity (CAT)
2.5. Proline Content Assay
2.6. Inorganic Element Content Analyses
2.7. Statistical Analyses
3. Results
3.1. Genetic Diversity among Rice Genotypes
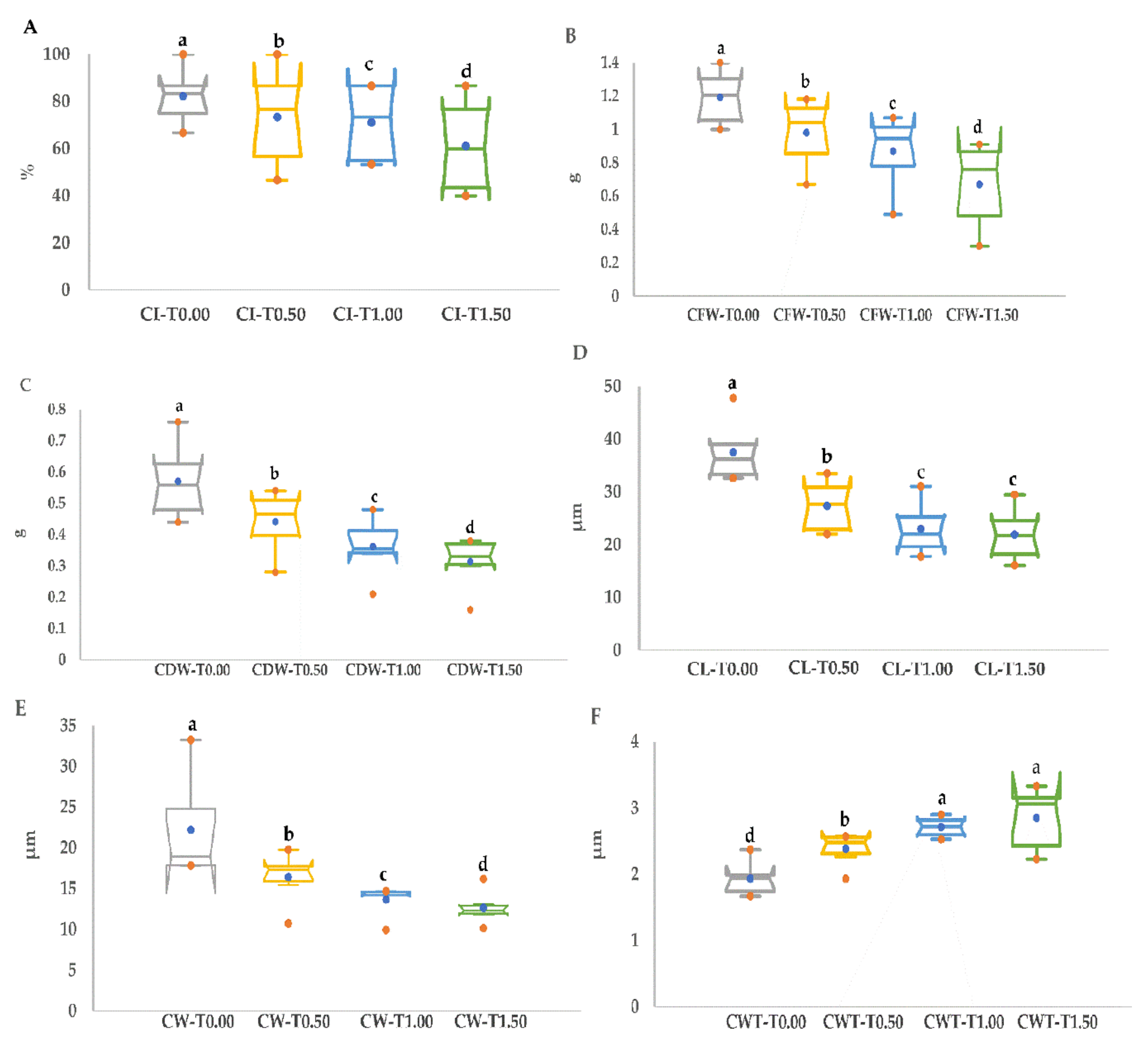
3.2. PEG Stress Tolerance Based on Callus Growth Traits
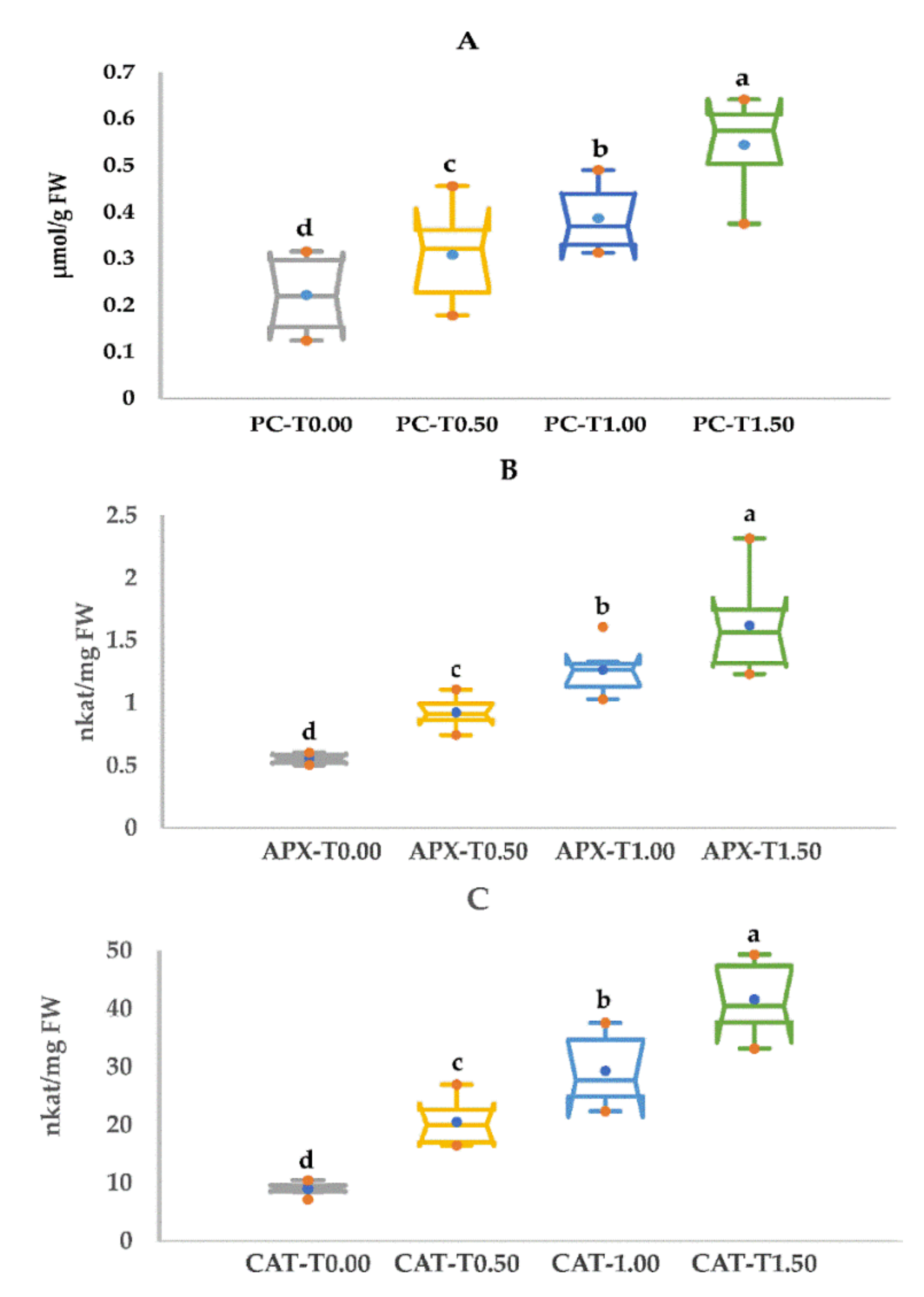
3.3. PEG Stress Tolerance Based on Proline Content and Antioxidant Enzyme Activity
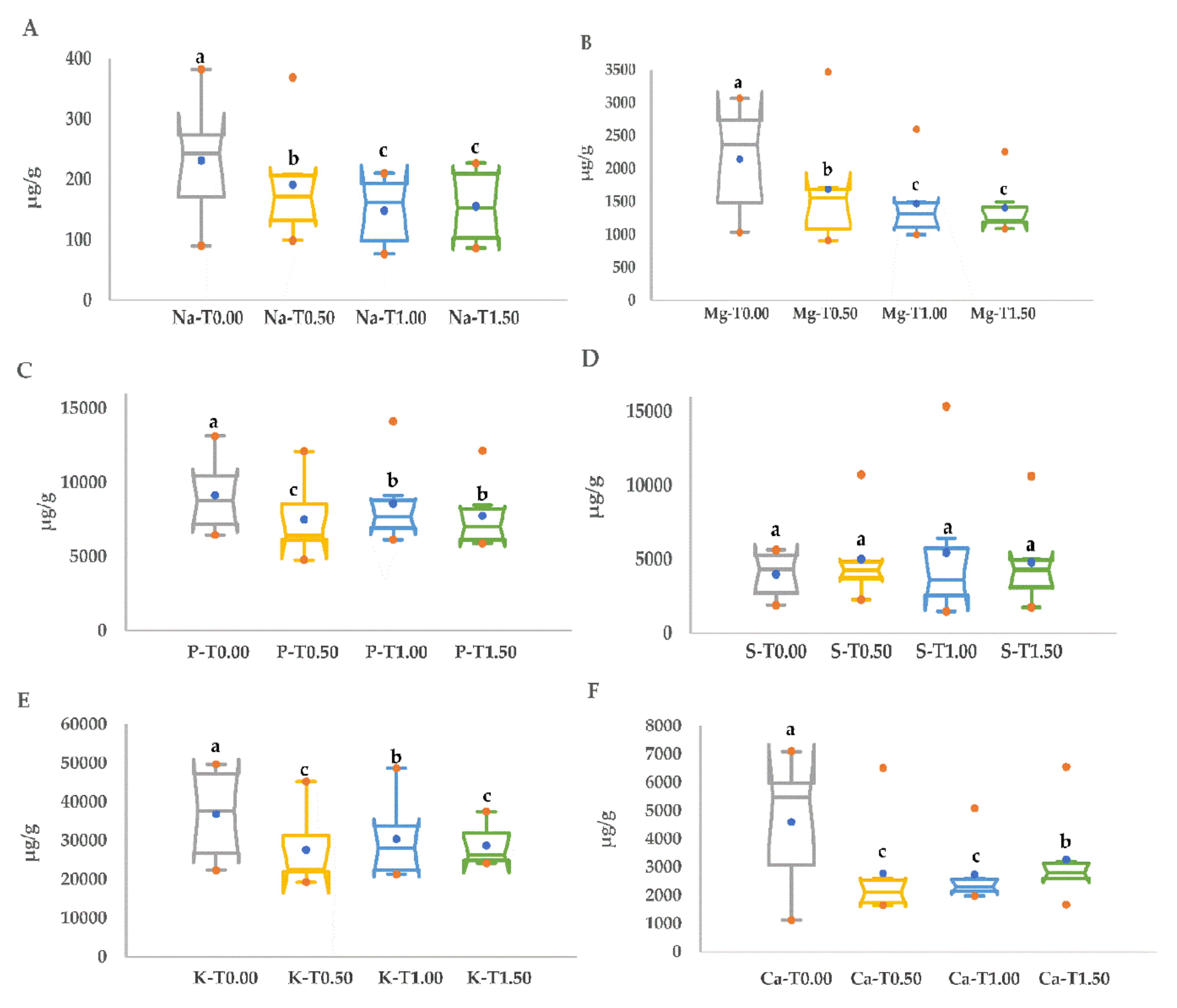
3.4. Inorganic Element Responses under Drought Stress
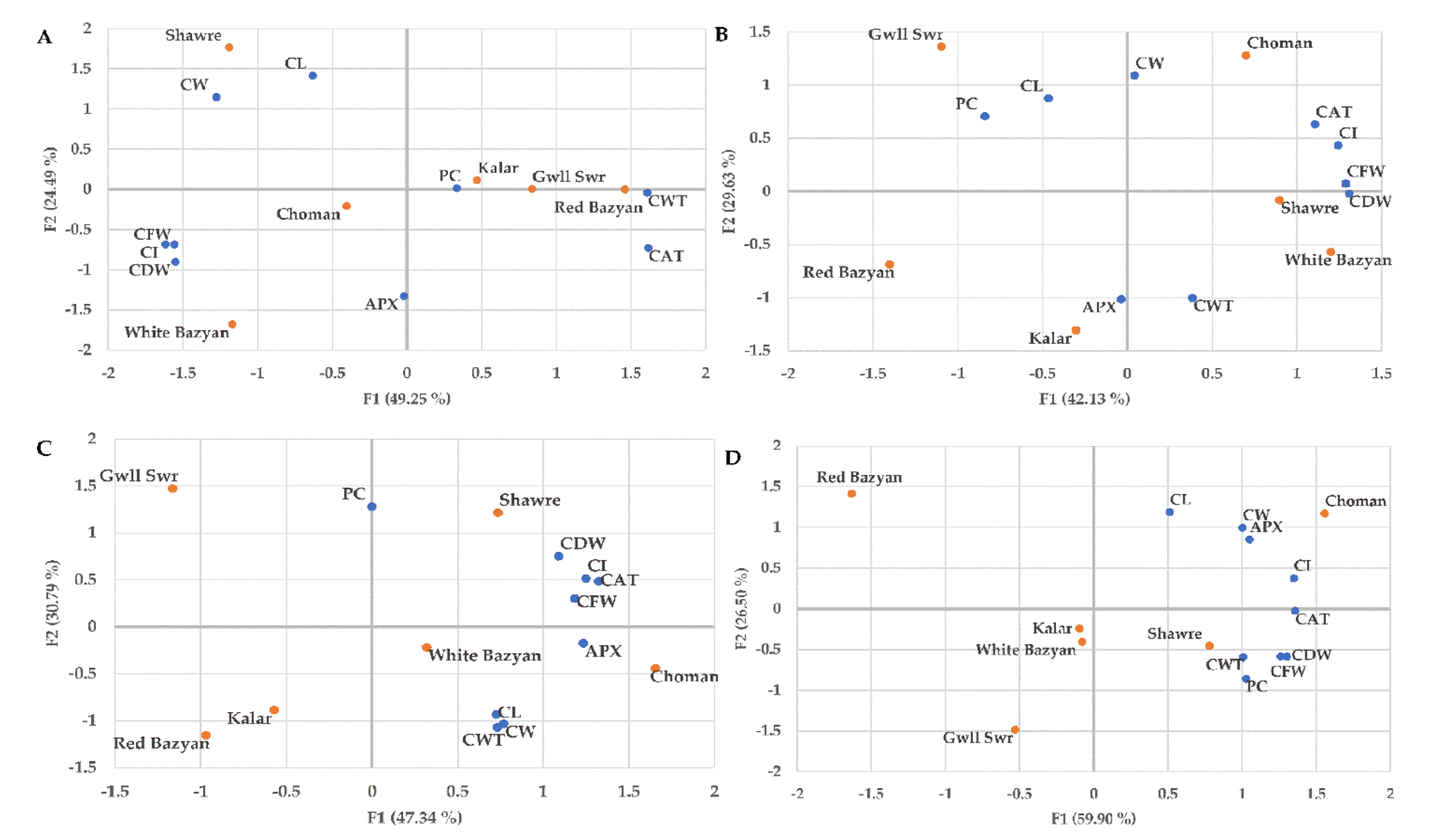
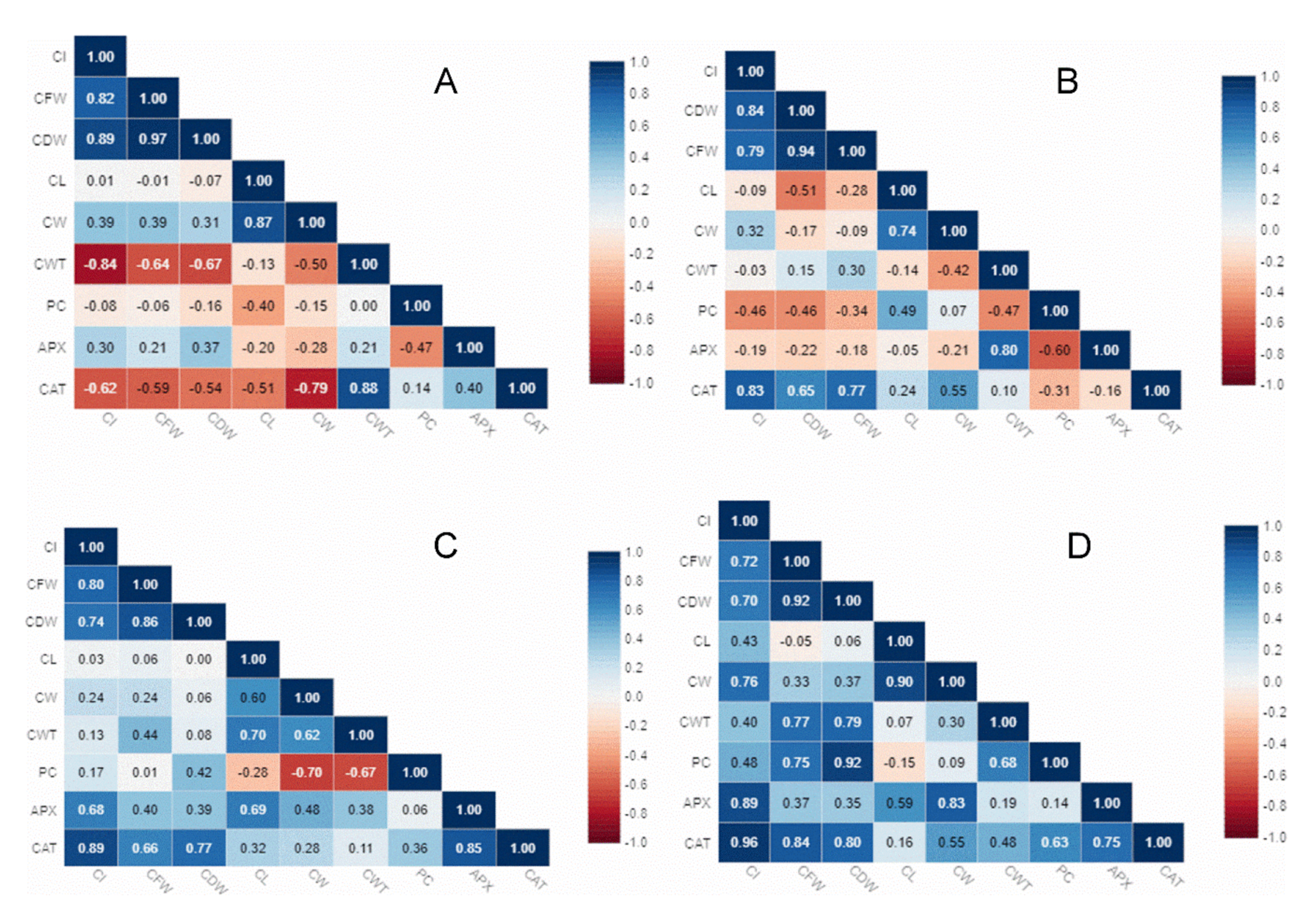
3.5. Relationship Analysis Between Phenotypic and Biochemical Traits under Optimum and Stress Conditions
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tripathi, K.K.; Warrier, R.; Govila, O.P.; Ahuja, T. Biology of Rice (Oryza sativa L.); Department of Biotechnology, Government of India and Ministry of Environment & Forests Government of India, India Government: New Delhi, India, 2011. [Google Scholar]
- William, B. IRAQ: Rice Production Drops by More Than Half Due to Lack of Irrigation; USDA-FAS, Office of Global Analysis: Washington, DC, USA, 2015; pp. 260–8109. [Google Scholar]
- Tahir, N.A. Genetic variability evaluation among Iraqi rice (Oryza sativa L.) varieties using RAPD markers and protein profiling. J. Biol. Sci. 2014, 7, 13–18. [Google Scholar]
- Khush, G.S.; Toenniessen, G.H. Rice Biotechnology; CAB International: Wallingford, UK, 1991; p. 320. [Google Scholar]
- Fadhil, A.M. Drought mapping using geoinformation technology for some sites in the Iraqi Kurdistan region. Int. J. Digit. Earth 2011, 4, 239–257. [Google Scholar] [CrossRef]
- Salehi-Lisar, S.Y.; Bakhshayeshan-Agdam, H. Drought Stress in Plants: Causes, Consequences, and Tolerance. In Drought Stress Tolerance in Plants; Physiology and Biochemistry; Hossain, M.A., Wani, S.H., Bhattacharjee, S., Burritt, D.J., Tran, L.-S.P., Eds.; Springer: Cham, Switzerland, 2016; Volume 1, pp. 1–16. [Google Scholar]
- Kumar, S.; Sachdeva, S.; Bhat, K.V.; Vats, S. Plant responses to drought stress: Physiological, biochemical and molecular basis. In Biotic and Abiotic Stress Tolerance in Plants; Springer: Singapore, 2018; pp. 1–2. [Google Scholar]
- Kamarudin, Z.S.; Yusop, M.R.; Mohamed, M.T.M.; Ismail, M.R.; Harun, A.R. Growth performance and antioxidant enzyme activities of advanced mutant rice genotypes under drought stress condition. Agronomy 2018, 8, 279. [Google Scholar] [CrossRef] [Green Version]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Muscolo, A.; Junker, A.; Klukas, C.; Weigelt-Fischer, K.; Riewe, D.; Altmann, T. Phenotypic and metabolic responses to drought and salinity of four contrasting lentil accessions. J. Exp. Bot. 2015, 66, 5467–5480. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Shukla, A.; Sairam, R.K. In vitro screening of rice genotypes for drought tolerance using polyethylene glycol. Acta Physiol. Plant 2011, 33, 2209–2217. [Google Scholar] [CrossRef]
- Islam, M.M.; Kayesh, E.; Zaman, E.; Urmi, T.A.; Haque, M.M. Evaluation of rice (Oryza sativa L.) genotypes for drought tolerance at germination and early seedling stage. Agriculturists 2018, 16, 44–54. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, Y.P.S. Biotechnology in rice improvement. In Biotechnology in Agriculture and Forestry; Springer: Berlin/Heidelberg, Germany, 1991; pp. 3–18. [Google Scholar]
- Yaqoob, U.; Jan, B.; Kaul, T.; Nawchoo, I.A. Tissue culture of rice: Problems, progress and prospects. Int. J. Innov. Res. Sci. Eng. Technol. 2016, 3, 581–613. [Google Scholar]
- Khan, S.; Javed, M.A.; Jahan, N.; Manan, F.A. A short review on the development of salt tolerant cultivars in rice. Int. J. Med. Sci. Public Health 2016, 5, 201–212. [Google Scholar]
- Al-Bahrany, A.M. Callus growth and proline accumulation in response to polyethylene glycol-induced osmotic stress in rice, Oryza sativa L. Pak. J. Biol. Sci. 2002, 5, 1294–1296. [Google Scholar]
- Islam, M.R.; Singh, R.K.; Salam, M.A.; Hassan, L.; Gregorio, G.B. Molecular diversity of stress tolerant rice genotypes using SSR markers. SABRAO J. Breed. Genet. 2008, 40, 127–139. [Google Scholar]
- Tahir, N.A.; Omer, D.A.; Lateef, D.D.; Ahmad, D.A.; Salih, S.H.; Khal, L.H. Diversity and population structure analysis of faba bean (Vicia faba L.) accessions using SSR markers. J. Agr. Sci. Technol. 2019, 21, 463–474. [Google Scholar]
- Youens-Clark, K.; Buckler, E.; Casstevens, T.; Chen, C.; Declerck, G.; Derwent, P.; Dharmawardhana, P.; Jaiswal, P.; Kersey, P.; Karthikeyan, A.S.; et al. Gramene database: Updates and extensions. Nucleic Acids Res. 2010, 39, D1085–D1094. [Google Scholar]
- Zahid, M.N.; Hasan, M.; Adil, M.; Hossain, M.M.; Mian, M.A.K. In vitro screening for salt tolerance in aromatic rice genotypes. J. Biosci. Bioeng. 2014, 1, 28–32. [Google Scholar]
- Ruzin, S.E. Plant Microtechnique and Microscopy; Oxford University Press: Oxford, UK, 1999; p. 322. [Google Scholar]
- Aebi, H. Catalase in vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar] [PubMed]
- Rao, M.V.; Paliyath, G.; Ormrod, D.P.; Murr, D.P.; Watkins, C.B. Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes. Salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol. 1997, 115, 137–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carillo, P.; Gibon, Y. Protocol: Extraction and determination of proline; PrometheusWiki: Clayton South, Australia, 2011. [Google Scholar]
- Ketata, H.Y.; Yau, S.K.; Nachit, M. Relative consistency performance across environments. In Proceedings of the International Symposium on Physiology and Breeding of Winter Cereals for Stressed Mediterranean Environments, Montpellier, France, 3–6 July 1989; pp. 391–400. [Google Scholar]
- Pour-Aboughadareh, A.; Yousefian, M.; Moradkhani, H.; Moghaddam Vahed, M.; Poczai, P.; Siddique, K.H.M. iPASTIC: An online toolkit to estimate plant abiotic stress indices. Appl. Plant Sci. 2019, 7, e11278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, T.; Kramer, D.M.; Raines, C.A. Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Curr. Opin. Biotech. 2012, 23, 215–220. [Google Scholar] [CrossRef]
- Donde, R.; Kumar, J.; Gouda, G.; Gupta, M.K.; Mukherjee, M.; Baksh, S.Y.; Mahadan, P.; Sahoo, K.K.; Behera, L.; Dash, S.K. Assessment of genetic diversity of drought tolerant and susceptible rice genotypes using microsatellite markers. Rice Sci. 2019, 26, 239–247. [Google Scholar] [CrossRef]
- Li, J.Z.; Chen, Y.H.; Zhang, D.J.; Zhang, G.F.; Lu, B.X. Genetic diversity analysis and DNA fingerprinting of main Japonica rice varieties in Heilongjiang Province. Qual. Assur. Saf. Crops Foods 2019, 11, 23–29. [Google Scholar] [CrossRef]
- Perez-Almeida, I.; Celi-Hearn, R.; Sanchez-Mora, F.; Paz-Carrasco, L.; Ramos-Viteri, B. Assessment of molecular diversity of Ecuadorian rice cultivars using simple sequence repeat markers. BioAgro 2019, 31, 3–12. [Google Scholar]
- Singh, R.K.; Sharma, R.K.; Singh, A.K.; Singh, V.P.; Dingh, N.K.; Tiwari, S.P.; Mohapatara, T. Suitability of mapped sequence tagged microsatellite markers for establishing distinctness, uniformity and stability in aromatic rice. Euphytica 2004, 135, 135–143. [Google Scholar] [CrossRef]
- Muto, C.; Ebana, K.; Kawano, K.; Bounphanousay, V.; Bouphanousay, C.; Kanyavong, K.; Inthapanya, P.; Boualaphanh, C.; Sato, T.; Ishikawa, R.; et al. Genetic variation in rice (Oryza sativa L.) germplasm from northern Laos. Breed. Sci. 2019, 69, 272–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wani, S.H.; Sofi, P.A.; Gosal, S.S.; Singh, N.B. In vitro screening of rice (Oryza sativa L.) callus for drought tolerance. Commun. Biometry Crop Sci. 2010, 5, 108–115. [Google Scholar]
- Nahar, S.; Sahoo, L.; Tanti, B. Screening of drought tolerant rice through morpho-physiological and biochemical approaches. Biocatal. Agric. Biotechnol. 2018, 15, 150–159. [Google Scholar] [CrossRef]
- Bunnag, S.; Suwanagul, A. Improvement of drought tolerance in Thai rice cultivar RD6 through somaclonal variation. Songklanakarin J. Sci. Technol. 2017, 39, 723–729. [Google Scholar]
- Inostroza, L.; Acuña, H.; Tapia, G. Relationships between phenotypic variation in osmotic adjustment, water use efficiency, and drought tolerance of seven cultivars of Lotus corniculatus L. Chik. J. Agr. Res. 2015, 75, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, P.; Amirnia, R.; Majidi, E.; Hadi, H.; Roustaii, M.; Nakhoda, B. Relationship between drought stress and some antioxidant enzymes with cell membrane and chlorophyll stability in wheat lines. Afr. J. Microbiol. Res. 2012, 6, 617–623. [Google Scholar]
- Swapna, S.; Shylaraj, K.S. Screening for osmotic stress responses in rice varieties under drought condition. Rice Sci. 2017, 24, 253–263. [Google Scholar] [CrossRef]
| Genotype Code | Genotype Name | Kurdistan Location |
|---|---|---|
| G1 | Choman | Penjwen |
| G2 | Gwll Swr | Hajiawa |
| G3 | Kalar | Kalar |
| G4 | Red Bazyan | Bazyan |
| G5 | Shawre | Raniyah |
| G6 | White Bazyan | Bazyan |
| SSR Marker | Chromosome Number | Amplicon Size (bp) | Number of Alleles | PIC | DI | PI |
|---|---|---|---|---|---|---|
| RM10793 | 1.00 | 140.00–160.00 | 3.00 | 0.51 | 0.57 | 0.25 |
| RM16 | 3.00 | 140.00–180.00 | 4.00 | 0.57 | 0.61 | 0.19 |
| RM241 | 4.00 | 120.00–140.00 | 2.00 | 0.38 | 0.50 | 0.38 |
| RM13 | 5.00 | 120.00–160.00 | 3.00 | 0.60 | 0.66 | 0.16 |
| RM400 | 6.00 | 140.00–160.00 | 3.00 | 0.54 | 0.61 | 0.22 |
| RM418 | 7.00 | 140.00–160.00 | 2.00 | 0.38 | 0.50 | 0.38 |
| RM443 | 8.00 | 240.00–300.00 | 5.00 | 0.77 | 0.78 | 0.04 |
| RM23818 | 9.00 | 100.00–140.00 | 3.00 | 0.52 | 0.53 | 0.19 |
| RM228 | 10.00 | 180.00–200.00 | 4.00 | 0.69 | 0.72 | 0.09 |
| RM287 | 11.00 | 80.00–120.00 | 4.00 | 0.84 | 0.84 | 0.02 |
| RM309 | 12.00 | 160.00–180.00 | 2.00 | 0.38 | 0.50 | 0.38 |
| Average | 3.18 | 0.56 | 0.62 | 0.21 |
| Genotype | Choman | Gwll Swr | Kalar | Red Bazyan | Shawre | White Bazyan |
|---|---|---|---|---|---|---|
| Choman | 0.00 | |||||
| Gwll Swr | 0.85 | 0.00 | ||||
| Kalar | 0.67 | 0.88 | 0.00 | |||
| Red Bazyan | 0.79 | 0.78 | 0.77 | 0.00 | ||
| Shawre | 0.71 | 0.63 | 0.71 | 0.63 | 0.00 | |
| White Bazyan | 0.33 | 0.85 | 0.68 | 0.67 | 0.78 | 0.00 |
| CI (%) | CFW (g) | CDW (g) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 |
| Choman | 86.67 ab | 86.67 a | 86.67 a | 86.67 a | 1.33 ab | 1.18 a | 1.07 a | 0.89 a | 0.64 b | 0.48 ab | 0.43 b | 0.38 a |
| Gwll Swr | 80.00 bc | 66.67 b | 53.33 b | 53.33 c | 1.01 c | 0.80 c | 0.74 c | 0.72 c | 0.46 d | 0.38 c | 0.34 c | 0.32 bc |
| Kalar | 73.33 bc | 53.33 bc | 60.00 b | 40.00 d | 1.19 b | 1.02 b | 0.90 b | 0.80 b | 0.54 cd | 0.45 b | 0.36 c | 0.34 b |
| Red Bazyan | 66.67 c | 46.67 c | 53.33 b | 40.00 d | 1.00 c | 0.67 c | 0.49 d | 0.40 d | 0.44 d | 0.28 d | 0.21 d | 0.16 d |
| Shawre | 86.67 ab | 86.67 a | 86.67 a | 80.00 ab | 1.22 b | 1.06 ab | 0.99 a | 0.91 a | 0.58 bc | 0.52 ab | 0.48 a | 0.38 a |
| White Bazyan | 100.00 a | 100.00 a | 86.67 a | 66.67 bc | 1.40 a | 1.15 ab | 1.02 a | 0.30 c | 0.76 a | 0.54 a | 0.35 c | 0.30 c |
| CL (µm) | CW (µm) | CWT (µm) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 |
| Choman | 38.90 b | 33.50 a | 31.07 a | 29.47 a | 26.37 b | 19.77 a | 14.67 a | 16.20 a | 2.00 b | 2.43 a | 2.90 a | 3.33 a |
| Gwll Swr | 33.57 c | 29.77 c | 17.77 f | 16.10 f | 17.83 d | 17.87 b | 9.93 b | 10.13 e | 1.93 b | 1.93 c | 2.53 c | 3.17 ab |
| Kalar | 32.63 c | 22.03 e | 23.43 c | 24.77 b | 17.83 d | 10.73 d | 14.20 a | 13.03 b | 1.97 b | 2.57 a | 2.83 a | 3.10 ab |
| Red Bazyan | 39.10 b | 31.20 b | 25.90 b | 23.97 c | 17.87 d | 17.47 b | 14.67 a | 12.17 cd | 2.37 a | 2.53 a | 2.67 bc | 2.23 d |
| Shawre | 47.80 a | 22.03 e | 20.53 d | 19.53 d | 33.23 a | 17.17 b | 14.17 a | 12.50 c | 1.67 c | 2.27 b | 2.57 c | 2.23 d |
| White Bazyan | 33.17 c | 25.63 d | 19.30 e | 17.80 e | 20.00 c | 15.47 c | 14.33 a | 11.83 d | 1.67 c | 2.57 a | 2.77 ab | 3.03 b |
| Stress Level | Index | Choman | Gwll Swr | Kalar | Red Bazyan | Shawre | White Bazyan |
|---|---|---|---|---|---|---|---|
| T0.50 | STI | 1.06 | 0.75 | 0.54 | 0.59 | 1.05 | 1.08 |
| AR | 2.18 | 3.64 | 5.82 | 4.73 | 3.55 | 1.09 | |
| Rank | 2 | 4 | 6 | 5 | 3 | 1 | |
| T1.00 | STI | 1.01 | 0.54 | 0.49 | 0.50 | 1.01 | 0.92 |
| AR | 1.45 | 4.73 | 5.55 | 4.64 | 2.09 | 2.55 | |
| Rank | 1 | 5 | 6 | 4 | 2 | 3 | |
| T1.50 | STI | 1.01 | 0.53 | 0.61 | 0.55 | 0.94 | 0.74 |
| AR | 1.18 | 5.64 | 3.18 | 4.36 | 2.73 | 3.91 | |
| Rank | 1 | 6 | 3 | 5 | 2 | 4 | |
| Overall | Sum of Ranks | 4 | 15 | 15 | 14 | 7 | 8 |
| Overall Rank | 1 | 5 | 5 | 4 | 2 | 3 |
| Proline Content (µmol/g FW) | APX (nkat/mg FW) | CAT (nkat/mg FW) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 |
| Choman | 0.30 a | 0.36 b | 0.40 b | 0.49 b | 0.57 b | 0.86 bc | 1.61 a | 2.32 a | 9.11 b | 26.98 a | 37.58 a | 48.91 a |
| Gwll Swr | 0.31 a | 0.46 a | 0.49 a | 0.62 a | 0.54 b | 0.75 c | 1.10 c | 1.23 d | 9.66 ab | 17.55 c | 25.93 c | 37.65 c |
| Kalar | 0.28 b | 0.35 b | 0.33 c | 0.56 b | 0.51 b | 0.93 abc | 1.03 c | 1.24 d | 8.99 b | 16.55 c | 22.35 d | 38.03 c |
| Red Bazyan | 0.12 d | 0.29 c | 0.34 c | 0.38 d | 0.59 a | 1.11 a | 1.27 b | 1.54 c | 10.49 a | 16.90 c | 24.56 cd | 33.21 d |
| Shawre | 0.16 c | 0.18 d | 0.45 a | 0.64 a | 0.52 b | 0.90 bc | 1.33 b | 1.80 b | 7.22 c | 22.63 b | 36.39 a | 49.35 a |
| White Bazyan | 0.15 c | 0.21 d | 0.31 c | 0.48 c | 0.61 a | 1.02 ab | 1.26 b | 1.59 c | 8.54 c | 22.60 b | 29.53 b | 42.96 b |
| Stress | Index | Choman | Gwll Swr | Kalar | Red Bazyan | Shawre | White Bazyan |
|---|---|---|---|---|---|---|---|
| T0.50 | STI | 2.94 | 2.06 | 1.82 | 2.14 | 1.96 | 2.32 |
| AR | 1.73 | 3.82 | 5.00 | 3.82 | 3.73 | 2.91 | |
| Rank | 1 | 4 | 5 | 4 | 3 | 2 | |
| T1.00 | STI | 4.13 | 3.03 | 2.42 | 3.07 | 3.15 | 3.03 |
| AR | 1.82 | 3.64 | 5.18 | 3.82 | 2.82 | 3.73 | |
| Rank | 1 | 3 | 6 | 5 | 2 | 4 | |
| T1.50 | STI | 5.41 | 4.35 | 4.07 | 4.12 | 4.28 | 4.38 |
| AR | 1.91 | 3.82 | 4.36 | 4.45 | 3.18 | 3.27 | |
| Rank | 1 | 4 | 5 | 6 | 2 | 3 | |
| Overall | Sum of Ranks | 3 | 11 | 16 | 15 | 7 | 9 |
| Overall Rank | 1 | 4 | 6 | 5 | 2 | 3 |
| Sodium-Na (µg/g) | Magnesium-Mg (µg/g) | Phosphorus-P (µg/g) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 |
| Choman | 277.18 ab | 200.36 b | 197.04 a | 226.60 a | 2176.58 ab | 945.64 c | 996.51 c | 1085.92 c | 9666.19 b | 6149.89 c | 6135.53 c | 6548.81 cd |
| Gwll Swr | 89.98 c | 128.92 b | 76.83 c | 88.50 b | 1032.93 b | 909.20 c | 1180.11 bc | 1207.69 bc | 6450.28 c | 4775.06 c | 6690.95 c | 6015.10 d |
| Kalar | 223.65 bc | 142.50 c | 142.01b | 158.82 ab | 2793.61 a | 1493.32 b | 1492.67 b | 1492.76 b | 10,710.00 b | 9133.18 b | 9133.02 b | 8473.59 b |
| Red Bazyan | 262.30 ab | 368.89 a | 210.28 a | 147.08 ab | 3066.46 a | 3466.10 a | 2596.67 a | 2256.66 a | 13,125.19 a | 12101.34 a | 14,121.74 a | 12,135.20 a |
| Shawre | 382.13 a | 99.03 c | 83.42 c | 86.48 b | 2546.66 a | 1606.44 b | 1445.75 b | 1180.56 bc | 6960.24 c | 6106.44 c | 7774.10 bc | 5880.94 d |
| White Bazyan | 153.07 bc | 208.59 b | 181.91 ab | 226.84 a | 1245.43 b | 1712.43 b | 1084.96 bc | 1212.17 bc | 7843.00 c | 6712.43 bc | 7606.30 bc | 7499.05 bc |
| Sulfur-S (µg/g) | Potassium-K (µg/g) | Calcium-Ca (µg/g) | ||||||||||
| Genotype | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 | T0.00 | T0.50 | T1.00 | T1.50 |
| Choman | 5646.40 a | 4767.39 b | 2280.43 b | 4924.31 b | 47,508.16 a | 21,935.74 b | 21,366.27 d | 24,449.70 b | 7097.42 a | 1869.90 c | 2144.98 b | 2980.92 b |
| Gwll Swr | 3964.31 b | 10,731.37 a | 15,361.57 a | 10,632.72 a | 22,382.91 b | 19,319.31 b | 21,886.44 d | 26,182.67 ab | 1127.05 b | 1648.26 c | 2161.53 b | 6551.41 a |
| Kalar | 4699.84 ab | 3773.94 b | 3773.38 b | 3670.19 b | 46,733.60 a | 34,232.55 ab | 34,232.13 b | 33,857.95 ab | 5034.94 a | 2595.58 b | 2595.08 b | 2612.69 b |
| Red Bazyan | 5473.38 a | 4886.95 b | 6428.98 b | 5002.00 b | 49,711.49 a | 45,255.60 a | 48,698.00 a | 37,482.84 a | 5902.47 a | 6507.31 a | 5083.59 a | 3185.23 b |
| Shawre | 1900.55 c | 3717.18 b | 1482.66 b | 1764.29 b | 28,554.20 b | 22,420.70 b | 32,474.17 bc | 24,190.05 b | 6014.87 a | 1706.08 c | 2448.78 b | 1670.19 b |
| White Bazyan | 2335.68 c | 2268.83 b | 3472.13 b | 2931.85 b | 26,115.37 b | 22,651.95 b | 23,770.32 cd | 26,389.31 ab | 2436.20 b | 2329.11 b | 1983.52 b | 2605.32 b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahim, D.; Kalousek, P.; Tahir, N.; Vyhnánek, T.; Tarkowski, P.; Trojan, V.; Abdulkhaleq, D.; Ameen, A.H.; Havel, L. In Vitro Assessment of Kurdish Rice Genotypes in Response to PEG-Induced Drought Stress. Appl. Sci. 2020, 10, 4471. https://doi.org/10.3390/app10134471
Rahim D, Kalousek P, Tahir N, Vyhnánek T, Tarkowski P, Trojan V, Abdulkhaleq D, Ameen AH, Havel L. In Vitro Assessment of Kurdish Rice Genotypes in Response to PEG-Induced Drought Stress. Applied Sciences. 2020; 10(13):4471. https://doi.org/10.3390/app10134471
Chicago/Turabian StyleRahim, Didar, Petr Kalousek, Nawroz Tahir, Tomáš Vyhnánek, Petr Tarkowski, Vaclav Trojan, Dana Abdulkhaleq, Ahmad Hama Ameen, and Ladislav Havel. 2020. "In Vitro Assessment of Kurdish Rice Genotypes in Response to PEG-Induced Drought Stress" Applied Sciences 10, no. 13: 4471. https://doi.org/10.3390/app10134471








