Abstract
The sea bottom acts as a key natural archive where the memory of long-term timescale environmental changes is recorded. This study discusses some ecological and chemical features of fjord sediments that were explored during the AREX cruise carried out in the Svalbard archipelago in the summer of 2021. The activity rates of the enzymes leucine aminopeptidase (LAP), beta-glucosidase (GLU), and alkaline phosphatase (AP) and community-level physiological profiles (CLPPs) were studied with the aim of determining the functional diversity of the benthic microbial community, while bacterial isolates were screened for their susceptibility to antibiotics in order to explore the role of these extreme environments as potential reservoirs of antibiotic resistance. Enzyme activity rates were obtained using fluorogenic substrates, and CLPPs were obtained using Biolog Ecoplates; antibiotic susceptibility assays were performed through the standard disk diffusion method. Spatial trends observed in the functional profiles of the microbial community suggested variability in the microbial community’s composition, presumably related to the patchy distribution of organic substrates. Complex carbon sources, carbohydrates, and amino acids were the organic polymers preferentially metabolized by the microbial community. Multi-resistance to enrofloxacin and tetracycline was detected in all of the examined samples, stressing the role of sediments as a potential reservoir of chemical wastes ascribable to antibiotic residuals. This study provides new insights on the health status of fjord sediments of West Spitsbergen, applying a dual ecological and biochemical approach. Microbial communities in the fjord sediments showed globally a good functional diversity, suggesting their versatility to rapidly react to changing conditions. The lack of significant diversification among the three studied areas suggests that microbial variables alone cannot be suitable descriptors of sediment health, and that additional measures (i.e., physical–chemical characteristics) should be taken to better define environmental status.
1. Introduction
The Svalbard archipelago is one of the regions on Earth most vulnerable to climate warming, but it is still unexplored regarding its microbiological features [1]. Fjord ecosystems are considered sensitive sentinels of climate change, affected by melting glaciers and warming seawater [2]. Fjords influenced by active glacier freshwater inputs are key hotspots for the burial of organic matter in sediments and subsequent microbial mineralization [3]. Both the freshwater supply and sedimentary processes drive the physical characteristics of fjords, making them natural laboratories where the effects of environmental changes can be detected and monitored. Particularly, fjord systems where temperature, salinity, nutrient, and light gradients occur can serve as suitable models for studying the response of microbial communities to environmental variability [4].
Being an extreme polar environment, the Svalbard archipelago has traditionally been considered as a pristine environment; however, in its continental area, mining, sewage wastes and dumpsites have been reported to be the main sources of terrestrial contamination, which runoff may discharge into the surrounding water bodies (including the sea), making the whole region particularly vulnerable to potential impacts of anthropic activities [5,6]. Marine sediments act as significant environmental archives, playing a key role as reservoirs of organic carbon [7] and contaminants that may accumulate into the benthic zone. Indeed, over the last few decades, the presence of pollutants in Svalbard marine sediments has been documented [8,9,10,11], providing a picture of a widespread phenomenon. Several studies indicate moderate enrichments of Svalbard marine sediments with heavy metals [12,13,14,15] and organic pollutants [8,16,17,18], resulting in bioaccumulation and biomagnification in the trophic chain [17,19,20]. The Svalbard archipelago receives pollutants mainly from global remote transport (air masses, ocean currents, ice drift, riverine discharge, etc.) (e.g., [21]). The rapid climate warming experienced by the Arctic region is expected to result in the mobilization/transport of freshwater through glacier systems, although currently only fragmentary data are available on the possible effects caused by the transport of contaminants stored in glaciers and permafrost. Local pollution sources connected mainly to human settlements as well as former and present coal mining can be important too [16,22,23]. The level of human impact on Svalbard fjords differs geographically. Kongsfjorden is influenced by high touristic activity (large cruise liners) [24], its marina, and its big research station. Isfjorden is exposed to intense human activity connected to marine and air traffic. Particularly, Adventfjorden is impacted by its town (having a permanent population of 2500 people, but being a very important touristic destination), harbors and docks’ shipments of coal [25], and the discharge of untreated sewage [26]. Hornsund is a less popular touristic destination with a small research station; this fjord is characterized by important sedimentary concentrations of some metals [15,27] discharged by melting glaciers as well as organic contaminants [20]. Not only legacy but also emerging pollutants have been found in Svalbard. The presence of pharmaceutical residues introduced mainly by untreated sewage in Kongsfjorden [28] and Isfjorden [29] has been reported. In Isfjorden, many pharmaceuticals (carbamazepine, diclofenac, caffeine, paracetamol, ciprofloxacin, and tetracycline) have been found to effectively accumulate in the tissues of biota, while some others (e.g., citalopram) have shown even biomagnification potential [30].
Anthropogenic activities, causing the release of wastewater effluents and chemical pollutants, may affect the abundance, distribution, and taxonomic and functional diversity of the bacterial community [22,31]. Chemical pollution generally leads to increased microbial abundance and diversity, altering community functioning, and causing, in turn, severe impacts on human health, for example through the spread of pathogens and antibiotic-resistant bacteria in polluted marine environments [32]. On the other hand, bacteria drive the transfer of toxic compounds to higher trophic levels, affecting the environmental fate of several pollutants [33].
In the benthic domain, microbial communities are major drivers of the cycling of nutrients, enabling efficient organic matter turnover and affecting the biogeochemical properties of sediments, like that reported in terrestrial soil [34]. Moreover, microorganisms respond quickly to environmental changes, modulating their composition and consequently their metabolic spectra. Thus, microbial enzymes’ activities—both in terms of activity rates and community profiles—can provide useful information for assessing environmental health [35,36]. As stated by Maher et al. [37], a healthy sediment “may be defined as one that supports an active and diverse biological population, and functions satisfactorily”. Sediments host biological communities, including algae, macrophytes, benthic invertebrates, and bacteria, and sediment health can be described through biological measurements, including species abundance and biomass, richness and similarity, and functional measurements such as the rates of organic matter production and decomposition, community respiration, or nutrient uptake. The concept of sediment health encompasses both biological and chemical properties, and the aforementioned authors [37] underlined how chemical measurements can be used as a surrogate for assessing sediment health. Insights gained at the microbial community level can be helpful for proposing new indicators of ecosystem health or developing new environmental health indices [38]. In this context, heterotrophic microorganisms are a significantly active fraction of the microbial community, playing a pivotal role in biogeochemical and energy fluxes [39]. The composition and structure of microbial communities in marine sediments is affected by several biotic and abiotic factors, such as temperature, the availability and quality of organic matter and nutrients, variations in the redox gradient, and the interactions and selective pressure among different microbial taxa [40]. In Svalbard marine sediments, a clear knowledge of the patterns of microbial taxonomical and functional diversity and their environmental drivers is still lacking, although assessing how microbial structure and function are modulated by freshwater, terrestrial, and marine organic carbon inputs would be critical for understanding the role of microbial communities in the whole fjord ecosystems’ functioning. Characterizing and quantifying the metabolic potential of microbial communities in benthic environments is of great interest for obtaining insights on the physiological role of prokaryotic assemblage in the carbon cycle. To this end, the Biolog Ecoplate method provides a simple and rapid tool for determining carbon substrates’ utilization patterns, and to get a picture of physiological profiles at a community level, as documented by previous studies performed in temperate and polar ecosystems [41,42,43]. This information on the biogeochemical role of microbial communities might be crucial for understanding how future changes in benthic communities’ dynamics may influence the efficiency of biological carbon pumping and, ultimately, the overall ecosystem functioning; in fact, evidence that sediment bacterial community structure and function are controlled through local processes such as increased transport of terrestrial organic matter and increased riverine discharge has recently been provided [44].
During the summer of 2021, a survey (AREX) was carried out in Svalbard by the Institute of Oceanology PAS (Sopot, Poland) and a multidisciplinary set of physical–chemical and biological variables were measured. In the light of the above-reported considerations and to get an updated picture of the environmental state, a quantitative and qualitative microbiological study was carried out in fjords differently affected by natural and anthropogenic factors. Particularly, two main hypotheses were tested:
- (i)
- Since there were differences in the intensity of human activity and the sediment pollution within the selected fjords [45], a higher abundance of antibiotic-resistant bacteria was expected to be found within the benthic microflora in human-impacted areas (e.g., Adventfjorden);
- (ii)
- As a glacier–marine transition occurs from the inner part to the outer part of the fjords, we hypothesized that this spatial gradient would be reflected in a greater complexity of carbon substrate utilization, with a potential shift from simple to complex carbon metabolism when moving from glacier to open-sea environments.
To verify this last hypothesis, the patterns of microbial metabolism were determined, focusing on the study of extracellular enzymatic activity rates and community-level physiological profiles (CLPPs); the potential impact of the pollution gradient on the abundance of antibiotic-resistant bacteria was tested by assessing the susceptibility of isolated bacterial strains to commonly used antibiotics, in order to detect the possible occurrence of (multi-)antibiotic-resistant bacteria.
Moreover, a presumptive identification of the benthic heterotrophic microbial community was performed, based on its phenotypic and biochemical characteristics.
2. Materials and Methods
2.1. Study Area and Sample Collection
Several fjords of the Svalbard archipelago on the western side of Spitsbergen covering the northernmost to the southernmost areas were investigated in this study: In the northern area, Krossfjorden (with stations KR1 to KR3, from the inner to the outer areas) and Kongsfjorden (stations KO1–KO3) were studied. In the central area, sampling was performed in Dicksonfjorden (station D1) and Isfjorden (stations IS1–IS3), with its branches of Nordfjorden (station N1), Sassenfjorden (station S1), Dicksonfjorden (station D1), Adventfjorden (station A1), and Grønfjorden (station G1). In the southern area, Hornsund (stations H1 to H3, from the inner part to the outer part of the fjord) was studied.
Krossfjorden and Kongsfjorden are two fjords located in the northern area on the western side of Spitsbergen. During summer, both receive sediment inputs from glacial meltwater, leading to decreased surface salinity and increased turbidity; all of these variables were found to affect the distribution of the bacterial community in the water column [1,46].
Isfjorden, with a mean width of 24 km, and a length of 107 km, is the largest fjord in western Spitsbergen; it receives freshwater from tidewater glaciers as well as from rivers fed by alpine glaciers and precipitation. Isfjorden seawater is of both Atlantic and Arctic origin. In this fjord, local processes produce the Local Water and the Winter- Cooled Water; mixed water masses such as the Intermediate Water and the Transformed Atlantic Water are also found [47].
Hornsund is a large 200 m deep fjord located in the southernmost area of Spitsbergen, opening to the Greenland Sea; it receives seasonal sediment inputs through melting water discharge from eight major tidewater glaciers. This fjord is affected by the Atlantic Core Water, Transformed Atlantic Water and Arctic Water [48].
The geographical coordinates of the sampled stations and the main characteristics of the sediments are shown in Table 1 and Figure 1. Surface (1–2 cm) sediment samples were collected using the Van Veen grab method by the researchers of the Institute of Oceanology PAS (Sopot, Poland); for the microbiological study, aliquots of the samples were stored in 50 mL sterile Falcon tubes at −20 °C for further processing, carried out upon their arrival at the CNR-ISP laboratories in Messina (Italy).

Table 1.
Geographical coordinates of the sampled stations and characteristics of the sediments.
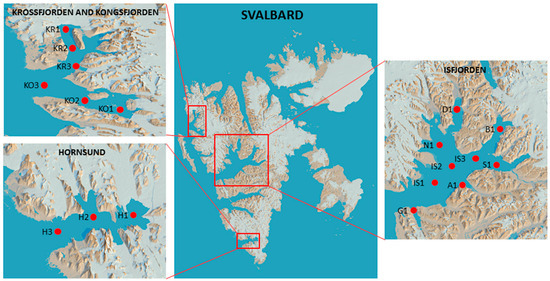
Figure 1.
Locations of the sampling sites (https://toposvalbard.npolar.no/ [49], accessed on 5 July 2024).
2.2. Heterotrophic (Marine and Non-Marine) Bacterial Abundance
Volumes (100 μL) of sediment suspension (obtained through sediment dilution in a 0.9% sodium chloride sterile physiological saline in a ratio of 1:10 w/v) were spread onto Marine Agar 2216 and Tryptic Soy Agar (Difco) + 2% sodium chloride plates in duplicate, which were incubated at +5 °C for at least 14 days in order to allow the growth of marine and non-marine bacteria, respectively. The colonies grown on both culture media were then counted. Bacterial strains were isolated by streaking them on plates of the same culture media until axenic cultures were obtained. During the isolation, colonies showing different phenotypic characteristics (i.e., morphology, size, and pigmentation), with at least one colony per each typology, were streaked in order to obtain a collection of bacterial isolates as representative as possible of the whole culturable bacterial community present in each sample.
2.3. Extracellular Enzymatic Activities (Leucine Aminopeptidase, LAP, Beta-Glucosidase, GLU, and Alkaline Phosphatase, AP)
Microbial ectoenzymatic activity measurements were performed to estimate the potential activity rates of leucine aminopeptidase (LAP), beta-glucosidase (GLU), and alkaline phosphatase (AP), enzymes involved in the decomposition mediated by the microbial community of proteins, polysaccharides, and organic phosphates, respectively [1]. These enzymatic assays relied on the hydrolysis of specific fluorogenic substrates, L-leucine-7 amido-4-methylcoumarin hydrochloride (Leu-MCA), 4-methylumbelliferyl-b-d-glucoside, and methylumbelliferyl phosphate, respectively, which are derivatives of methylcoumarin (MCA) and of methylumbelliferone (MUF), following the method reported by Hoppe [50]. Increasing amounts (from 20 to 400 µmol) of substrates were added to 10 mL sub-volumes of sediment supernatant and fluorometric measurements were performed at the initial time and after incubation at an “in situ” temperature for 2 h. Through calibration with the standard curves obtained with known amounts of MCA for LAP and MUF for GLU and AP, the enzymatic values were expressed in terms of maximum velocity of hydrolysis (Vmax, in nmol of substrate hydrolyzed per gram and per hour, nmol/g/h).
2.4. Community-Level Physiological Profiles (CLPPs) via Biolog Ecoplates
Microbial community metabolism was studied through the analysis of the carbon substrate utilization patterns [51,52,53]. Biolog Ecoplates (Rigel Life Sciences, Rome, Italy) were used to determine the differences in the metabolic potentials of microbial assemblages hosted in the samples. Each plate was a 96-well microtiter plate containing 31 carbon substrates and a control in triplicate together with the redox dye tetrazolium violet. The Biolog plates were inoculated with 150 μL of sample and incubated at 22 °C in the dark under aerobic conditions. The oxidation of the carbon substrates into formazan was quantified through absorbance measurements at 590 nm using the compact plate reader Byonoy Absorbance 9 (Hamburg, Germany). The optical density (OD) of the reaction product was measured at time zero (T0, namely immediately after inoculation), and at fixed time intervals until 35 days of incubation. The color development as a violet pigment in each plate well was expressed in terms of the averaged well color development (AWCD), which was calculated using the following formula:
where R is the averaged absorbance of three wells with the substrate and C is the averaged absorbance of the control wells (without substrate) (according to Sala et al. [52]).
AWCD = Σ [(R − C)/31],
The absorbance percentages of each substrate were determined in agreement with Sala et al.’s method [53], setting a value of 2% absorbance of the total absorbance measured per plate as a threshold for substrate utilization.
The carbon substrates were grouped into the following six guilds: complex carbon sources (polymers); carbohydrates; phosphate–carbon sources; carboxylic and acetic acids; amino acids; and amines.
2.5. Phenotypic and Biochemical Characterization of Bacterial Isolates
Bacterial isolates were phenotypically characterized for their presumptive identification and classified according to cell morphology, Gram’s staining, pigment production, oxidase production, colony morphology, and catalase testing [54]. Biochemical characterization of the isolates, using methods such as sugar fermentation tests on Triple Sugar Iron agar for glucose, lactose, and sucrose utilization, was also performed.
2.6. Antibiotic Susceptibility Profiles of Bacterial Isolates
The screening of bacterial isolates for their susceptibility to antibiotics was performed according to the procedure reported by Laganà et al. [55]. Briefly, the bacterial isolates were screened for their antibiotic susceptibility using the Bauer’s test [56], performed with three replicates per isolate. The isolates were grown for 48 h on plates of Tryptic Soy Agar (TSA, Oxoid, Thermo Fisher Diagnostics S.p.A., Rodano, Milan, Italy), harvested, and then suspended in sterile water adjusted to a 0.5 McFarland turbidity standard (bioMérieux, Marcy-l’Etoile, France), corresponding to 1.5 × 108 CFU mL−1. The inoculum was streaked onto plates of Mueller–Hinton agar using a cotton swab; the produced diameters of inhibition were measured after 48 h of incubation at 5 °C and averaged. Commercially available antibacterial disks (Oxoid) were used to determine the resistance patterns of the isolates against 34 different antibacterials (dose/disk as found in the common reagents), grouped into the following categories according to their mechanisms of action:
- -
- Cell wall antibiotics such as beta-lactams, including penicillins [ampicillin (10 μg, code CT0003B)];
- -
- Nucleic acid inhibitors, including fluoroquinolones [enrofloxacin (5 μg, CT0639B)];
- -
- Protein synthesis inhibitors, including (1) aminoglycoside antibiotics [gentamycin (10 μg, CT0024B)], (2) lincosamides [clindamycin (2 μg, CT0064B)], and (3) tetracyclines [tetracycline (30 μg, CT0054B)].
The diameter of the zone of inhibition around each antibiotic disk was measured with a precision caliper (Mitutoyo, Andover, UK). Each bacterial isolate was classified as resistant (R) or sensitive (S) according to the breakpoints established by EUCAST [57].
2.7. Statistical Analyses
Pearson correlation coefficients among the abundance, metabolic, and biodiversity indices were computed after logarithmic transformation of the data that failed to follow a normal distribution. Non-parametric Multi-Dimensional Scaling (nMDS) analysis was performed on the data resemblance matrix generated according to the Euclidean distance. ANOSIM (analysis of similarities) and SIMPER (similarity percentage) assessments were computed on the whole dataset, including average values of bacterial abundance, enzyme activity rates, and CLPPs absorbance values, to detect whether there were significant spatial differences among the benthic microbial communities. Based on the Euclidean distance matrix (D), the variables responsible for the significant dissimilarities among the stations were identified. Moreover, the dataset was verified through two-way PERMANOVA analyses using 2 fixed variables (areas, stations) to assess the statistical significance of the differences observed among different areas (northern, central, and southern) and among the different stations within each area. Only the Pseudo-F values reaching a significance threshold of at least p = 0.05 were considered to be statistically significant. All of these analyses were carried out using the software Primer version 7 (Plymouth Marine Laboratory, Rodborough, UK).
3. Results
According to the spatial scale, the results of this microbiological characterization of fjord sediments are reported referring to northern, central, and southern areas separately.
3.1. Culturable Heterotrophic Bacterial Abundance and Organic Matter Decomposition by Enzyme Activities
Heterotrophic bacterial abundance varied between 9.6 × 103 and 7.64 × 104 CFU/g of sediment for marine bacteria (MA) and between 4.05 × 103 and 7.44 × 104 CFU/g of sediment for non-marine bacteria (TSA), with mean ± standard deviation values of 4.56 × 104 ± 1.71 × 104 CFU/g and 2.43 × 104 ± 2.28 × 104 CFU/g, respectively (Figure 2).
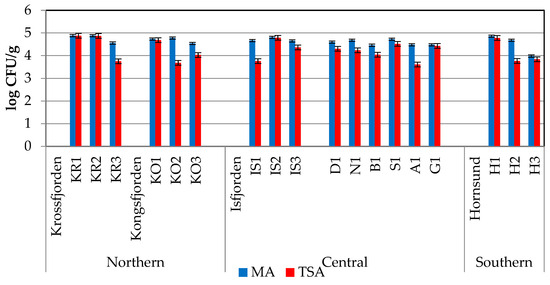
Figure 2.
Marine (MA) and non-marine (TSA) bacterial concentrations (in Colony-Forming Units per g of sediment) recorded in the sediments collected in northern, central, and southern Svalbard regions, separately.
The spatial distribution of marine and non-marine bacteria pointed out high abundances of microorganisms in both the northern and central areas (increasing from B1 to IS2), while lower abundances were detected in the Hornsund region. Within Krossfjorden and Kongsfjorden, non-marine bacteria increased from the outer zone to the inner zone of the fjords (stations KR1 and KO1, respectively). While marine and non-marine bacteria occurred in similar quantitative ratios in the sediments of the northern and central areas (1.07 ± 0.08 and 1.06 ± 0.05, respectively), an abrupt decrease in the abundance of marine bacteria was observed in the southern Hornsund fjord (with a ratio of 0.74 ± 0.07), especially at its outer fjord station H3.
The measured enzyme activity rates (Figure 3a–c) were generally in the following order of magnitude: LAP > AP > GLU, with average values ± sd of 4.618 ± 6.058 µmol/g/h, 1.574 ± 4.198 µmol/g/h, and 0.021 ± 0.027 µmol/g/h, respectively.
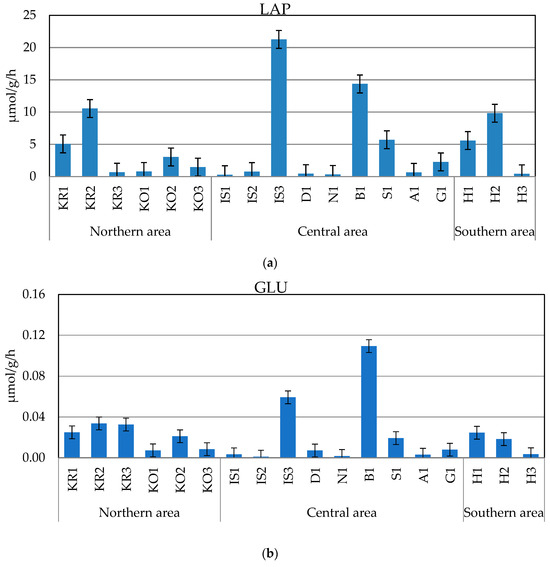
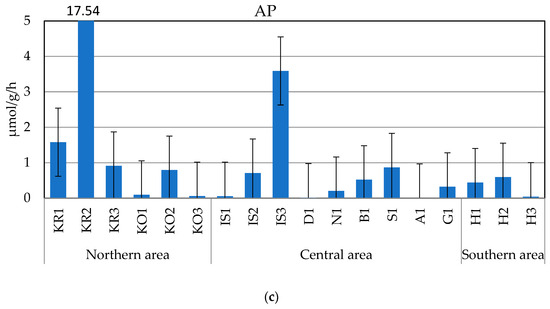
Figure 3.
(a–c) Leucine aminopeptidase (LAP, (a)), beta-glucosidase (GLU, (b)), and alkaline phosphatase (AP, (c)) activity rates measured in the sampled sediments.
LAP ranged from 0.296 to 21.27 µmol/g/h, measured at stations IS1 and IS3, respectively. Enhanced proteolytic activity was observed in the central area at the inner Isfjorden stations B1 and IS3, while similar levels of proteolytic activity were found in the Krossfjorden–Kongsfjorden and Hornsund fjords, where LAP peaked at station KR2 and at the inner station (H2), respectively. An abrupt decrease in microbial metabolism was recorded in the central area at stations IS1, IS2, A1, N1, and D1.
The GLU activity rates were comprised between 0.0011 and 0.109 µmol/g/h, recorded at stations IS2 and B1, respectively. Like LAP, the spatial patterns of this glycolytic enzyme depicted high metabolic rates in the central area at the inner Isfjorden stations B1 and IS3, and a decreasing trend moving towards stations IS2 and N1. Low activity rates were measured in the southern Hornsund fjord, where GLU decreased across a western–eastern gradient, as well as in the northern area at Kongsfjorden. At the entrance and in the central part of Krossfjorden, GLU was still more active, while this enzyme reached its minimum at the stations in the central area (IS1, A1, D1).
AP varied from 0.008 to 17.54 µmol/g/h, measured, respectively, at stations A1 and KR2. Except for the high activity rates observed in the northern and central areas at stations KR2 and IS3, respectively, AP values were generally <1 µmol/g/h, with minimal enzyme activity at stations IS1, A1, and D1. In the southern Horsund area, the eastern station was characterized by the lowest AP activity, as was observed for LAP and GLU.
Significant Pearson correlations were found in the northern area between LAP, AP, and bacterial abundance (Supplementary Table S2). In the central area, the number of significant correlations decreased and only LAP correlated significantly with GLU and AP.
3.2. Community-Level Physiological Profiles (CLPPs) via Biolog Ecoplates
The average well color development (AWCD) values, expressed as absorbance values (Supplementary Figure S1a–c), showed that during the incubation, the absorbance values gradually increased, reaching after 25 days a peak value sometimes followed by a plateau until the end of the readings (35 days).
To compare the levels of microbial metabolism recorded at the different stations, only the peak values of absorbance recorded during the incubation were considered. The CLPPs pointed out a generalized spatial variability (Figure 4a–c), with the lowest activity levels recorded at station D1 and the highest ones recorded at stations G1, A1, and IS1. In the northern area (Kongsfjorden), microbial metabolism decreased from inner to outer stations. In Hornsund, the inner station displayed the highest microbial metabolism. Considering the main guilds of carbon substrates, complex carbon sources, carbohydrates, and amino acids were the substrate sources preferentially metabolized, while amines, carboxylic acids, and phosphate–carbon compounds were less utilized (Figure 4a–c).
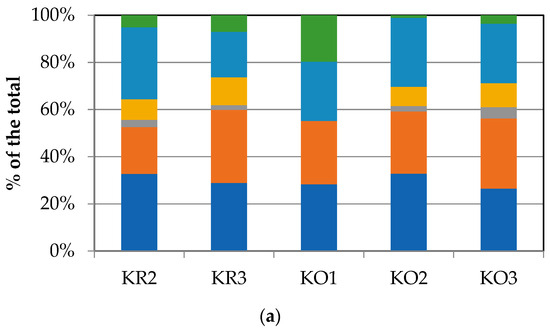
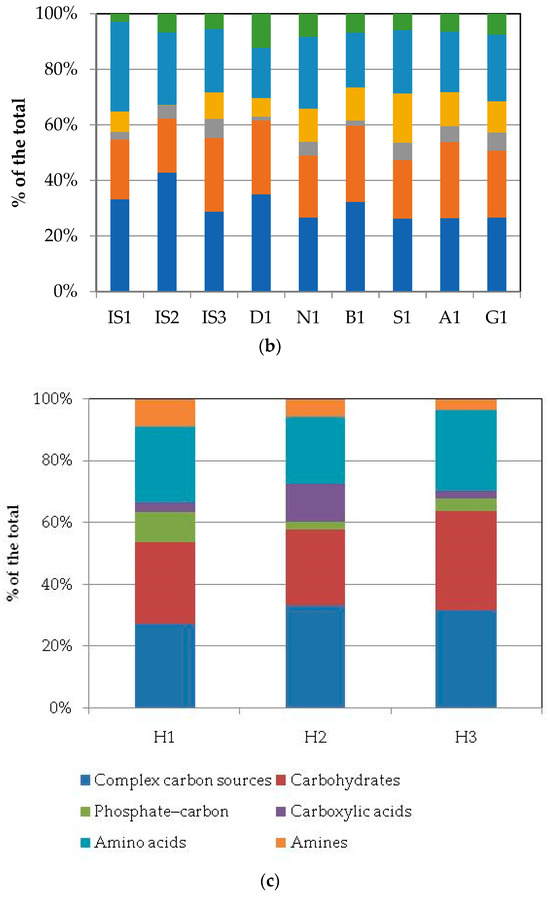
Figure 4.
(a–c) Community-level physiological profiles (as % of the total) of the sediments collected from the northern (a), central (b), and southern areas (c) of the Svalbard archipelago.
Heatmap graphs (Figure 5a,b) highlighted the occurrence of peaks in the utilization of complex C sources at stations IS2, H3, and KO1, as well as in the utilization of carbohydrates at stations H3 and KO1. Amino acid utilization was moderate in all three areas, peaking at stations H2 and KO2. Carboxylic acids were metabolized at stations KR2 and KO2, everywhere in the central area (Isfjorden), and at station H2. Amines were also utilized with high efficiency at stations KO1 and D1, while a limited utilization of phosphate–carbon compounds was recorded, with these compounds being utilized mostly at stations H1, IS3, and S1. In the northern area, marine heterotrophic bacteria correlated significantly with the utilization of complex carbon sources and amino acids; LAP activity also correlated with amino acid utilization. In the southern area, significant negative relationships between the utilization of carbohydrates and complex carbon sources and LAP and GLU were observed (Supplementary Table S2).
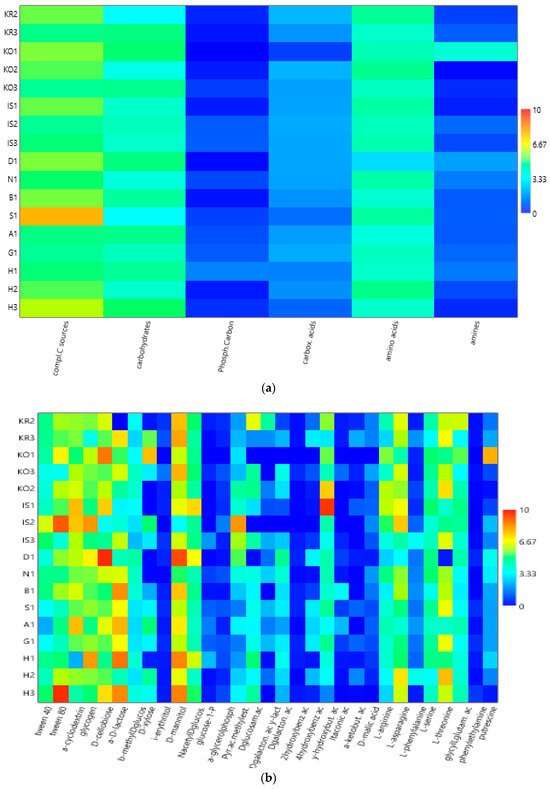
Figure 5.
(a,b) Heatmaps showing the utilization patterns of different groups of organic substrates (carbohydrates, phosphate–carbon sources, carboxylic and acetic acids, amino acids, and amines) (a) and of individual substrates (b) assayed using Biolog Ecoplates in the sediments of the examined areas in the Svalbard archipelago.
At a single-substrate level, within complex carbon sources, tween 80 was the preferentially metabolized substrate, with peaks of utilization at stations KO1, IS2, and H3 in the northern, central, and southern areas, respectively. Within the carbohydrates, D-cellobiose was utilized mostly at stations KO1 and D1, while a-D-lactose was utilized mostly at station H1. With respect to amino acids, L-threonine was the amino acid preferentially utilized at stations KO2 and H2 in the northern and southern areas, respectively, while L-asparagine was the one utilized in the central area at station IS2. Within the phosphate–carbon compounds, glucose-1-phosphate was the most metabolized substrate, especially at stations IS2 and IS3 in the central area and H1 in the southern one. Within the carboxylic acids, y-hydroxy butyric acid was actively used at stations KO2 and IS1 in the northern and central areas, respectively. Putrescine was the most metabolized amine at station VI in the northern area.
The diversity indices calculated from the CLPPs (Table 2) suggested that microbial communities in the sediments were highly metabolically active, being able to utilize more than 29 carbon sources in the northern area (in Krossfjorden and at Kongsfjorden station KO3), as well as in the inner and outer parts of the central area (Isfjorden stations S1, G1, and IS3). High nutritional versatility was observed at the inner station of the southern Hornsund area (station H2). The Shannon–Weaver diversity index reflected this trend too.

Table 2.
Diversity indices. Richness indicates the total number of substrates that could be metabolized.
3.3. Statistical Elaboration of the Whole Abundance and Metabolic Dataset
The outputs of the ANOSIM pointed out that the three areas examined in this study were not significantly different, as shown by a Global R of 0.1 (with a significance level of 0.45) and pairwise test R statistics values of 0.001, 0.012, and 0.014 computed when comparing the northern area vs. the central area, the northern area vs. the southern one, and the central area vs. the southern area, with significance levels of 42.7, 40.5, and 45.9%, respectively.
The nMDS analysis confirmed the lack of significant spatial discrimination among the stations; five main clusters were identified, with three of them having a heterogeneous composition (Cluster 1, mostly composed by outer stations KO3, K3, S1, and G1, plus the central stations AI and H2; Cluster 2, with the central stations KR2, KO2, IS1, and N1, the inner ones KR1 and H1, and one outer station, namely H3; Cluster 3, consisting of inner and outer stations H1 and H3, respectively) (Figure 6). Consistent results were obtained from the PERMANOVA analyses (Table 3); this non-parametric assessment underlined that there were no significant differences among the areas and the stations for each of the tested variables, with the exception of carbohydrates. Within this category, pairwise comparisons computed between the central and southern areas showed that the utilization patterns of D-cellobiose, a-D-lactose, and N-acetyl-D-glucosamin were significantly different.
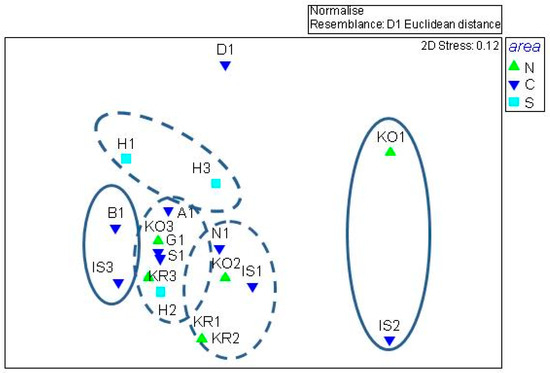
Figure 6.
Output of the nMDS analysis; the continuous lines group inner stations only. N, northern; C, central; and S, southern areas.

Table 3.
Results of two-way PERMANOVA tests for differences in sediment characteristics among the three areas and among the stations within each area. Tests were performed on log-transformed data for each measured parameter. The Pseudo-F (PsF) and Probability (P, in brackets) values are reported. Significant effects are marked with asterisks (***, p < 0.001; **, p < 0.01).
Nevertheless, the SIMPER outputs (Table 4) showed that main distinctive traits characterized some stations, such as high levels of AP (at station KO3), putrescine (at station KO1), N-acetyl-D-glucosamin, complex carbon sources, and LAP (at stations IS1, IS2, and IS3, respectively). GLU, amino acids (i.e., threonine), and MA were the variables more frequently contributing to the total variance at stations B1, D1, and H1, respectively.

Table 4.
SIMPER analysis showing the percentage contribution of each variable within each station. D, Euclidean distance.
3.4. Antibiotic Susceptibility Profiles of Bacterial Isolates
Phenotypic and biochemical tests of the bacterial isolates (n = 66 in total) pointed out that the bacterial isolates were mostly oxidase- and catalase-positive microorganisms. Glucose-fermenting strains were also found (accounting for <30% of the total isolates). From the presumptive identification test, most of the bacterial isolates were assigned to Pseudomonas spp. glucose-nonfermenting bacteria), followed by Flavobacterium spp. (pigmented bacteria) and Vibrio spp. (glucose-fermenting bacteria); only six strains produced hydrogen sulfide and were identified as Pseudomonas putrefaciens (Figure 7).
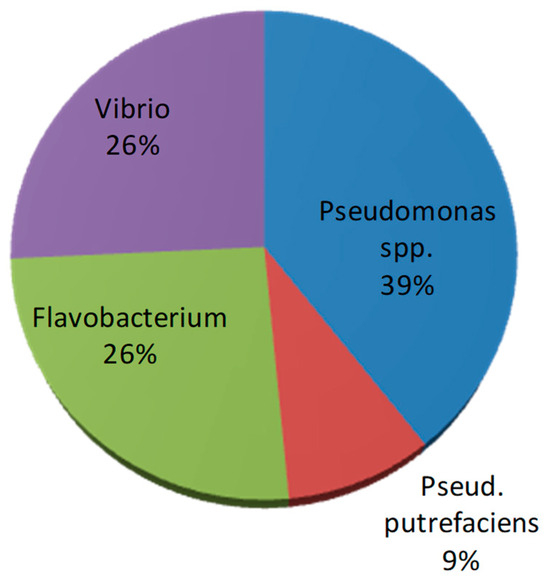
Figure 7.
Presumptive identification of retrieved bacterial isolates (n = 66).
With respect to the spatial distribution of bacterial species, a lower diversity of species composition was observed in Krossfjorden compared to the other sites (Table 5).

Table 5.
Presumptive identification of the bacterial strains isolated in each sampling area according to phenotypic and biochemical tests.
The antibiotic susceptibility profiles of the bacterial isolates are shown in Supplementary Table S1 and Figure 8 where the percentage of the strains resistant to the assayed antibiotics was calculated. Most of the strains were sensitive to ampicillin, clindamycin, and gentamycin; all of the isolates were resistant to enrofloxacin; and a great percentage (95% of the total) were also resistant to tetracycline. Regarding the geographical distribution of antibiotic-resistant bacteria, the highest occurrence of strains resistant to tetracycline was observed in the central area, particularly in Adventfjorden (100% of the total isolates), followed by Dicksonfjorden (80%) and Sassenfjorden (60%). Multi-resistance to all of the tested antibiotics was detected in the bacterial strains isolated from the Hornsund sediments.
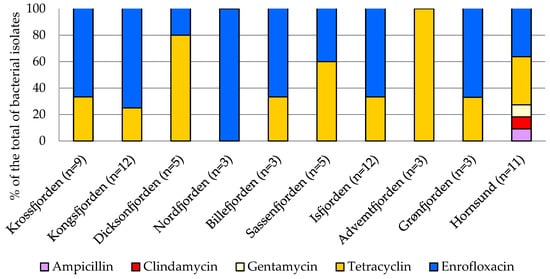
Figure 8.
Percentage of antibiotic-resistant bacteria isolated from the examined areas.
4. Discussion
This study’s contribution aims to depict for several different fjords of west Spitsbergen a comprehensive picture of the benthic microbial community and to elucidate its potential role as a descriptor of fjord sediment health. To this end, a dual ecological and biochemical approach was used, looking at the metabolic activity rates and physiological profiles of the benthic microbial community and at the presence and distribution of antibiotic-resistant bacteria as potential bioindicators of pollution. The microflora hosted in sediments is driven by complex processes affecting both the microbial community’s composition and its structure [58]. Besides ocean currents [59], important factors in shaping benthic microbial communities include the sediment age and its diagenetic state [3], as well as terrestrial inputs and human activity [60]. The role of marine sediments as organic carbon reservoirs is crucial in fjords with active glacier discharges, which act as hotspots for organic matter burial in the benthic domain and subsequent microbial mineralization [61]. Glacial bays in inner fjords receive terrestrial organic carbon inputs from soils and rocks eroded by glaciers. A high contribution (up to 40%) of glacier-derived petrogenic organic matter (ice-rafted debris and coal-derived matter) was found to be buried in marine sediments of the Kongsfjorden–Krossfjorden system [62]. An even higher contribution of petrogenic carbon to the sedimentary carbon pool (up to 84%) was found in Hornsund [63].
Bacterial communities associated with Arctic sediments show different physiological properties and metabolic functions, driven by the availability of substrates [64,65]. Through microbial extracellular enzymatic activity measurements and measurements of carbon substrate utilization (with Biolog Ecoplates), new quantitative and qualitative insights into the metabolism of the benthic microbial community were obtained in our research.
Moreover, our survey gave us the opportunity to assess whether microbial variables were suitable descriptors of the health of Svalbard marine sediments, according to the bioindicators suggested by Nielsen and Winding [66]. Particularly, in our study, we focused on three potential bioindicators, namely the enzyme activity rate (as a surrogate measure of carbon cycling), the CLPPs (as an indicator of community functional diversity), and the abundance of antibiotic-resistant bacteria (as a bioindicator of contaminants’ bioavailability). To this regard, it is important to bear in mind that caution must be taken in data interpretation, since the definition of a healthy sediment ecosystem relies, generally, on a comparison with a reference sediment (control) chosen among a pristine or minimally impacted area [37]. Also, the use of microbes as bioindicators of environmental health involves many related potential advantages and limitations that cannot be neglected, like those reported for soils [67].
4.1. Culturable Heterotrophic Bacterial Abundance and Organic Matter Decomposition by Enzyme Activities
A fairly homogeneous abundance of viable heterotrophic bacteria was observed in the fjord sediments examined in this study, suggesting that these fjords host a heterotrophic component capable of utilizing a broad range of organic substrates. The abundance of viable heterotrophic bacteria measured in our sediment samples (9.6 × 103–7.64 × 104 CFU/g) was about two orders of magnitude lower than that found within Kongsfjorden by Conte et al. [5] (mean value of 60.7 × 105 CFU/g, range of 1.1–184.8 × 105 CFU/g). The increase in non-marine bacterial abundance towards the inner parts of Krossfjorden and Kongsfjorden, although not statistically significant, suggests the presence of allochthonous organic matter as a substrate for these microorganisms.
Svalbard fjord sediments are affected by the different physical–chemical properties (temperature, nutrient content, light, and dissolved oxygen) and hydrodynamic patterns of the overlaying water column, which in turn affect the species composition of the bacterial community [68,69]. In the water column of the Hornsund fjord, bacterial numbers and biomasses higher than those found in Kongsfjorden have been reported [70], suggesting a better adaptation of Arctic bacteria to the low water temperatures and low availability of organic substrates. Unlike the findings of our study, where bacterial distribution did not show statistically significant spatial variability, in a previous study [71], the quantitative differences found at the sediment level were explained to be due to an Arctic-derived microbial community in Hornsund compared to the one inhabiting the Kongsfjorden sediments, more affected by the Atlantic influence [2,72].
Benthic microbial communities play a critical role in driving biogeochemical processes, including carbon, nitrogen, phosphorus, and sulfur cycles [73,74]. Therefore, measuring the organic matter turnover mediated by hydrolytic enzymes, although in terms of potential activities, is a key step towards understanding the organic matter processing carried out by these microbial assemblages [1]. The fjord sediments examined in the present study were characterized by a predominance of LAP and AP compared to GLU. The spatial enzyme activity patterns recorded in these sediments suggested that in the central area, especially at the inner Isfjorden stations (stations IS3 and B1), the organic matter pool was rich in proteins and mucopolysaccharides, as shown by high LAP and GLU activity rates compared to those observed in the other stations within Isfjorden. Inner areas of Arctic fjords were found to be characterized by high levels of dissolved organic matter (DOM) and a prevalence of Bacteroidetes [1,75]; this microbial group, especially with Cytophaga–Flavobacteria members, is known to contribute effectively to the turnover of detritus and, more generally, to carbon cycling within aquatic ecosystems [76]. Diversified spectra of polysaccharide hydrolases were detected in Svalbard sediments by Teske et al. [77], who attributed the occurrence of these enzyme activities to members of the Chloroflexi genus. Conversely, low GLU activities (around 0.01 mmol ml−1 d−1) were measured in surface sediments from the LTER Hausgarten station, in the Fram Strait [78].
Organic phosphate esters were more abundant in the northern (Krossfjorden station KR2) and central areas of Svalbard. The southern Hornsund area and the outer stations of Isfjorden (A1 and G1) were characterized by low activity rates for all of the three examined enzymes; this could indicate that the sediment organic pool was quite refractory or hardly prone to active microbial decomposition, as was also suggested by the low heterotrophic bacterial abundance found in this area.
4.2. Microbial Community Metabolism (CLPPs)
While extracellular enzyme activities provide information on the microbial ability to hydrolyze complex molecules, Biolog Ecoplates give additional information on the utilization of single monomers or oligomers (i.e., amino acids, sugars, complex carbon substrates) at a community level, providing an index of the metabolic complexity of the microbial community [52,53]. Complex carbon sources, carbohydrates, and amino acids were the main categories of organic substrates preferentially metabolized by the benthic microbial assemblage in our fjord sediments. Complex carbon sources were actively metabolized in all three areas, particularly in the central and southern areas; amino acids were mainly metabolized in the northern and southern areas and carbohydrates were mainly metabolized in the southern Hornsund fjord. Variable metabolic patterns suggested an active role played by the microbial community in driving carbon and nutrient cycling, as well as the quality of the benthic organic matter pool. Indeed, in Arctic fjords, terrestrial runoff, water mass exchanges, and glaciers’ melting waters coexist, creating contrasting environmental conditions, and their interplay is complex, especially during the summer season, when biological activities are more enhanced and detritus released from melting ice accumulates into deep matrices [1,46,79]. Furthermore, on a spatial scale, the metabolic diversification probably reflected a variable community structure, as it has been reported in other Svalbard fjord sediments, strongly affected by changing glacial, marine, and terrestrial inputs [5].
The diversity indices suggested that in the sediments of the central area (Isfjorden, stations S1 and G1) and in the southern area (Hornsund fjord, station H2), the microbial community was able to metabolize the highest number of carbon substrates, although no statistically significant differences were detected among the areas and stations in the PERMANOVA assessment. The presence of a muddy sediment was a common trait of these three stations; nevertheless, even where clay sediments were found (i.e., in Krossfjorden, at station KO2 in Kongsfjorden, and at station B1 in Isfjorden), the CLPPs showed the presence of a metabolically active microbial community in the sediments regardless of their granulometry, in agreement with the results reported by Hargrave et al. [80]. Moreover, functional spectra of the benthic microbial community did not seem to relate to a glacial–marine transition pattern; indeed, the functional diversity of the sediments collected from outer fjord stations did not differ significantly from that of sediments collected from inner ones.
The use of a wide range of organic compounds as carbon sources—as shown by the Biolog Ecoplate data—confirmed the metabolic complexity and versatility of the microbial community; on the other hand, the latter is closely linked to the synthesis of extracellular enzymes. From an ecological point of view, the presence of a complex metabolism indicates the high nutritional versatility of the microbial community and its large adaptation to diversified ecological niches; conversely, a simple metabolism suggests that microorganisms have a limited capacity of adaptation and thus can colonize only specific ecological niches [81].
Looking at the single substrates metabolized by the microbial community, within the complex carbon sources, the widespread utilization of tween 80 was related to the presence of specific enzymes like lipases or esterases; lipases are, after proteinases, one of the most widespread enzymes in marine ecosystems [82]. Within the carbohydrates, α-D-glucose was preferentially used by the microbial community; this result was not surprising, since this sugar is the principal substrate utilized to obtain pyruvate [81]. Other well-metabolized carbohydrates were mannitol and a-D-lactose.
Within carboxylic acids, which are normal constituents of hydrophobic molecules, hydroxybutyric acid is involved in the metabolism of fatty acids and amino acids. With respect to amino acids, L-threonine and L-asparagine were the most metabolized molecules; both of these amino acids play a key role in cell metabolism.
The diversity indices (both richness and Shannon–Weaver) suggested that the microbial assemblage was metabolically well adapted to cope with changing conditions such as variable nutrient inputs coming from terrestrial runoff and glacier melting. Nevertheless, comparing the inner stations with the outer stations, the PERMANOVA results showed that there were no distinctive spatial characteristics; the presence of refractory substrates, not available to the microbial community, could explain this finding.
4.3. Antibiotic-Resistant Bacteria
The screening of bacterial isolates for susceptibility to antibiotics showed that in the examined fjord sediments, bacterial resistance to tetracycline (in a percentage ranging from 33 to 100% of the total isolates) and enrofloxacin (in all of the isolates) was a widespread phenomenon. Since the first detection of multidrug-resistant bacteria in the Arctic [83], reports on the spread of antibiotic resistance have exponentially increased. Bacterial isolates showing multi-antibiotic resistance patterns have been detected in the Pasvik river (Arctic Norway [55]).
A high prevalence of antibiotic resistance was observed in aerobic heterotrophic bacteria and coliform bacteria isolated from the water and sediment of Kongsfjorden, suggesting that drug-resistant mutants were preferentially selected in sediments compared to in water [84]. Kalinowska et al. [85] evidenced the role of wastewater discharge in the dissemination of antibiotic-resistant Enterococcus spp. in two Arctic lakes (a water supply system affected by birds and a wastewater close to the Polish Polar Station) in the Hornsund area. In our study, the low percentage of tetracycline-resistant bacteria found in Hornsund compared to that found in the central area was probably related to the lower human impact found in this fjord. In ice, water, and sediment samples from Longyearbyen and Adventfjorden, the most populated area of Svalbard, class 1 integrons—as markers of anthropogenic pollution—were detected in ARB isolates. Antibiotic resistance and virulence genes were found in the variable regions of integrons; the relative abundance of intI1 genes was higher in the wastewater outflow site and correlated also with an abundance of heavy-metal genes, suggesting that the Svalbard resistome was affected by both natural (melting glaciers) and human (wastewater discharge) drivers [85].
In bacterial strains (237 in total) isolated from the sediments of Kongsfjorden and Krossfjorden, Visnupriya et al. [86] reported high percentages of bacteria resistant to the Extended-Spectrum β-lactam antibiotic (ESBL), including ceftazidime (45.56%) followed by trimethoprim (27%) and sulphamethizole (24.05%); resistance against tetracycline and gentamycin was observed in only a limited percentage of strains (2.53% and 2.95% of the total isolates, respectively). Unlike our results, the distribution of antibiotic-resistant bacteria was reported to differ significantly between inner and outer stations of Kongsfjorden and Krossfjorden. The authors related this finding to the potential release into the fjords of antibiotic-resistant bacteria from the preserved permafrost due to the melting of glaciers, horizontal gene transfer, and human presence.
The presence of ARB and integrons in the marine environment may potentially cause a threat due to the potential transfer of ARB and integrons to humans and the environment.
Antibiotic resistance genes have frequently been found in natural environments due to a range of both anthropogenic and natural selective pressures causing the development and dissemination of more antibiotic-resistant bacteria [87,88]. Extreme environments represent important pristine ecosystems for studying the evolution of antibiotic resistance genes (ARGs) and the spread of antibiotic-resistant bacteria (ARB) in the presence/absence of pollution from antibiotics [89]. Several studies have reported the occurrence in polar environments of both ARB and ARGs; this has been related to the mobilization of antibiotic residues as chemical contaminants accumulated in sediments or sea ice because of climate warming [84,90]. Due to the melting of glaciers, ice cover, and permafrost, contaminants accumulated on glacier surfaces over many decades can be discharged to marine ecosystems. Ciprofloxacin is the only antibiotic that has been detected in the Kongsfjorden sediments, with concentrations ranging from 6.85 to 684.5 ng/g, indicating the presence of antibiotics in the Arctic marine ecosystem [91]. Moreover, plastic debris, frequently sinking into sediments, may act as a reservoir of ARB and ARGs [92], and wild migratory birds can introduce many drug-resistant strains over long distances [93].
Contaminants may be transported in the Arctic from several remote and local sources across different pathways, including biological transport driven by migrating animals, in addition to sea ice, riverine, and groundwater discharge, and atmospheric transport pathways [94]; however, data on the presence and impact of contaminants in polar regions are still fragmentary. Marine sediments represent natural reservoirs of pollutants originating from global (mostly oceanic transport and atmospheric deposition) and local sources. Antibiotic residuals present in sediments may directly influence the structure of natural microbial communities, often causing a reduction in microbial biodiversity or functional stability [95]. Most antibiotics can also undergo transformation processes along the trophic web, accumulating into organisms. Previous studies from Svalbard and Bjørnøya have documented pollutants’ presence in sediment cores going back more than 100 years in time [96,97]. The co-occurrence of antibiotic and metal resistances has also been shown [98]. A better understanding of the distribution of contaminants within fjord environments is important since global changes are affecting the strength and directions of contaminant transport processes. On the other hand, the ability of microorganisms to metabolize xenobiotics, including antibiotics, makes them suitable candidates for the remediation of polluted environments [99].
Although antibiotic resistance is a global problem, it appears evident that compared to other environments, in Arctic regions, the diversity and spread of ARB and ARGs, and, generally, of the aquatic resistome, are strongly understudied [100]. Even though Norway is characterized by a lower use of antibiotics for human health, animal husbandry, and food production compared with many other countries [101,102], there is still a lack of current data on the usage of antimicrobial agents in humans and animals [103], making it not possible to predict the effects of these pollutants on the environment and the local wildlife.
5. Conclusions
The metabolic potential of microbial communities is a helpful criterion for understanding their role in complex environments like Arctic marine sediments. Globally, the CLPPs determined in our research indicate a nutritional versatility of the benthic microbial community to adapt to changing scenarios, and this characteristic is of paramount importance to enable microbial survival under extreme conditions. Although the functional spectra of sediment bacterial communities occurring in the fjords of the Svalbard archipelago did not seem to clearly reflect a glacial–marine transition, on a spatial scale, community metabolism was more diversified in the inner central Isfjorden area, probably in response to the organic detritus made available by glacier melting and terrestrial inputs.
Multi-resistance to enrofloxacin and tetracycline was detected in all of the sampled sediments, and in the southern Hornsund area, multi-resistance to all of the assayed antibiotics was observed, stressing the role of sediments as a potential reservoir of chemical wastes ascribable to antibiotic residuals; although, no evident environmental risks were pointed out from this preliminary screening given the widespread occurrence of a resistome in many natural environments. A higher percentage of tetracycline-resistant bacteria characterized the benthic microflora of the central area in comparison to the other ones, although no statistically significant differences were found among the areas. This finding was therefore not consistent with our first working hypothesis, suggesting the need for further studies to investigate potential links between human-related activities and the spread of antibiotic resistance.
From this combined analysis of microbial enzyme activity rates, community functional profiles, and antibiotic-resistance patterns, no solid discrimination between healthy and unhealthy sediments was achieved. This probably depended on the local availability of resources, affecting microbial dynamics, as well as the ability of microbes to adapt and grow well in environmental unhealthy conditions. Taking into consideration that the concept of sediment health varies with the specific environmental context, our findings provide evidence that microbial variables alone cannot be universally used to assess sediment health and that additional criteria—such as organic carbon pool characterization or physical properties affecting microbial functions—could be helpful for the Svalbard environment, stressing the need for further studies addressing this specific issue.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/environments11070148/s1: Supplementary Figures S1a–c: Trends in the Average Well Color Development (as absorbance values) recorded during the incubation period in the sediments collected from the northern (a), central (b), and southern (c) areas of the Svalbard archipelago. Supplementary Table S1: Results of the screening of bacterial strains for susceptibility to antibiotics. For each assayed antibiotic, sensitive bacteria are scored as 0, and resistant bacteria are scored as 1. Supplementary Table S2: Pearson correlation coefficients computed in the three examined Svalbard areas. Values reported in bold are significant at a p < 0.05 level.
Author Contributions
Conceptualization, G.C., A.Z.; methodology, G.C., G.M., A.C.R.; software, G.C., A.C.R.; validation, G.C., G.M., G.Z.; formal analysis, A.C., G.Z.; investigation, M.S., A.Z.; resources, G.C., A.Z.; data curation, G.C.; writing—original draft preparation, G.C., G.M.; writing—review and editing, G.C., G.M., A.Z.; visualization, G.Z.; supervision, G.C.; project administration, A.Z.; funding acquisition, A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by IO PAN activities (Theme 2.2), and project n. 2020/39/B/ST10/01504 funded by the National Science Centre, Poland. The APC was funded by G.C.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Caruso, G.; Madonia, A.; Bonamano, S.; Miserocchi, S.; Giglio, F.; Maimone, G.; Azzaro, F.; Decembrini, F.; La Ferla, R.; Piermattei, V.; et al. Microbial Abundance and Enzyme Activity Patterns: Response to Changing Environmental Characteristics along a Transect in Kongsfjorden (Svalbard Islands). J. Mar. Sci. Eng. 2020, 8, 824. [Google Scholar] [CrossRef]
- Svendsen, H.; Beszczynska-Møller, A.; Hagen, J.O.; Lefauconnier, B.; Tverberg, V.; Gerland, S.; Ørbaek, J.B.; Bischof, K.; Papucci, C.; Zajaczkowski, M.; et al. The physical environment of Kongsfjorden–Krossfjorden, an Arctic fjord system in Svalbard. Polar Res. 2002, 21, 133–166. [Google Scholar] [CrossRef]
- Pelikan, C.; Jaussi, M.; Wasmund, K.; Seidenkrantz, M.-S.; Pearce, C.; Kuzyk, Z.Z.A.; Herbold, C.W.; Røy, H.; Kjeldsen, K.U.; Loy, A. Glacial runoff promotes deep burial of sulfur cycling-associated microorganisms in marine sediments. Front. Microbiol. 2019, 10, 2558. [Google Scholar] [CrossRef]
- Tobias-Hünefeldt, S.P.; Wing, S.R.; Baltar, F.; Morales, S.E. Changes in microbial community phylogeny and metabolic activity along the water column uncouple at near sediment aphotic layers in fjords. Sci. Rep. 2021, 11, 19303. [Google Scholar] [CrossRef]
- Conte, A.; Papale, M.; Amalfitano, S.; Mikkonen, A.; Rizzo, C.; De Domenico, E.; Michaud, L.; Lo Giudice, A. Bacterial community structure along the subtidal sandy sediment belt of a high Arctic fjord (Kongsfjorden, Svalbard Islands). Sci. Total Environ. 2018, 619–620, 203–211. [Google Scholar] [CrossRef]
- Granberg, M.; Winberg von Friesen, L.; Bach, L.; Collard, F.; Strand, J.; Gabrielsen, G.W. Anthropogenic Microlitter in Wastewater and Marine Samples from Ny-Ålesund, Barentsburg and Signehamna, Svalbard; Report Number C 373; IVL Swedish Environmental Research Institute: Stockholm, Sweden, 2019; pp. 1–28. ISBN 978-91-7883-020-6. [Google Scholar]
- Hedges, J.I.; Keil, R.G. Sedimentary organic matter preservation: An assessment and speculative synthesis. Mar. Chem. 1995, 49, 81–115. [Google Scholar] [CrossRef]
- Evenset, A.; Leknes, H.; Christensen, G.N.; Warner, N.; Remberger, M.; Gabrielsen, G.W. Screening of New Contaminants in Samples from the Norwegian Arctic: Silver, Platinum, Sucralose, Bisphenol A, Tetrabrombisphenol A, Siloxanes, Phtalates (DEHP), Phosphororganic Flame Retardants; SPFO-Report: 1049/2009; Norwegian Pollution Control Authority: Oslo, Norway, 2009. [Google Scholar]
- Jiao, L.; Zheng, G.J.; Minh, T.B.; Richardson, B.; Chen, L.; Zhang, Y.; Yeung, L.W.; Lam, J.C.W.; Yang, X.; Lam, P.K.S.; et al. Persistent toxic substances in remote lake and coastal sediments from Svalbard, Norwegian Arctic: Levels, sources and fluxes. Environ. Pollut. 2009, 157, 1342–1351. [Google Scholar] [CrossRef]
- Sapota, G.P.; Wojtasik, B.; Burska, D.; Nowiński, K. Persistent organic pollutants (POPs) and polycyclic aromatic hydrocarbons (PAHs) in surface sediments from selected fjords, tidal plains and lakes of the North Spitsbergen. Pol. Polar Res. 2009, 30, 59–76. [Google Scholar]
- Kallenborn, R.; Borgå, K.; Christensen, J.H.; Dowdall, M.; Evenset, A.; Odland, J.Ø.; Ruus, A.; Aspmo Pfaffhuber, K.; Pawlak, J.; Reiersen, L.-O. Combined Effects of Selected Pollutants and Climate Change in the Arctic Environment; Arctic Monitoring and Assessment Programme (AMAP): Oslo, Norway, 2011; pp. 1–108. [Google Scholar]
- Choudhary, S.; Nayak, G.N.; Khare, N. Source, mobility, and bioavailability of metals in fjord sediments of Krossfjord-Kongsfjord system, Arctic, Svalbard. Environ. Sci. Pollut. Res. 2020, 27, 15130–15148. [Google Scholar] [CrossRef]
- Vishnu Sagar, M.K.; Kannan, V.M.; Gopikrishna, V.G.; Krishnan, K.P.; Mohan, M. Geochemistry and distribution of Metals in the Sediments of Kongsfjorden, Svalbard, Arctic. Reg. Stud. Mar. Sci. 2021, 44, 101729. [Google Scholar] [CrossRef]
- Rudnicka-Kępa, P.; Bełdowska, M.; Zaborska, A. Enhanced heavy metal discharges to marine deposits in glacial bays of two Arctic fjords (Hornsund and Kongsfjorden). J. Mar. Syst. 2024, 241, 103915. [Google Scholar] [CrossRef]
- Evenset, A.; Christensen, G.N.; Palerud, R. Environmental Toxins in Marine Sediments in Isfjorden, Svalbard 2009; Research Outside Longyearbyen, Barentsburg, Pyramiden and Coles Bay. Akvaplan-niva report 4707-1, 1-134; Akvaplan-NIVA: Tromsø, Norway, 2009. (In Norwegian) [Google Scholar]
- Van den Heuvel-Greve, M.J.; Szczybelski, A.S.; van den Brink, N.W.; Kotterman, M.J.J.; Kwadijk, C.J.A.F.; Evenset, A.; Murk, A.J. Low organotin contamination of harbour sediment in Svalbard. Polar Biol. 2016, 39, 1699–1709. [Google Scholar] [CrossRef]
- Evenset, A.; Hallanger, I.G.; Tessmann, M.; Warner, N.; Ruus, A.; Borgå, K.; Gabrielsen, G.W.; Christensen, G.; Renaud, P.E. Seasonal variation in accumulation of persistent organic pollutants in an Arctic marine benthic food web. Sci. Tot. Environ. 2016, 542, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Pouch, A.; Zaborska, A.; Pazdro, K. Concentrations and origin of polychlorinated biphenyls (PCBs) and polycyclic aromatic hydrocarbons (PAHs) in sediments of western Spitsbergen fjords (Kongsfjorden, Hornsund, and Adventfjorden). Environ. Monit. Assess. 2017, 189, 175. [Google Scholar] [CrossRef] [PubMed]
- Jæger, I.; Hop, H.; Gabrielsen, G.W. Biomagnification of mercury in selected species from an Arctic marine food web in Svalbard. Sci. Total Environ. 2009, 407, 4744–4751. [Google Scholar] [CrossRef] [PubMed]
- Pouch, A.; Zaborska, A.; Legeżyńska, J.; Deja, K.; Pazdro, K. Assessment of exposure of benthic organisms to selected organochlorine pollutants in the west Spitsbergen fjords. Sci. Total Environ. 2023, 896, 165262. [Google Scholar] [CrossRef]
- Macdonald, R.W.; Harner, T.T.; Fyfe, J. Recent climate change in the Arctic and its impact on contaminants pathway and interpretation on temporal trend data. Sci. Total Environ. 2005, 342, 5–86. [Google Scholar] [CrossRef] [PubMed]
- Ager, D.; Evans, S.; Li, H.; Lilley, A.K.; van der Gast, C.J. Anthropogenic disturbance affects the structure of bacterial communities. Environ. Microbiol. 2010, 12, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Granberg, M.E.; Ask, A.; Gabrielsen, G.G. Local Contamination in Svalbard. Overview and Suggestions for Remediation Actions; Report no. 044; Norsk Polarinstitutt: Tromsø, Norway, 2017. [Google Scholar]
- Zhan, J.; Gao, Y.; Li, W.; Chen, L.; Lin, H.; Lin, Q. Effects of ship emissions on summertime aerosols at Ny–Alesund in the Arctic. Atmos. Pollut. Res. 2014, 5, 500–510. [Google Scholar] [CrossRef]
- Khan, A.L.; Dierssen, H.; Schwarz, J.P.; Schmitt, C.; Chlus, A.; Hermanson, M.; Painter, T.H.; McKnight, D.M. Impacts of coal dust from an active mine on the spectral reflectance of Arctic surface snow in Svalbard, Norway. J. Geophys. Res. Atmos. 2017, 122, 1767–1778. [Google Scholar] [CrossRef]
- Kalinowska, A.; Szopińska, M.; Chmiel, S.; Kończak, M.; Polkowska, Ż.; Artichowicz, W.; Jankowska, K.; Nowak, A.; Łuczkiewicz, A. Heavy Metals in a High Arctic Fiord and Their Introduction with the Wastewater: A Case Study of Adventfjorden-Longyearbyen System, Svalbard. Water 2020, 12, 794. [Google Scholar] [CrossRef]
- Zaborska, A.; Strzelewicz, A.; Rudnicka, P.; Moskalik, M. Processes driving heavy metal distribution in the seawater of an Arctic fjord (Hornsund, southern Spitsbergen). Mar. Pollut. Bull. 2020, 161A, 111719. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Kitae, K.; Deokwon, K.; Moon, H.-B.; Jeon, J. Ny-Ålesund-oriented organic pollutants in sewage effluent and receiving seawater in the Arctic region of Kongsfjorden. Environ. Pollut. 2020, 258, 113792. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, N.E.; Aksu, A.; Karacık, B.; Bayırhan, İ.; Çağlar, N.; Gazioğlu, C.; Özsoy, B. Presence of some commonly used pharmaceutical residues in seawater and net plankton: A case study of Spitsbergen, Svalbard Archipelago. Int. J. Environ. Geoinform. 2022, 9, 1–10. [Google Scholar] [CrossRef]
- Sokołowski, A.; Mordec, M.; Caban, M.; Øverjordet, I.B.; Wielogórska, E.; Włodarska-Kowalczuk, M.; Balazy, P.; Chełchowski, M.; Lepoint, G. Bioaccumulation of pharmaceuticals and stimulants in macrobenthic food web in the European Arctic as determined using stable isotope approach. Sci. Total Environ. 2024, 909, 168557. [Google Scholar] [CrossRef]
- Jokanović, S.; Kajan, K.; Perović, S.; Ivanić, M.; Mačić, V.; Orlić, S. Anthropogenic influence on the environmental health along Montenegro coast based on the bacterial and chemical characterization. Environ. Pollut. 2021, 271, 116383. [Google Scholar] [CrossRef] [PubMed]
- Nogales, B.; Lanfranconi, M.P.; Piña-Villalonga, J.M.; Bosch, R. Anthropogenic perturbations in marine microbial communities. FEMS Microbiol. Rev. 2011, 35, 275–298. [Google Scholar] [CrossRef] [PubMed]
- Gillan, D.C.; Danis, B.; Pernet, P.; Joly, G.; Dubois, P. Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment. Appl. Environ. Microbiol. 2005, 71, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Varma, A. Role of Enzymes in Maintaining Soil Health. In Soil Enzymology. Soil Biology; Shukla, G., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 22. [Google Scholar] [CrossRef]
- Alkorta, I.; Aizpurua, A.; Riga, P.; Albizu, I.; Amézaga, I.; Garbisu, C. Soil Enzyme Activities as Biological Indicators of Soil Health. Rev. Environ. Health 2003, 18, 65–73. [Google Scholar] [CrossRef]
- Rao, M.A.; Scelza, R.; Acevedo, F.; Diez, M.C.; Gianfreda, L. Enzymes as useful tools for environmental purposes. Chemosphere 2014, 107, 145–162. [Google Scholar] [CrossRef]
- Maher, W.; Batley, G.E.; Lawrence, I. Assessing the health of sediment ecosystems: Use of chemical measurements. Freshw. Biol. 1999, 41, 361–372. [Google Scholar] [CrossRef]
- Astudillo-García, C.; Hermans, S.M.; Stevenson, B.; Buckley, H.L.; Lear, G. Microbial assemblages and bioindicators as proxies for ecosystem health status: Potential and limitations. Appl. Microbiol. Biotechnol. 2019, 103, 6407–6421. [Google Scholar] [CrossRef]
- Caruso, G.; Genovese, L.; Mancuso, M.; Modica, A. Effects of fish farming on microbial enzyme activities and densities: Comparison between three Mediterranean sites. Lett. Appl. Microbiol. 2003, 37, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Chen, Z.; Deng, Y.; Zhao, D.; Gao, P.; Zhang, L.; Tu, Q.; Qu, L.; Zheng, L.; Zhang, Y.; et al. Contrasting archaeal and bacterial community assembly processes and the importance of rare taxa along a depth gradient in shallow coastal sediments. Sci. Total Environ. 2022, 852, 158411. [Google Scholar] [CrossRef]
- Caruso, G.; Maimone, G.; Rappazzo, A.C.; Dell’Acqua, O.; Laganà, P.; Azzaro, M. Microbial Biofilm Colonizing Plastic Substrates in the Ross Sea (Antarctica): First Overview of Community-Level Physiological Profiles. J. Mar. Sci. Eng. 2023, 11, 1317. [Google Scholar] [CrossRef]
- Rutgers, M.; Wouterse, M.; Drost, S.M.; Breure, A.M.; Mulder, C.; Stone, D.; Creamer, R.E.; Winding, A.; Bloem, J. Monitoring soil bacteria with community-level physiological profiles using Biolog™ ECO-plates in the Netherlands and Europe. Appl. Soil Ecol. 2016, 97, 23–35. [Google Scholar] [CrossRef]
- Wan, L.; Caruso, G.; Cao, X.; Song, C.; Maimone, G.; Rappazzo, A.C.; Laganà, P.; Zhou, Y. Microbial Response to Coastal-Offshore Gradients in Taiwan Straits: Community Metabolism and Total Prokaryotic Abundance as Potential Proxies. Microb. Ecol. 2023, 85, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Handler, E.R.; Andersen, S.D.J.; Gradinger, R.; McGovern, M.; Vader, A.; Poste, A.E. Seasonality in land-ocean connectivity and local processes control sediment bacterial community structure and function in a High Arctic tidal flat. FEMS Microb. Ecol. 2024, 100, fiad162. [Google Scholar] [CrossRef]
- Zaborska, A.; Włodarska-Kowalczuk, M.; Legeżyńska, J.; Jankowska, E.; Winogradow, A.; Deja, K. Sedimentary organic matter sources, benthic consumption and burial in west Spitsbergen fjords—Signs of maturing of Arctic fjordic systems? J. Mar. Syst. 2018, 180, 112–123. [Google Scholar] [CrossRef]
- Piquet, A.M.-T.; Scheepens, J.F.; Bolhuis, H.; Wiencke, C.; Buma, A.G.J. Variability of protistan and bacterial communities in two Arctic fjords (Spitsbergen). Polar Biol. 2010, 33, 1521–1536. [Google Scholar] [CrossRef][Green Version]
- Skogseth, R.; Olivier, L.L.A.; Nilsen, F.; Falck, E.; Fraser, N.; Tverberg, V.; Ledang, A.B.; Vader, A.; Jonassen, M.O.; Søreide, J.; et al. Variability and decadal trends in the Isfjorden (Svalbard) ocean climate and circulation—An indicator for climate change in the European Arctic. Progr. Oceanogr. 2020, 187, 102394. [Google Scholar] [CrossRef]
- Prominska, A.; Cisek, M.; Walczowski, W. Kongsfjorden and Hornsund hydrography—Comparative study based on a multiyear survey in fjords of West Spitsbergen. Oceanologia 2017, 59, 397–412. [Google Scholar] [CrossRef]
- Norwegian Polar Institute. TOPOSVALBARD. Available online: https://toposvalbard.npolar.no/ (accessed on 3 June 2024).
- Hoppe, H.G. Use of fluorogenic model substrates for extracellular enzyme activity (EEA) measurement of bacteria. In Handbook of Methods in Aquatic Microbial Ecology; Kemp, P.F., Sherr, B.F., Sherr, E.B., Cole, J.J., Eds.; Lewis Publisher: London, UK, 1993; pp. 423–431. [Google Scholar]
- Garland, J.L.; Mills, A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [CrossRef] [PubMed]
- Sala, M.M.; Arin, L.; Balagué, V.; Felipe, J.; Guadayol, O.; Vaqué, D. Functional diversity of bacterioplankton assemblages in Western Antarctic seawaters during late spring. Mar. Ecol. Progr. Ser. 2005, 292, 13–21. [Google Scholar] [CrossRef]
- Sala, M.M.; Estrada, M.; Gasol, J.M. Seasonal changes in the functional diversity of bacterioplankton in contrasting coastal environments of the NW Mediterranean. Aquat. Microb. Ecol. 2006, 44, 1–9. [Google Scholar] [CrossRef]
- Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T.; Williams, S.T. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Williams & Wilkins: Baltimore, MD, USA, 1994; pp. 1–787. [Google Scholar]
- Laganà, P.; Votano, L.; Caruso, G.; Azzaro, M.; Lo Giudice, A.; Delia, S. Bacterial isolates from the Arctic region (Pasvik River, Norway): Assessment of biofilm production and antibiotic susceptibility profiles. Environ. Sci. Pollut. Res. 2018, 25, 1089–1102. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turk, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- EUCAST (European Committee on Antimicrobial Susceptibility Testing). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 3 June 2024).
- Petro, C.; Starnawski, P.; Schramm, A.; Kjeldsen, K.U. Microbial community assembly in marine sediments. Aquat. Microb. Ecol. 2017, 79, 177–195. [Google Scholar] [CrossRef]
- Hamdan, L.J.; Coffin, R.B.; Sikaroodi, M.; Greinert, J.; Treude, T.; Gillevet, P.M. Ocean currents shape the microbiome of Arctic marine sediments. ISME J. 2013, 7, 685–696. [Google Scholar] [CrossRef]
- Delpech, L.M.; Vonnahme, T.R.; McGovern, M.; Gradinger, R.; Præbel, K.; Poste, A.E. Terrestrial inputs shape coastal bacterial and archaea communities in a high Arctic fjord (Isfjorden, Svalbard). Front. Microbiol. 2021, 12, 614634. [Google Scholar] [CrossRef]
- Jørgensen, B.B.; Laufer, K.; Michaud, A.B.; Wehrmann, L.M. Biogeochemistry and microbiology of high Arctic marine sediment ecosystems—Case study of Svalbard fjords. Limnol. Oceanogr. 2020, 66, S273–S292. [Google Scholar] [CrossRef]
- Kim, H.; Kwon, S.Y.; Lee, K.; Lim, D.; Han, S.; Kim, T.W.; Joo, Y.J.; Lim, J.; Kang, M.H.; Nam, S.I. Input of terrestrial organic matter linked to deglaciation increased mercury transport to the Svalbard fjords. Sci. Rep. 2020, 10, 3446. [Google Scholar] [CrossRef]
- Kim, J.-H. Large ancient organic matter contributions to Arctic marine sediments (Svalbard). Limnol. Oceanogr. 2011, 56, 1463–1474. [Google Scholar] [CrossRef]
- Thomas, F.A.; Mohan, M.; Krishnan, K.P. Bacterial diversity and their metabolic profiles in the sedimentary environments of Ny-Ålesund, Arctic. Antonie van Leeuwenhoek 2021, 114, 1339–1360. [Google Scholar] [CrossRef]
- Vishnupriya, S.; Jabir, T.; Krishnan, K.P.; Mohamed Hatha, A.A. Bacterial community structure and functional profiling of high Arctic fjord sediments. World J. Microbiol. Biotechnol. 2021, 37, 133. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.N.; Winding, A. Microorganisms as Indicators of Soil Health; Technical Report No. 388; National Environmental Research Institute: Roskilde, Denmark, 2002; pp. 1–85. [Google Scholar]
- Fierer, N.; Wood, S.A.; Bueno de Mesquita, C.P. How microbes can, and cannot, be used to assess soil health. Soil Biol. Biochem. 2021, 153, 108111. [Google Scholar] [CrossRef]
- Fang, X.M.; Zhang, T.; Li, J.; Wang, N.-F.; Wang, Z.; Yu, L.-Y. Bacterial community pattern along the sediment seafloor of the Arctic fjorden (Kongsfjorden, Svalbard). Antonie van Leeuwenhoek 2019, 112, 1121–1136. [Google Scholar] [CrossRef]
- Venkatachalam, S.; Gopinath, A.; Krishnan, K.P. Fjords of the western and northern regions of Svalbard harbour distinct bacterioplankton community structures. World J. Microbiol. Biotechnol. 2023, 39, 57. [Google Scholar] [CrossRef]
- Kalinowska, A.; Ameryk, A.; Jankowska, K. Microbiological survey in two Arctic fjords: Total bacterial number and biomass comparison of Hornsund and Kongsfjorden. In Impact of Climate Changes on Marine Environments, GeoPlanet: Earth and Planetary Sciences; Zielinski, T., Weslawski, M., Kuliński, K., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 115–126. [Google Scholar] [CrossRef]
- Børsheim, K.Y.; Drinkwater, K.F. Different temperature adaptation in Arctic and Atlantic heterotrophic bacteria in the Barents Sea Polar Front region. J. Mar. Syst. 2014, 130, 160–166. [Google Scholar] [CrossRef]
- Beszczynska-Moller, A.; Weslawski, J.M.; Walczowski, W.; Zajaczkowski, M. Estimation of glacial meltwater discharge into Svalbard coastal waters. Oceanologia 1997, 39, 289–299. [Google Scholar]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.S.; Elliott, M. Ecology of Marine Sediments: From Science to Management; Oxford University Press: Oxford, UK, 2009; pp. 1–216. [Google Scholar] [CrossRef]
- Brakstad, O.G.; Nonstad, I.; Faksness, L.G.; Brandvik, P.J. Responses of Microbial Communities in Arctic Sea Ice After Contamination by Crude Petroleum Oil. Microb. Ecol. 2008, 55, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Cottrell, M.T.; Kirchman, D.L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 2000, 66, 1692–1697. [Google Scholar] [CrossRef] [PubMed]
- Teske, A.; Durbin, A.; Ziervogel, K.; Cox, C.; Arnosti, C. Microbial community composition and function in permanently cold seawater and sediments from an Arctic fjord of Svalbard. Appl. Environ. Microbiol. 2011, 77, 2008–2018. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, K.; Hassenrück, C.; Salman-Carvalho, V.; Holtappels, M.; Bienhold, C. Response of bacterial communities to different detritus compositions in arctic deep-sea sediments. Front. Microbiol. 2017, 8, 266. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, S.; Kerhervé, P.; Calleja, M.L.; Many, G.; Morata, N. Glacier inputs influence organic matter composition and prokaryotic distribution in a high Arctic fjord (Kongsfjorden, Svalbard). J. Mar. Syst. 2016, 164, 112–127. [Google Scholar] [CrossRef]
- Hargrave, B.T.; Holmer, M.; Newcombe, C.P. Towards a classification of organic enrichment in marine sediments based on biogeochemical indicators. Mar. Pollut. Bull. 2008, 56, 810–824. [Google Scholar] [CrossRef]
- Fenice, M.; Gallo, A.; Juarez-Jimenez, B.; Gonzalez-Lopez, J. Screening for extracellular enzyme activities by bacteria isolated from samples collected in the Tyrrhenian Sea. Ann. Microbiol. 2007, 57, 93–99. [Google Scholar] [CrossRef]
- Davey, K.E.; Kirby, R.R.; Turley, C.M.; Weightman, A.J.; Fry, J.C. Depth variation of bacterial extracellular enzyme activity and population diversity in the northeastern North Atlantic Ocean. Deep Sea Res. Part II 2001, 48, 1003–1017. [Google Scholar] [CrossRef]
- Sjölund, M.; Bonnedahl, J.; Hernandez, J.; Bengtsson, S.; Cederbrant, G.; Pinhassi, J.; Kahlmeter, G.; Olsen, B. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg. Infect. Dis. 2008, 14, 70–72. [Google Scholar] [CrossRef]
- Kalinowska, A.; Jankowska, K.; Fudala-Ksiazek, S.; Pierpaoli, M.; Luczkiewicz, A. The microbial community, its biochemical potential, and the antimicrobial resistance of Enterococcus spp. in Arctic lakes under natural and anthropogenic impact (West Spitsbergen). Sci. Total Environ. 2021, 763, 142998. [Google Scholar] [CrossRef] [PubMed]
- Visnupriya, S.; Jabir, T.; Akhil Prakash, E.; Mohamed Hatha, A.A. Antibiotic resistance of heterotrophic bacteria from the sediments of adjoining high Arctic fjords, Svalbard. Braz. J. Microbiol. 2024. [Google Scholar] [CrossRef]
- Allen, H.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Tan, L.; Li, L.; Ashbolt, N.; Wang, X.; Cui, Y.; Zhu, X.; Xu, Y.; Yang, Y.; Mao, D.; Luo, Y. Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci. Tot. Environ. 2018, 621, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.C.; Lee, N.; Aw, T.G. Antibiotic resistance in minimally human-impacted environments. Int. J. Environ. Res. Public Health 2020, 17, 3939. [Google Scholar] [CrossRef] [PubMed]
- Mohamed Hatha, A.A.; Neethu, S.; Nikhil, S.M.; Mujeeb Rahiman, K.M.; Krishnan, K.P.; Saramma, A.V. Relatively high antibiotic resistance among heterotrophic bacteria from Arctic fjord sediments than water—Evidence towards better selection pressure in the fjord sediments. Polar Sci. 2015, 9, 382–388. [Google Scholar] [CrossRef]
- AMAP (Arctic Monitoring and Assessment Programme). AMAP Assessment 2018: Biological Effects of Contaminants on Arctic Wildlife and Fish; Arctic Monitoring and Assessment Programme (AMAP): Tromsø, Norway, 2018; pp. vii+84. [Google Scholar]
- Rauseo, J.; Spataro, F.; Pescatore, T.; Patrolecco, L. Multiresidue determination and predicted risk assessment of emerging contaminants in sediments from Kongsfjorden, Svalbard. Sci. Tot. Environ. 2024, 922, 171156. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, G.; Song, W.; Ye, C.; Lin, H.; Li, Z.; Liu, W. Plastics in the marine environment are réservoirs for antibiotic and metal resistance genes. Environ. Int. 2019, 123, 79–86. [Google Scholar] [CrossRef]
- Agnew, A.; Wang, J.; Fanning, S.; Bearhop, S.; McMahon, B.J. Insights into antimicrobial resistance among long distance migratory East Canadian High Arctic light-bellied Brent geese (Branta bernicla hrota). Ir. Vet. 2015, 69, 13. [Google Scholar] [CrossRef]
- Pouch, A.; Zaborska, A. Climate change influence on migration of contaminants in the Arctic marine environment. GeoPlanet Earth Planet. Sci. 2015, 22, 75–90. [Google Scholar] [CrossRef]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Zaborska, A.; Beszczyńska-Moller, A.; Włodarska-Kowalczuk, M. History of heavy metal accumulation in the Svalbard area: Distribution, origin and transport pathways. Environ. Poll. 2017, 231, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Evenset, A.; Christensen, G.N.; Carroll, J.; Zaborska, A.; Berger, U.; Herzke, D.; Gregor, D. Historical trends in persistent organic pollutants and metals recorded in sediment from Lake Ellasjøen, Bjørnøya, Norwegian Arctic. Environ. Pollut. 2007, 146, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-G.; Xia, Y.; Zhang, T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 2017, 11, 651–662. [Google Scholar] [CrossRef]
- Gianfreda, L.; Rao, M.A. Interactions Between Xenobiotics and Microbial and Enzymatic Soil Activity. Crit. Rev. Environ. Sci. Technol. 2008, 38, 269–310. [Google Scholar] [CrossRef]
- Makowska-Zawierucha, N.; Mokracka, J.; Małecka, M.; Balazy, P.; Chełchowski, M.; Ignatiuk, D.; Zawierucha, K. Quantification of class 1 integrons and characterization of the associated gene cassettes in the high Arctic—Interplay of humans and glaciers in shaping the aquatic resistome. Ecol. Indic. 2022, 145, 109633. [Google Scholar] [CrossRef]
- Astrup, E. Antibiotic Resistance in Norway- NIPH. Public Health Report. 2014. Available online: https://www.fhi.no/en/op/public-health-report-2014/health--disease/antibiotic-resistance-in-norway---p/ (accessed on 14 September 2023).
- Norwegian Ministries. National Strategy against Antibiotic Resistance 2015–2020; Norwegian Ministry of Health and Care Services: Oslo, Norway, 2015; pp. 1–36. [Google Scholar]
- NORM/NORM-VET 2021. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Tromsø, Norway. September 2022, pp. 1–64. Available online: www.vetinst.no (accessed on 14 September 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).