Abstract
Environmental DNA (eDNA) is widely used for detecting target species, including monitoring endangered species and detecting the presence of invasive species. Detecting targeted species using the eDNA approach is typically carried out with species-specific qPCR assays. Amazon frogbit (Limnobium laevigatum) is classified as a State-Prohibited Matter Weed in NSW, Australia. It is a fast-growing perennial aquatic weed that outcompetes native aquatic plants, leading to a reduction in the habitats of aquatic animals. Early detection is crucial for the effective management of this species. In this study, we developed a qPCR assay for L. laevigatum based on the rpoB gene sequence. This assay was validated against 25 non-target aquatic and terrestrial species. It was found to be species-specific, with the positive signal exclusively detected in L. laevigatum. The assay was highly sensitive with the modelled detection limits of 3.66 copies of DNA/µL. Furthermore, our assay was validated using environmental samples collected from field sites with and without the presence of L. laevigatum. Our assay is an effective tool for targeted eDNA detection of L. laevigatum, which will enhance efforts to monitor and control this invasive aquatic weed.
1. Introduction
Environmental DNA (eDNA) refers to genetic materials obtained from environmental samples without information about the source organisms [1]. Since the first application of eDNA in macro-organisms in 2005 [2], it has been widely used for monitoring species in various environments, particularly in aquatic ecosystems [3,4]. EDNA technologies offer a non-invasive, sensitive and cost-efficient approach compared with traditional labor-intensive surveys.
EDNA has great potential for biosecurity surveillance of non-indigenous species (NIS), which are often rare and challenging to detect through visual observation. Nowadays, eDNA has been extensively applied to monitor target and non-target NIS using qPCR [4], digital PCR [5], or metabarcoding [6]. Although metabarcoding allows the simultaneous monitoring of multiple NIS, several studies reported that qPCR and digital PCR methods could provide higher sensitivities [7,8,9,10]. In addition, metabarcoding often faces challenges identifying plant DNA to the species level due to general lack of a single universally informative gene target suitable for plant DNA barcoding, and genetic overlap among some species at targeted genes [11].
Despite being broadly applied in monitoring of animal species, the application of eDNA in weed biosecurity is limited [12,13]. Several studies used a metabarcoding approach for biodiversity analyses to investigate the plant communities using soil [14], dust [15] and honey [16] samples. Additionally, qPCR-based targeted species detection methods have been developed for various aquatic weeds, such as Hydrilla verticillata (L.f.) Royle, Egeria densa Planch. and Myriophyllum spicatum L. [17,18,19]. These studies suggest that eDNA is a useful tool in addition to the traditional monitoring approaches for weed biosecurity.
Limnobium laevigatum (Humb. & Bonpl. ex Willd.) Heine, Amazon frogbit, is listed as a Prohibited Matter Weed in New South Wales (NSW) [20]. Native to Central and South America [21,22], this species has been widely introduced beyond its native range. Limnobium laevigatum is recognised as a noxious weed in numerous countries, including Australia, Indonesia, Japan, USA, Zambia and Zimbabwe. It was sold as an ornamental aquatic plant in Australia and first found outside of cultivation in 2011 in Queensland (QLD), and has since been reported in NSW, Western Australia (WA), Northern Territory (NT), and Victoria (VIC).
Limnobium laevigatum is a fast-growing, freshwater, perennial species that outcompetes native aquatic plant species, obstructs waterways and reduces shelter for aquatic animals [20]. Limnobium laevigatum is adapted to grow in tropical and subtropical climates [20]. It grows on the surface of freshwater, either free-floating or with its roots anchored in underwater substrates [20]. Limnobium laevigatum reproduces sexually through seed and asexually through stolon segments. Each plant can produce multiple seed pods, with each pod containing 20–30 seeds [20]. The seed bank can last for at least three years. The floating rosettes and seeds can be easily dispersed by water [20].
Currently, surveys for new incursions of L. laevigatum in Australia rely heavily on labour-intensive visual detection with the assistance of weed officers, contractors and volunteers. New approaches, such as remote sensing, are often limited by heavy canopy coverage and expensive operational costs. Visual monitoring of L. laevigatum is further hindered by the presence of a morphologically similar aquatic plant, Hydrocharis dubia (Blume) Backer, also commonly known as frogbit.
The use of eDNA offers an approach for early detection of weed species across all stages of their development. By so doing, it can enable proactive management efforts to identify and control novel weed emergences or, alternatively, monitor the spread of those that have established. In this study, we aimed to develop species-specific qPCR assays for L. laevigatum and validate the assays using environmental samples. Our goal is to establish a robust protocol for the detection and monitoring of L. laevigatum. The developed protocol will enhance weed management strategies and contribute to the preservation of aquatic ecosystems.
2. Materials and Methods
2.1. Specimens
The plant tissues used in this study comprised herbarium and fresh samples, including L. laevigatum (N = 16) from various localities and 25 non-targeted species (N = 64) used as negative controls in specificity testing of the assay (Supplementary Table S1). DNA extraction from sampled specimens was carried out using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The species identity of each sample was genetically verified through their rbcL sequences (Supplementary Table S1). The sequences of these specimens were deposited in GenBank under accession numbers PP001306-PP001385.
2.2. Assay Development
We obtained chloroplast genomic sequences of L. laevigatum and closely related species from GenBank. The sequences were aligned using MAFFT [23] and manually adjusted using BioEdit [24]. We selected highly diverse gene regions for designing primers and probes.
For L. laevigatum, we designed the diagnostic assay Ll26823. The assay targeted a 125 bp region of the chloroplast rpoB gene. This region genetically differs by over 12% between L. laevigatum and its most closely related species in Australia, H. dubia [25] (Figure 1). Primers and probes (Table 1) of assay Ll26823 were visually designed, and we used Multiple Primer Analyzer [26] to check them for the potential presence of confounding secondary structures. The primers and probes were synthesised by Sigma Aldrich (Woodlands, USA) with HPLC purification. G-Block DNA was synthesised by Integrated DNAs Technologies (Coralville, USA; Table 1) and used for assay optimisation.
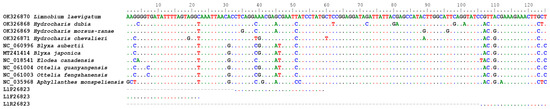
Figure 1.
Primer annealing sites in Limnobium laevigatum, targeting the rpoB gene.

Table 1.
Primers and probe designed for Limnobium laevigatum, amplifying a 125 bp region of the rpoB genes. Tm, melting temperature.
We optimised and tested the Ll26823 assay using the synthetic gBlock DNA. A standard curve of the Ll26823 assay was calculated using 10-fold serial dilutions of the gBlock, ranging from 106 copies/µL to 1 copy/µL with three replicates per dilution. The qPCR reactions were conducted in a 10 µL solution, consisting of 1 × PerfecTa Tough Mix (QuantaBio, Beverly, MA, USA), 900 nM of each primer, 250 nM of the probe and 2.5 µL of DNA template. The qPCR runs were performed on a Magnetic Induction Cycler (MIC, Bio Molecular Systems, Upper Coomera, Australia) with an initial denaturation for 5 min at 95 °C and 40 cycles of denaturing at 95 °C for 30 s and annealing at 58 °C for 30 s. We conducted an additional eight qPCR replicates using gBlock DNA at 5, 2.5, 1 and 0.5 copies/µL and evaluated the limits of detection (LOD) and quantification (LOQ) of the Ll26823 assay using the R script described by Klymus, et al. [27]. The specificity of the assays was tested using replicates of L. laevigatum and 25 non-targeted species (Supplementary Table S1).
2.3. Assay Multiplexing with QC1292
QC1292 is a quality control qPCR assay designed to target generic plant and bacterial DNA in the environment and can be used as a control to monitor the entire eDNA workflow [28]. We multiplexed Ll26823 with QC1292 to detect any potential false negatives resulting from processing errors and/or PCR inhibitors. The efficiency and sensitivity of the multiplexed assays were evaluated using a 1:1 mixture of gBlock DNA of the Ll26823 and the QC1292 assays. The gBlock DNA concentrations and qPCR reactions were conducted under the same reaction conditions we reported for the singleplex reactions with the addition of 100 nM of forward and reverse QC1292 primers and 28 nM of the QC1292 probe.
2.4. Field Application
To evaluate the applicability of these assays to field samples, we tested freshwater environmental samples collected from various locations in NSW, including locations with active L. laevigatum infestations, historical L. laevigatum infestations, and locations without historical records of L. laevigatum (Table 2 and Supplementary Table S2). Sample sites from Park Road, Bellambi, were located 500 m upstream from the infested property. Samples from Cawley Street, Bellambi, and Bellambi Lagoon were located 150 and 500 m downstream of the infested property, respectively (Supplementary Table S2). At most locations, samples were collected using a Smith-Root eDNA sampler with a self-preserving 5 µm PES filter unit (EnviroDNA, Brunswick, Victoria, Australia). Up to 2.0 L of water was filtered for each sample, with a targeted flow rate of 1.0 L/min, minimum flow rate of 0.3 L/min, and pressure limit of 10 psi. Two to ten samples were collected from each location, depending on the size of the water body and the volume of water that could be filtered (Supplementary Table S2). Prior to sampling at each location, a negative control was obtained by filtering 1.0 L ultrapure water using the Smith-Root eDNA sampler, as described above. Filters were stored at room temperature until returning to the laboratory, where they were removed from the filter housings and cut in half. One half was stored at −20 °C for later use, while the other half was cut into approximately 5 mm squares, and the eDNA was extracted using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) as described [28].

Table 2.
Results of analysis with the field-collected eDNA samples using the Limnobium laevigatum qPCR assay Ll268233 multiplexed with the QC1292 assay. All values are shown as mean ± standard error of the mean. Further details of these environmental samples are included in Supplementary Table S2. N/A: No Cq or eDNA concentration was recorded when there was no amplification in the Ll268233 assay for any sample or replicate.
Samples from Bulahdelah were obtained using a syringe-based eDNA sampling kit with a 5 µm PES filter unit (EnviroDNA, Brunswick, Victoria, Australia) following the manufacturer’s instructions. Two replicates and a negative control were collected. In the laboratory, 20 µL of proteinase K (20 mg/mL, Qiagen, Hilden, Germany) was added directly to the syringe filter unit without removing the preservative buffer and incubated overnight at 56 °C with the syringe attached. After incubation, the syringe was used to remove the lysate from the filter, and eDNA was extracted from the lysate using the DNeasy Blood and Tissue Kit. Each sample was tested using multiplex Ll26823 + QC1292 assays using the aforementioned reaction systems with three technical replicates. The individual qPCR efficiency [29] of each reaction was calculated using the built-in software in MIC. The concentration (copy number/µL) of DNA extractions was calculated from the standard curve generated by qPCR of gBlock, using the software in MIC. Samples that yielded a negative result in the QC1292 assay or had an efficiency lower than 85% in all qPCR replicates were excluded from the analysis. The concentration (copy number/L) of the eDNA in the environment was calculated using the formula (a × b)/(c × d), where:
- a represents the eDNA concentration of the DNA extraction (copy number/µL),
- b denotes the elution volume of DNA extraction (µL),
- c is the total volume of water filtered for each sample (L), and
- d indicates the proportion of filter extracted.
3. Results
3.1. Assay Development
Assay Ll26823 (Table 1) was developed based on a region of the chloroplast coding gene rpoB, and this region exhibited high levels of genetic variation among closely related species (Figure 1). The assay was specific to its target species and successfully amplified all 16 L. laevigatum samples in all replicates, while all 25 non-target species yielded negative results in the qPCR.
3.2. Assay Sensitivity
Synthetic gBlock DNA was used to evaluate the sensitivity of the Ll26823 assay. Our assay demonstrated high efficiency and sensitivity (Table 3). The qPCR efficiency of the Ll26823 assay was 99.06% when run as a singleplex reaction. When multiplexed with quality control assay QC1292, the Ll26823 assay maintained a high qPCR efficiency at 94.54%. The modelled limit of detection (LOD) and the limit of quantification (LOQ) of Ll26823 changed from 3.66 and 86 copies/µL when singleplexed to 16.5 and 16.5 copies/µL when multiplexed, respectively (Table 3).

Table 3.
The performance of the species-specific assay of Limnobium laevigatum when run as singleplex or multiplex (*) reactions. Showing the efficiency, modelled limit of detection (LOD) and modelled limit of quantification (LOQ) of Ll26823.
3.3. Field Application
The field application of multiplexed Ll26823 and QC1292 assays successfully detected L. laevigatum in all samples collected from the sites where the species was recorded in the three months prior to sampling, with Cq values ranging from 22.47 to 31.43 and eDNA concentrations from 1.92 × 104 to 1.11 × 106 copies/L (Table 2). The assay also successfully detected L. laevigatum at approximately 150 m and 500 m downstream from a source infestation (Supplementary Table S2). With increasing distance, the mean Cq value increased from 31.71 to 34.63, and eDNA concentrations decreased from 9.09 × 104 to 4.48 × 103, respectively. In contrast, no L. laevigatum eDNA was detected in the negative controls, at the control sites where the species had never been recorded, at one historic site where L. laevigatum had not been recorded for eight months prior to sampling, or upstream from an infestation (Table 2 and Supplementary Table S2). Endogenous DNA was detected in all environmental samples with individual qPCR efficiency over 85%, except as indicated in Supplementary Table S2, suggesting the absence of systematic errors in the eDNA workflow and minimal inhibition of the reaction.
4. Discussion
In this study, we developed a qPCR assay for the detection of L. laevigatum eDNA. The assay targets the chloroplast rpoB gene, which is normally present in hundreds to thousands of copies in plant leaf cells. Given the high sensitivity of this assay, we expect that it will assist in L. laevigatum monitoring for early detection of the weed and confirmation of eradication.
EDNA has been widely applied to animal and insect monitoring for biosecurity [30] and conservation purposes [31]. However, the application of eDNA in plant biosecurity is less common and poses additional challenges. Due to the highly fragmented nature of eDNA, the target gene region must be short, usually less than 280 bp [1]. A high level of between-species genetic diversity is needed in this target fragment for species-specific analysis, not only for molecular diagnostics but also for assay development. However, there is no generic gene region for plant molecular identification. The rbcL and matK genes commonly used for plant DNA barcoding are usually not diverse enough for species-specific primer and probe design. In the case of L. laevigatum, the genetic distance to the closest species, H. dubia, in Australia is 95.6% and 98.9% for matK and rbcL genes (Based on Blast result of sequences AB002574 and NC_061221, and AB004894 and AB004892), respectively. It is very difficult to design a species-specific qPCR assay based on these low levels of genetic distance. Sequence information, especially for some weed species, is limited in public databases, such as GenBank. Limnobium laevigatum, for example, only has 11 sequences available in GenBank (accessed 26 December 2023). It is challenging to find a gene region that has been sequenced among closely related species and is sufficiently diverse for assay development. In this study, we compared the chloroplast genome of L. laevigatum against other closely related species from the Family of Hydrocharitaceae [25] and discovered the highly diverse rpoB gene (Figure 1). As a coding region, this region is generally more conserved than non-coding regions within the same species. Subsequently, intraspecific polymorphisms in the gene are rare, and chances of obtaining false negatives due to primer and or probe mismatch are low.
The amount of DNA released from an individual plant can vary significantly depending on its developmental stages, and this can present a challenge to the detection of plant eDNA [19]. Kuehne, et al. [19] reported very low eDNA concentrations of Egeria densa and Myriophyllum spicatum. The amount of eDNA detected only weakly corresponded with plant abundance but increased significantly during plant senescence [19]. Often, due to the release of pollen, the observed plant eDNA peaks during the flowering period. Surprisingly, Kuehne, et al. [19] did not report such a peak in their study. Other research has reported higher eDNA concentration during the growth period of Hydrilla verticillata [32].
In our study, we did not measure the relationship between eDNA abundance and the L. laevigatum growth stage or its biomass. Unlike biodiversity conservation research, biosecurity research targets NIS, in this case, a Prohibited Matter Weed, which is required to be eradicated from the environment. The detection of the species triggers the same management responses regardless of how many plants are found. Similarly, the detection of eDNA raises the alarm that the target species could potentially be present in the environment. Paired with research on eDNA degradation (research in progress), we anticipate that future studies may enable us to use eDNA abundance to estimate the potential distribution of L. laevigatum at various spatial scales, as well as determine the frequency of field surveys required.
In this study, we detected L. laevigatum from all locations where the species was recently recorded. Compared with other studies of aquatic plants based on qPCR of cpDNA markers, the eDNA concentration detected in our study was similar to that measured earlier, ranging from 4.48 × 103 to 1.11 × 106 copies/L. This compares with a maximum of around 3.5 × 104 copies/L of invasive Egeria densa in Japan [33] and up to 2.12 × 105 copies/L of the native Hydrilla verticillata in Japan [17]. We found a decrease of more than an order of magnitude between samples taken ~150 m and ~500 m downstream from an infestation and no detectable eDNA upstream from an infestation. This suggests that eDNA samples taken at regular intervals along a watercourse or riverine drainage system could be used to more effectively direct manual surveillance efforts. For instance, if downstream eDNA is detected, focusing upstream surveillance efforts may reveal sites likely containing patches of an invasive weed. The relationships between eDNA concentration and plant abundance or biomass, and between eDNA concentration and distance from source plants, is poorly understood (see above), so this may need to be assessed on a species-by-species basis. Concentrations of eDNA may be influenced by variables such as plant developmental stage, with some studies showing a peak in eDNA concentration during plant senescence [19,33] and others showing a peak during the growth period [32]. Given that all the sample sites in our study had been controlled with herbicides, we may expect high eDNA concentrations due to forced “senescence”, a result of the control efforts. Further studies under controlled laboratory conditions, as well as repeated sampling from the same field sites over time, would help to parameterise relationships between eDNA concentration, plant abundance, and growth stage for L. laevigatum.
It’s worth mentioning that eDNA of L. laevigatum was detected not only in the locations where live plants were visually observed but also in previous infestation sites where no live plants were observed. For example, we detected L. laevigatum eDNA at Ogden Road, Oakville, where the plans were most recently observed 3 months earlier. This finding highlighted an important issue of legacy DNA, which can persist in the environment for a very long period under optimal conditions after the source species have been extirpated, e.g., ancient DNA. The persistence of legacy DNA in the environment is a case-by-case issue depending on the type of environment, temperature, pH, target species, etc [34,35,36,37]. Therefore, the detection of eDNA does not necessarily equate to the presence of target species. This issue could possibly be resolved with environmental RNA (eRNA), which degrades much faster than eDNA [37]. A positive detection of eRNA is normally considered to be the detection of a living target [38]. However, the nature of rapid degradation poses a considerable challenge in eRNA research, which is currently much less studied compared with eDNA. The eRNA analysis for L. laevigatum is currently under development, which could potentially further assist in the monitoring of L. laevigatum and eliminate the potential issue of false positives caused by the persistence of legacy DNA in the environment.
Notably, in our study, we incorporated a quality control qPCR QC1292 in the assay to monitor false negative results [28]. The eDNA workflow consists of multiple steps, from sample collection to data analysis [39]. A negative result could arise not only from the absence or low abundance of the target DNA but also from systematic errors in the workflow, such as a broken filter. The introduction of a quality control assay targeting endogenous eDNA enables the monitoring of the entire workflow and enhances the reliability of interpreting the eDNA results. In addition, the target assay Ll26823 exhibited a similar LOD when multiplexed with the quality control assay, which was similar to a previous study [28].
In this study, we demonstrate that L. laevigatum infestations can be detected through eDNA using a qPCR assay. Further research using this assay under controlled laboratory conditions, as well as the development of eRNA methods, will improve our understanding of the detectability of L. laevigatum in the field. This will provide a useful surveillance tool for delimitation of new infestations of L. laevigatum and for assessment of the effectiveness of eradication efforts.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/environments11040066/s1, Table S1: Plant samples used in this study for species specification test of Ll26823 assay. Table S2. Details of environmental samples collected in this study. N: number of replicates per location.
Author Contributions
Conceptualization, X.Z. and K.L.B.; methodology, X.Z. and K.L.B.; data analysis, X.Z.; investigation, X.Z. and K.L.B.; resources, X.Z. and K.L.B.; data curation, X.Z.; writing—original draft preparation, X.Z.; writing—review and editing, X.Z., K.L.B., H.W. and D.G.; supervision, H.W. and D.G.; project administration, H.W.; funding acquisition, H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the NSW Department of Primary Industries.
Data Availability Statement
Sequence data that is published with this manuscript is available from GenBank under accession numbers PP001306-PP001385.
Acknowledgments
We acknowledge the Queensland Department of Agriculture and Fisheries, Queensland Herbarium and Biodiversity Sciences, Australian National Herbarium, NSW Department of Primary Industries, and many weed officers and local councils who provided plant samples. Camden, Shoalhaven, Illawarra, Mid Coast and Hawkesbury River County councils are acknowledged for their assistance with the collection of environmental samples. Bernie Dominiak, Fiona Schneiders and Graham Charles (New South Wales Department of Primary Industries) reviewed a pre-submission version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Brauwer, M.; Chariton, A.; Clarke, L.; Cooper, M.; Dibattista, J.; Furlan, E.; Giblot-Ducray, D.; Gleeson, D.; Harford, A.; Herbert, S.; et al. Environmental DNA Test Validation Guidelines; National eDNA Reference Centre: Canberra, Australia, 2022. [Google Scholar]
- Martellini, A.; Payment, P.; Villemur, R. Use of eukaryotic mitochondrial DNA to differentiate human, bovine, porcine and ovine sources in fecally contaminated surface water. Water Res. 2005, 39, 541–548. [Google Scholar] [CrossRef]
- Wilkes Walburn, J.; Rourke, M.L.; Furlan, E.; DiBattista, J.D.; Broadhurst, M.K.; Fowler, A.M.; Hughes, J.M.; Fielder, S. Robust environmental DNA assay development and validation: A case study with two vulnerable Australian fish. Aquat. Conserv. Mar. Freshw. Ecosyst. 2022, 32, 1225–1231. [Google Scholar] [CrossRef]
- Prabhakaran, G.K.; Sunkara, M.; Raghavan, R.; Umapathy, G. Development of a species-specific qPCR assay for the detection of invasive African sharptooth catfish (Clarias gariepinus) using environmental DNA. Biol. Invasions 2023, 25, 975–982. [Google Scholar] [CrossRef]
- Doi, H.; Takahara, T.; Minamoto, T.; Matsuhashi, S.; Uchii, K.; Yamanaka, H. Droplet Digital Polymerase Chain Reaction (PCR) Outperforms Real-Time PCR in the Detection of Environmental DNA from an Invasive Fish Species. Environ. Sci. Technol. 2015, 49, 5601–5608. [Google Scholar] [CrossRef] [PubMed]
- Mychek-Londer, J.G.; Balasingham, K.D.; Heath, D.D. Using environmental DNA metabarcoding to map invasive and native invertebrates in two Great Lakes tributaries. Environ. DNA 2020, 2, 283–297. [Google Scholar] [CrossRef]
- McColl-Gausden, E.F.; Weeks, A.R.; Coleman, R.; Song, S.; Tingley, R. Using hierarchical models to compare the sensitivity of metabarcoding and qPCR for eDNA detection. Ecol. Inform. 2023, 75, 102072. [Google Scholar] [CrossRef]
- Yu, Z.; Ito, S.I.; Wong, M.K.; Yoshizawa, S.; Inoue, J.; Itoh, S.; Yukami, R.; Ishikawa, K.; Guo, C.; Ijichi, M.; et al. Comparison of species-specific qPCR and metabarcoding methods to detect small pelagic fish distribution from open ocean environmental DNA. PLoS ONE 2022, 17, e0273670. [Google Scholar] [CrossRef]
- Harper, L.R.; Lawson Handley, L.; Hahn, C.; Boonham, N.; Rees, H.C.; Gough, K.C.; Lewis, E.; Adams, I.P.; Brotherton, P.; Phillips, S.; et al. Needle in a haystack? A comparison of eDNA metabarcoding and targeted qPCR for detection of the great crested newt (Triturus cristatus). Ecol. Evol. 2018, 8, 6330–6341. [Google Scholar] [CrossRef] [PubMed]
- Chandelier, A.; Hulin, J.; San Martin, G.; Debode, F.; Massart, S. Comparison of qPCR and metabarcoding methods as tools for the detection of airborne inoculum of forest fungal pathogens. Phytopathology 2021, 111, 570–581. [Google Scholar] [CrossRef]
- Kress, W.J. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef]
- Anglès d’Auriac, M.B.; Strand, D.A.; Mjelde, M.; Demars, B.O.L.; Thaulow, J. Detection of an invasive aquatic plant in natural water bodies using environmental DNA. PLoS ONE 2019, 14, e0219700. [Google Scholar] [CrossRef]
- Batchelor, K.L.; Bell, K.L.; Campos, M.; Webber, B.L. Can honey bees be used to detect rare plants? Taking an eDNA approach to find the last plants in a weed eradication program. Environ. DNA 2023, 5, 1516–1526. [Google Scholar] [CrossRef]
- Vasar, M.; Davison, J.; Moora, M.; Sepp, S.-K.; Anslan, S.; Al-Quraishy, S.; Bahram, M.; Bueno, C.G.; Cantero, J.J.; Fabiano, E.C.; et al. Metabarcoding of soil environmental DNA to estimate plant diversity globally. Front. Plant Sci. 2023, 14, 1106617. [Google Scholar] [CrossRef]
- Lennartz, C.; Kurucar, J.; Coppola, S.; Crager, J.; Bobrow, J.; Bortolin, L.; Comolli, J. Geographic source estimation using airborne plant environmental DNA in dust. Sci. Rep. 2021, 11, 16238. [Google Scholar] [CrossRef]
- Jones, L.; Brennan, G.L.; Lowe, A.; Creer, S.; Ford, C.R.; de Vere, N. Shifts in honeybee foraging reveal historical changes in floral resources. Commun. Biol. 2021, 4, 37. [Google Scholar] [CrossRef]
- Matsuhashi, S.; Doi, H.; Fujiwara, A.; Watanabe, S.; Minamoto, T. Evaluation of the environmental DNA method for estimating distribution and biomass of submerged aquatic plants. PLoS ONE 2016, 11, e0156217. [Google Scholar] [CrossRef] [PubMed]
- Gantz, C.A.; Renshaw, M.A.; Erickson, D.; Lodge, D.M.; Egan, S.P. Environmental DNA detection of aquatic invasive plants in lab mesocosm and natural field conditions. Biol. Invasions 2018, 20, 2535–2552. [Google Scholar] [CrossRef]
- Kuehne, L.M.; Ostberg, C.O.; Chase, D.M.; Duda, J.J.; Olden, J.D. Use of environmental DNA to detect the invasive aquatic plants Myriophyllum spicatum and Egeria densa in lakes. Freshw. Sci. 2020, 39, 521–533. [Google Scholar] [CrossRef]
- van de Witte, Y. Limnobium laevigatum (South American Spongeplant); CABI Compendium: Oxford, UK, 2022. [Google Scholar]
- Simon, J.B.; Panetta, F.D.; Kylie, E.G. Progress towards the Eradication of Mikania Vine (Mikania micrantha) and Limnocharis (Limnocharis flava) in Northern Australia. Invasive Plant Sci. Manag. 2008, 1, 296–303. [Google Scholar] [CrossRef]
- Howard, G.; Hyde, M.; Bingham, M. Alien Limnobium laevigatum (Humb. & Bonpl. ex Willd.) Heine (Hydrocharitaceae) becoming prevalent in Zimbabwe and Zambia. BioInvasions Rec. 2016, 5, 221–225. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Li, Z.-Z.; Lehtonen, S.; Gichira, A.W.; Martins, K.; Efremov, A.; Wang, Q.-F.; Chen, J.-M. Plastome phylogenomics and historical biogeography of aquatic plant genus Hydrocharis (Hydrocharitaceae). BMC Plant Biol. 2022, 22, 106. [Google Scholar] [CrossRef]
- Anonymous. Multiple Primer Analyzer. Available online: https://www.thermofisher.com/au/en/home/brands/thermo-scientific/molecular-biology/molecular-biology-learning-center/molecular-biology-resource-library/thermo-scientific-web-tools/multiple-primer-analyzer.html (accessed on 4 December 2023).
- Klymus, K.E.; Merkes, C.M.; Allison, M.J.; Goldberg, C.S.; Helbing, C.C.; Hunter, M.E.; Jackson, C.A.; Lance, R.F.; Mangan, A.M.; Monroe, E.M.; et al. Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA 2020, 2, 271–282. [Google Scholar] [CrossRef]
- Zhu, X.; Bell, K.L.; Rourke, M.L.; Wu, H.; Gopurenko, D. Generic qPCR assays for quality control in environmental DNA research. Environ. DNA 2024, accepted. [Google Scholar]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-González, A.; Thuo, D.N.; Divi, U.; Sparks, K.; Wallenius, T.; Gleeson, D. Detection of Khapra beetle environmental DNA using portable technologies in Australian biosecurity. Front. Insect Sci. 2022, 2, 795379. [Google Scholar] [CrossRef]
- Lunghi, E.; Valle, B.; Guerrieri, A.; Bonin, A.; Cianferoni, F.; Manenti, R.; Ficetola, G.F. Environmental DNA of insects and springtails from caves reveals complex processes of eDNA transfer in soils. Sci. Total Environ. 2022, 826, 154022. [Google Scholar] [CrossRef]
- Matsuhashi, S.; Minamoto, T.; Doi, H. Seasonal change in environmental DNA concentration of a submerged aquatic plant species. Freshw. Sci. 2019, 38, 654–660. [Google Scholar] [CrossRef]
- Kodama, T.; Miyazono, S.; Akamatsu, Y.; Tsuji, S.; Nakao, R. Abundance estimation of riverine macrophyte Egeria densa using environmental DNA: Effects of sampling season and location. Limnology 2022, 23, 299–308. [Google Scholar] [CrossRef]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of environmental DNA in marine systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef] [PubMed]
- Joseph, C.; Faiq, M.E.; Li, Z.; Chen, G. Persistence and degradation dynamics of eDNA affected by environmental factors in aquatic ecosystems. Hydrobiologia 2022, 849, 4119–4133. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental Conditions Influence eDNA Persistence in Aquatic Systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Kagzi, K.; Hechler, R.M.; Fussmann, G.F.; Cristescu, M.E. Environmental RNA degrades more rapidly than environmental DNA across a broad range of pH conditions. Mol. Ecol. Resour. 2022, 22, 2640–2650. [Google Scholar] [CrossRef] [PubMed]
- Pochon, X.; Zaiko, A.; Fletcher, L.M.; Laroche, O.; Wood, S.A. Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications. PLoS ONE 2017, 12, e0187636. [Google Scholar] [CrossRef]
- Furlan, E.M.; Gleeson, D. Improving reliability in environmental DNA detection surveys through enhanced quality control. Mar. Freshw. Res. 2017, 68, 388–395. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).