Runs of Homozygosity Preliminary Investigation in Pig Breeds
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Population and Genomic Data
2.2. ROH Detection
2.3. Genetic Differentiation and Selection Signatures
3. Results
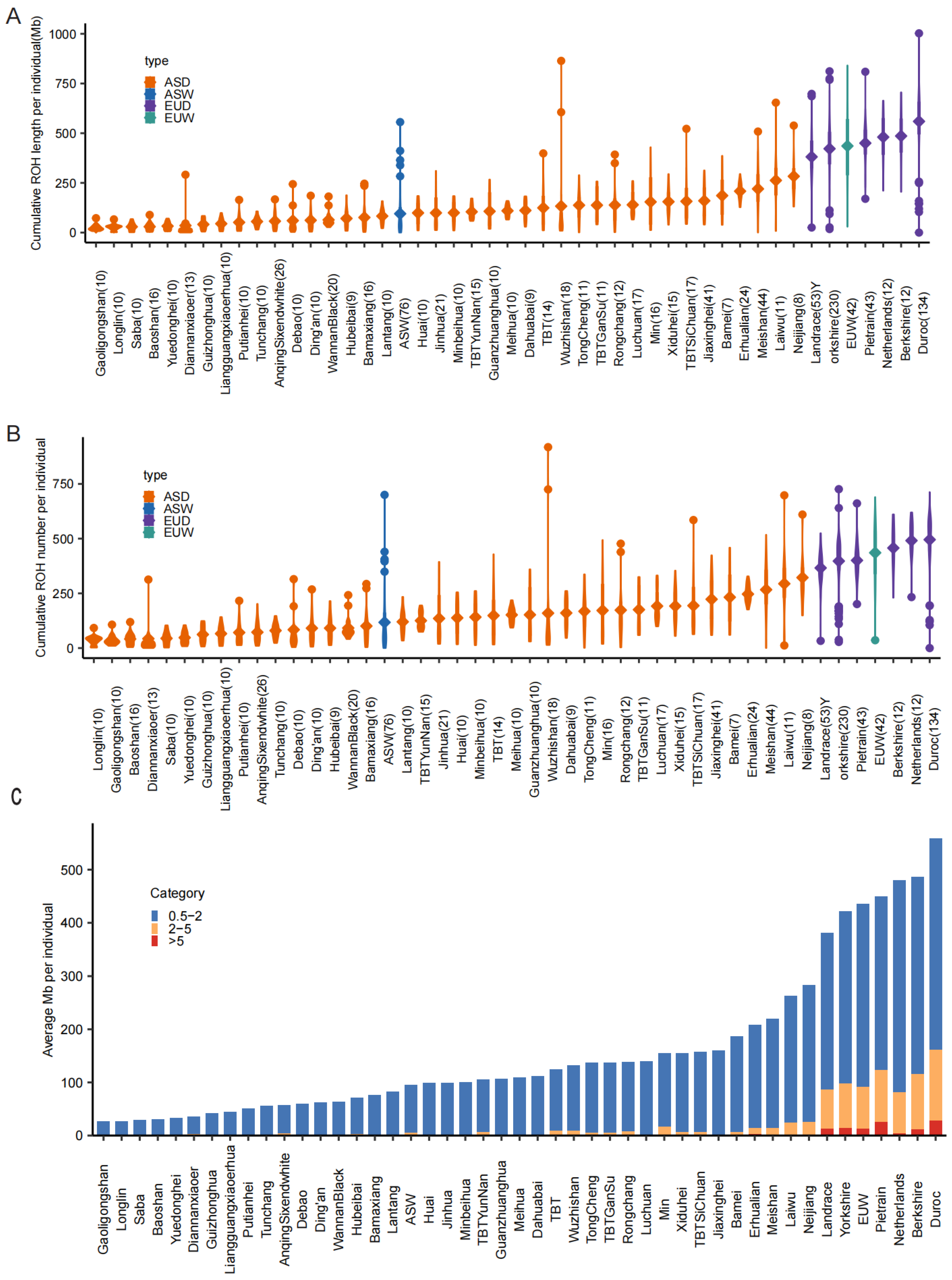
3.1. ROH Distribution Across Global Pig Populations
3.2. Hybridization Reduces ROH Burden
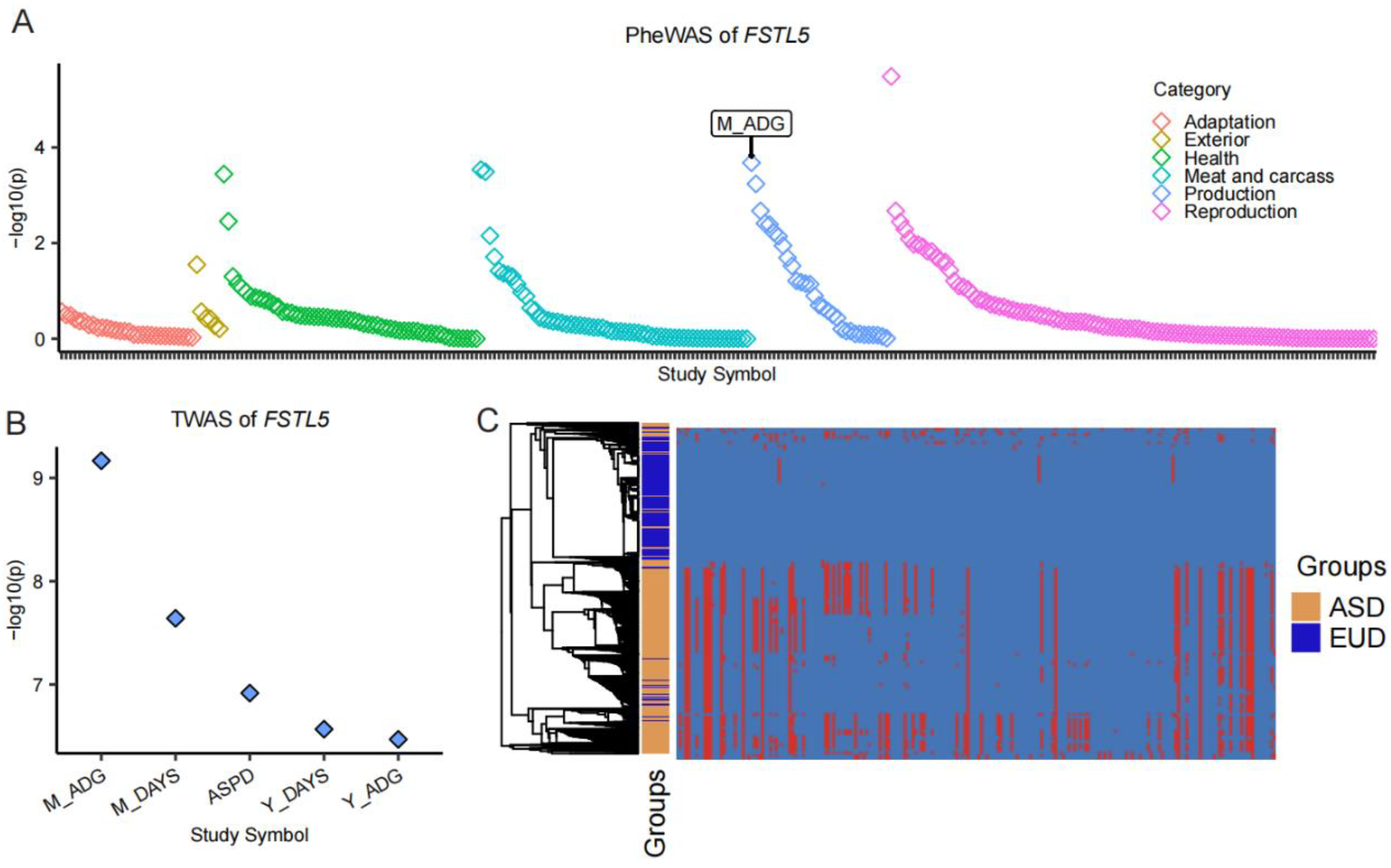
3.3. Selection Pressures and Functional Significance of ROH Islands
3.4. FSTL5: A Key Gene Under Selection in European Pigs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Broman, K.W.; Weber, J.L. Long homozygous chromosomal segments in reference families from the centre d’Etude du polymorphisme humain. Am. J. Hum. Genet. 1999, 65, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhao, G.; Yang, L.; Zhu, B.; Chen, Y.; Zhang, L.; Gao, X.; Gao, H.; Liu, G.E.; Li, J. Genomic Patterns of Homozygosity in Chinese Local Cattle. Sci. Rep. 2019, 9, 16977. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, G.; Lin, X.; Zhang, J.; Hou, G.; Zhang, L.; Liu, D.; Li, Y.; Li, J.; Xu, L. Genomic inbreeding and runs of homozygosity analysis of indigenous cattle populations in southern China. PLoS ONE 2022, 17, e0271718. [Google Scholar] [CrossRef]
- Di Gregorio, P.; Perna, A.; Di Trana, A.; Rando, A. Identification of ROH Islands Conserved through Generations in Pigs Belonging to the Nero Lucano Breed. Genes 2023, 14, 1503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Xiao, Q.; Sun, H.; Gao, H.; Yang, Y.; Chen, J.; Li, Z.; Xue, M.; Ma, P.; et al. Distribution of runs of homozygosity in Chinese and Western pig breeds evaluated by reduced-representation sequencing data. Anim. Genet. 2018, 49, 579–591. [Google Scholar] [CrossRef]
- Chen, C.; Zhu, B.; Tang, X.; Chen, B.; Liu, M.; Gao, N.; Li, S.; Gu, J. Genome-Wide Assessment of Runs of Homozygosity by Whole-Genome Sequencing in Diverse Horse Breeds Worldwide. Genes 2023, 14, 1211. [Google Scholar] [CrossRef]
- Abdoli, R.; Mirhoseini, S.Z.; Ghavi Hossein-Zadeh, N.; Zamani, P.; Moradi, M.H.; Ferdosi, M.H.; Sargolzaei, M.; Gondro, C. Runs of homozygosity and cross-generational inbreeding of Iranian fat-tailed sheep. Heredity 2023, 130, 358–367. [Google Scholar] [CrossRef]
- Bertolini, F.; Cardoso, T.F.; Marras, G.; Nicolazzi, E.L.; Rothschild, M.F.; Amills, M.; AdaptMap Consortium. Genome-wide patterns of homozygosity provide clues about the population history and adaptation of goats. Genet. Sel. Evol. 2018, 50, 59. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yuan, M.; Zhan, F.; Song, M.; Shang, P.; Yang, F.; Li, X.; Qiao, R.; Han, X.; et al. Genome-Wide Association Studies and Runs of Homozygosity to Identify Reproduction-Related Genes in Yorkshire Pig Population. Genes 2023, 14, 2133. [Google Scholar] [CrossRef]
- Wang, S.; Yang, J.; Li, G.; Ding, R.; Zhuang, Z.; Ruan, D.; Wu, J.; Yang, H.; Zheng, E.; Cai, G.; et al. Identification of Homozygous Regions with Adverse Effects on the Five Economic Traits of Duroc Pigs. Front. Vet. Sci. 2022, 9, 855933. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, W.; Wang, Z.; Pan, Y.; Wang, Q.; Zhang, Z. Integration of ssGWAS and ROH analyses for uncovering genetic variants associated with reproduction traits in Large White pigs. Anim. Genet. 2024, 55, 714–724. [Google Scholar] [CrossRef]
- Xu, Z.; Sun, H.; Zhang, Z.; Zhao, Q.; Olasege, B.S.; Li, Q.; Yue, Y.; Ma, P.; Zhang, X.; Wang, Q.; et al. Assessment of Autozygosity Derived from Runs of Homozygosity in Jinhua Pigs Disclosed by Sequencing Data. Front. Genet. 2019, 10, 274. [Google Scholar] [CrossRef]
- Wang, Z.; Zhong, Z.; Xie, X.; Wang, F.; Pan, D.; Wang, Q.; Pan, Y.; Xiao, Q.; Tan, Z. Detection of Runs of Homozygosity and Identification of Candidate Genes in the Whole Genome of Tunchang Pigs. Animals 2024, 14, 201. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Q.; Xu, Y.J.; Chen, Z.T.; Li, H.; Zhang, Z.; Wang, Q.S.; Pan, Y.C. Genome-wide detection of runs of homozygosity and heterozygosity in Tunchang pigs. Animal 2024, 18, 101236. [Google Scholar] [CrossRef] [PubMed]
- Peripolli, E.; Munari, D.P.; Silva, M.; Lima, A.L.F.; Irgang, R.; Baldi, F. Runs of homozygosity: Current knowledge and applications in livestock. Anim. Genet. 2017, 48, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Gusev, A.; Lowe, J.K.; Stoffel, M.; Daly, M.J.; Altshuler, D.; Breslow, J.L.; Friedman, J.M.; Pe’er, I. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009, 19, 318–326. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. High-resolution detection of identity by descent in unrelated individuals. Am. J. Hum. Genet. 2010, 86, 526–539. [Google Scholar] [CrossRef]
- Teng, J.; Gao, Y.; Yin, H.; Bai, Z.; Liu, S.; Zeng, H.; Pig, G.C.; Bai, L.; Cai, Z.; Zhao, B.; et al. A compendium of genetic regulatory effects across pig tissues. Nat. Genet. 2024, 56, 112–123. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Szpiech, Z.A. selscan 2.0: Scanning for sweeps in unphased data. Bioinformatics 2024, 40, btae006. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhang, W.; Lin, Q.; Gao, Y.; Teng, J.; Xu, Z.; Cai, X.; Zhong, Z.; Wu, J.; Liu, Y.; et al. PigBiobank: A valuable resource for understanding genetic and biological mechanisms of diverse complex traits in pigs. Nucleic Acids Res. 2024, 52, D980–D989. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.W.; Fletcher, A.J.; Hill, J.P.; McNabb, W.C. Modeling the Contribution of Meat to Global Nutrient Availability. Front. Nutr. 2022, 9, 766796. [Google Scholar] [CrossRef]
- Zhao, F.; Zhang, P.; Wang, X.; Akdemir, D.; Garrick, D.; He, J.; Wang, L. Genetic gain and inbreeding from simulation of different genomic mating schemes for pig improvement. J. Anim. Sci. Biotechnol. 2023, 14, 87. [Google Scholar] [CrossRef]
- Larson, G.; Liu, R.; Zhao, X.; Yuan, J.; Fuller, D.; Barton, L.; Dobney, K.; Fan, Q.; Gu, Z.; Liu, X.H.; et al. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc. Natl. Acad. Sci. USA 2010, 107, 7686–7691. [Google Scholar] [CrossRef]
- Larson, G.; Dobney, K.; Albarella, U.; Fang, M.; Matisoo-Smith, E.; Robins, J.; Lowden, S.; Finlayson, H.; Brand, T.; Willerslev, E.; et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 2005, 307, 1618–1621. [Google Scholar] [CrossRef]
- Humble, E.; Stoffel, M.A.; Dicks, K.; Ball, A.D.; Gooley, R.M.; Chuven, J.; Pusey, R.; Remeithi, M.A.; Koepfli, K.P.; Pukazhenthi, B.; et al. Conservation management strategy impacts inbreeding and mutation load in scimitar-horned oryx. Proc. Natl. Acad. Sci. USA 2023, 120, e2210756120. [Google Scholar] [CrossRef]
- Groenen, M.A.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.J.; et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef]
- Barratt, J.; Weitz, I. Complement Factor D as a Strategic Target for Regulating the Alternative Complement Pathway. Front. Immunol. 2021, 12, 712572. [Google Scholar] [CrossRef]
- Hemmann, P.; Kloppenburg, L.; Breinbauer, R.; Ehnert, S.; Blumenstock, G.; Reumann, M.K.; Erne, F.; Jazewitsch, J.; Schwarz, T.; Baumgartner, H.; et al. AZU1: A new promising marker for infection in orthopedic and trauma patients? EXCLI J. 2024, 23, 53–61. [Google Scholar] [CrossRef]
- Ye, J.; Liu, W.; Yu, X.; Wu, L.; Chen, Z.; Yu, Y.; Wang, J.; Bai, S.; Zhang, M. TRAF7-targeted HOXA5 acts as a tumor suppressor in prostate cancer progression and stemness via transcriptionally activating SPRY2 and regulating MEK/ERK signaling. Cell Death Discov. 2023, 9, 378. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Li, H.; Xiao, M.; Zhou, C.; Liu, J.; Weng, S.; Wei, R. CCNF is a potential pancancer biomarker and immunotherapy target. Front. Oncol. 2023, 13, 1109378. [Google Scholar] [CrossRef]
- Steklov, M.; Pandolfi, S.; Baietti, M.F.; Batiuk, A.; Carai, P.; Najm, P.; Zhang, M.; Jang, H.; Renzi, F.; Cai, Y.; et al. Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science 2018, 362, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bu, X.; Chu, C.; Dai, X.; Asara, J.M.; Sicinski, P.; Freeman, G.J.; Wei, W. PRMT1 mediated methylation of cGAS suppresses anti-tumor immunity. Nat. Commun. 2023, 14, 2806. [Google Scholar] [CrossRef]
- Li, C.; Dai, L.; Zhang, J.; Zhang, Y.; Lin, Y.; Cheng, L.; Tian, H.; Zhang, X.; Wang, Q.; Yang, Q.; et al. Follistatin-like protein 5 inhibits hepatocellular carcinoma progression by inducing caspase-dependent apoptosis and regulating Bcl-2 family proteins. J. Cell. Mol. Med. 2018, 22, 6190–6201. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Fu, Y.; Zhu, F.; Mu, C.; Li, R.; Song, W.; Shi, C.; Ye, Y.; Wang, C. Transcriptomic analysis of Portunus trituberculatus reveals a critical role for WNT4 and WNT signalling in limb regeneration. Gene 2018, 658, 113–122. [Google Scholar] [CrossRef]
- Fadel, I.M.; Ragab, M.H.; Eid, O.M.; Helmy, N.A.; El-Bassyouni, H.T.; Mazen, I. IGF1R, IGFALS, and IGFBP3 gene copy number variations in a group of non-syndromic Egyptian short children. J. Genet. Eng. Biotechnol. 2021, 19, 109. [Google Scholar] [CrossRef]
- Alizadeh, F.; Moradian, F.; Farhadi, A. Association of allelic polymorphisms of IGFALS gene with growth traits in Makouei and Ghezel sheep breeds. Trop. Anim. Health Prod. 2020, 52, 3027–3034. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Li, G.; Ayalew, W.; Zhong, Z.; Liu, X.; Sun, J.; Li, J. Runs of Homozygosity Preliminary Investigation in Pig Breeds. Animals 2025, 15, 988. https://doi.org/10.3390/ani15070988
Liu Y, Li G, Ayalew W, Zhong Z, Liu X, Sun J, Li J. Runs of Homozygosity Preliminary Investigation in Pig Breeds. Animals. 2025; 15(7):988. https://doi.org/10.3390/ani15070988
Chicago/Turabian StyleLiu, Yuqiang, Guangzhen Li, Wondossen Ayalew, Zhanming Zhong, Xiaohong Liu, Jiajie Sun, and Jiaqi Li. 2025. "Runs of Homozygosity Preliminary Investigation in Pig Breeds" Animals 15, no. 7: 988. https://doi.org/10.3390/ani15070988
APA StyleLiu, Y., Li, G., Ayalew, W., Zhong, Z., Liu, X., Sun, J., & Li, J. (2025). Runs of Homozygosity Preliminary Investigation in Pig Breeds. Animals, 15(7), 988. https://doi.org/10.3390/ani15070988





