SIRT5 Regulates Lipid Deposition in Goat Preadipocytes via PI3K-Akt and MAPK Signaling Pathways
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. The Isolation, Culture, and Cryopreservation of Caprine Intramuscular Preadipocytes
2.3. Cloning and Biological Analysis of the Goat SIRT5 Gene
2.4. Construction of Goat SIRT5 Gene Time Series Expression Profile
2.5. Vector Construction, Chemical Synthesis of siRNA, and Transfection
2.6. Oil Red O Staining and Triglyceride Assay
2.7. Total RNA Extraction and Quantitative Real-Time PCR (RT-qPCR)
2.8. RNA Sequencing (RNA-seq)
2.9. Cell Counting Kit-8 (CCK-8) Assay
2.10. Flow Cytometry for Apoptosis Analysis
2.11. Western Blot Analysis
2.12. Statistical Analysis
3. Results
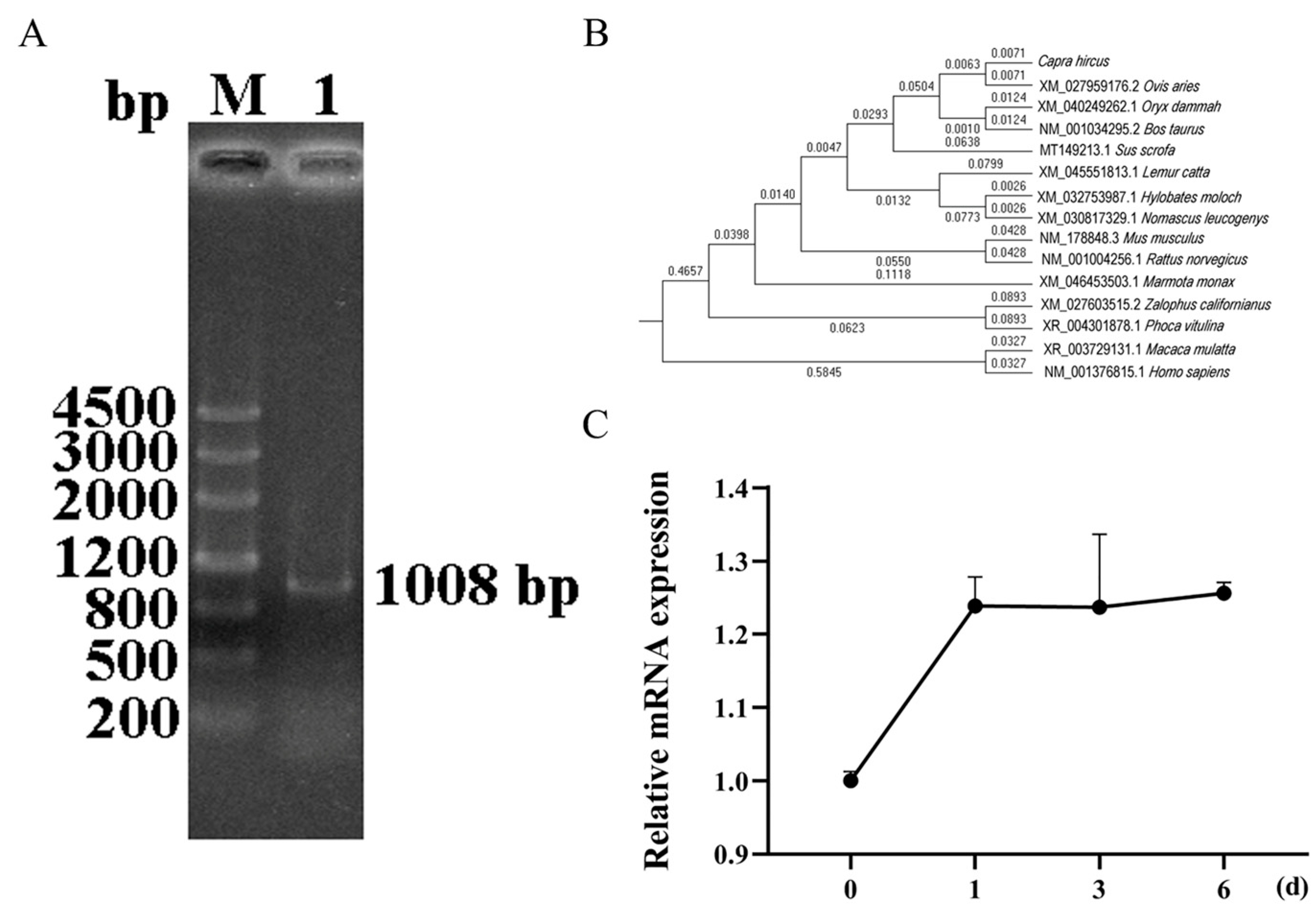
3.1. Cloning of Goat SIRT5 Gene and Its Temporal Expression Profile During Differentiation of Intramuscular Preadipocytes
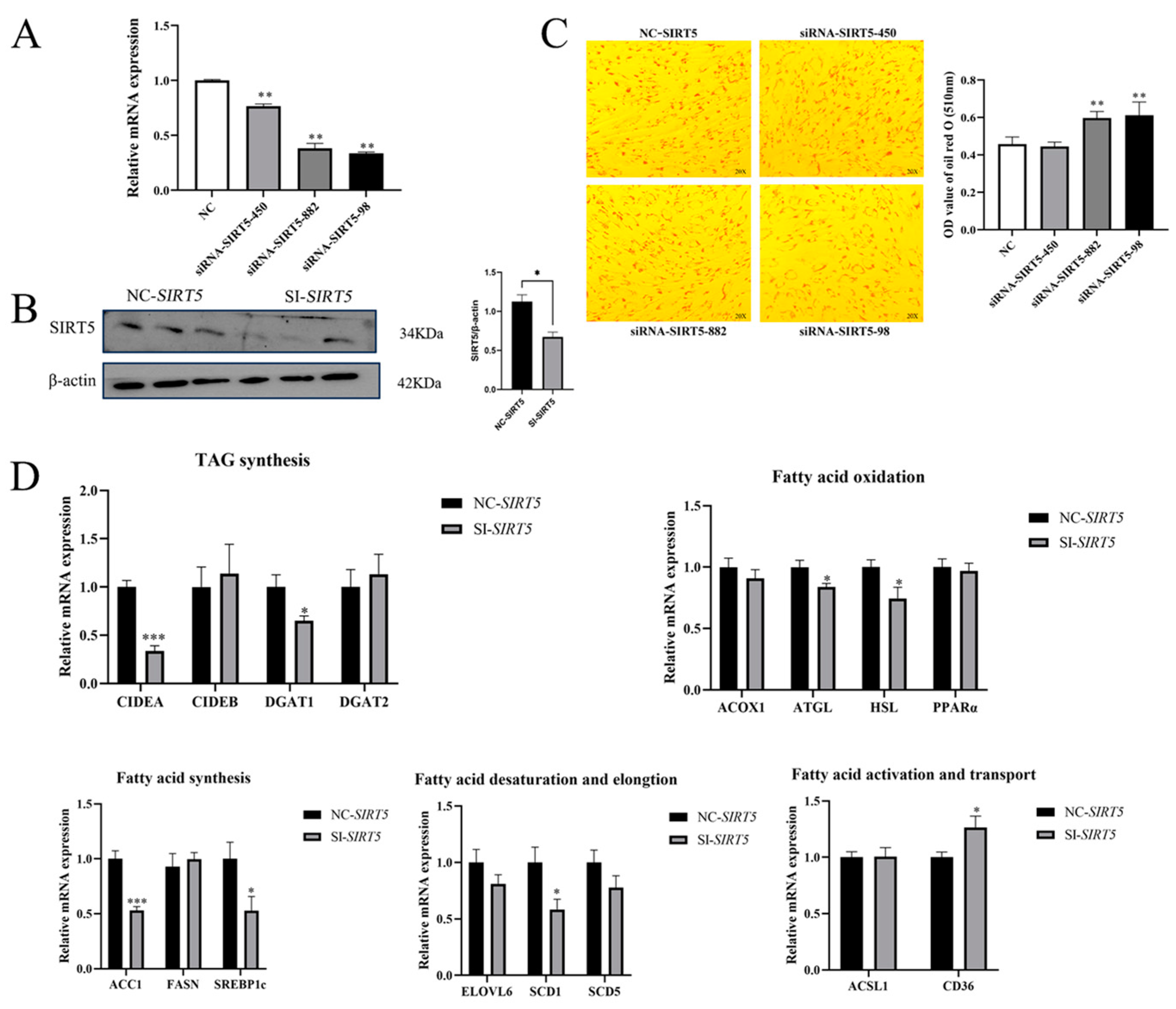
3.2. Effect of Interference with the SIRT5 Gene on Lipid Deposition in Goat Intramuscular Precursor Adipocytes
3.3. Inhibition of Proliferation of Goat Intramuscular Precursor Adipocytes After Interference with the SIRT5 Gene
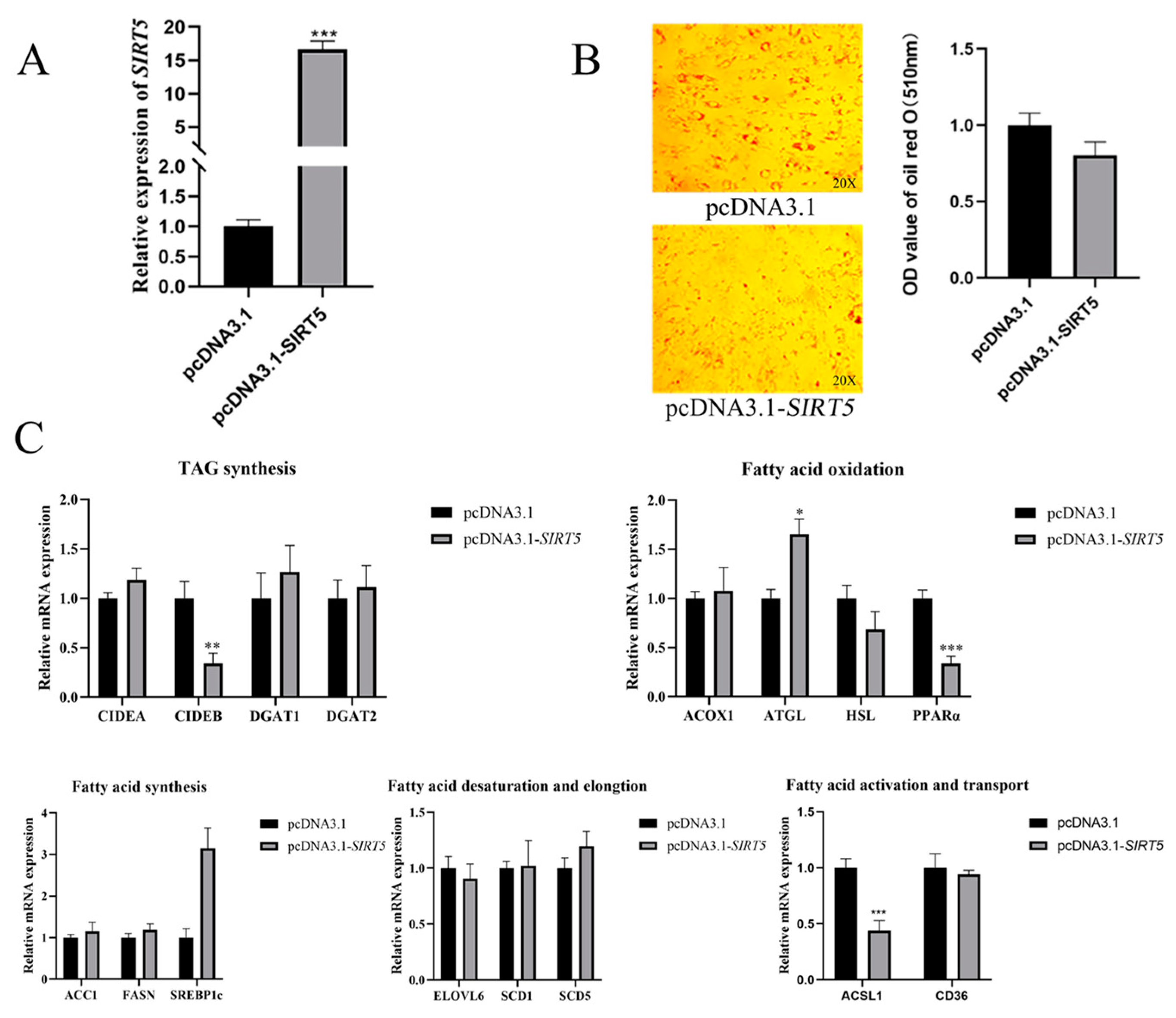
3.4. Effect of SIRT5 Gene Overexpression on Lipid Deposition in Goat Intramuscular Precursor Adipocytes
3.5. Promotion of Proliferation of Goat Intramuscular Precursor Adipocytes After Overexpression of the SIRT5 Gene
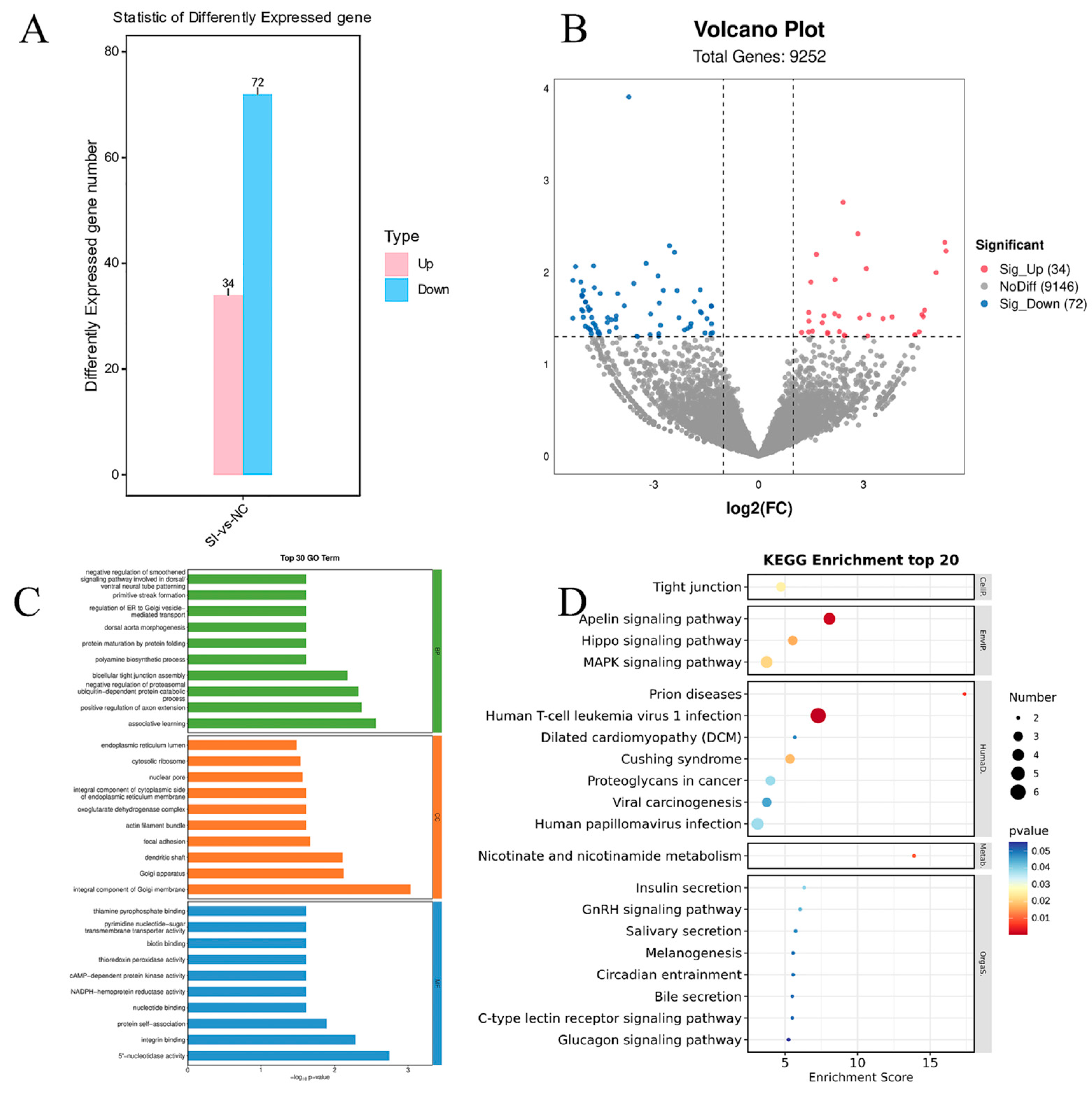
3.6. Interfering with FATP4 to Alter Gene Expression Profiles in Goat Precondition Adipocytes
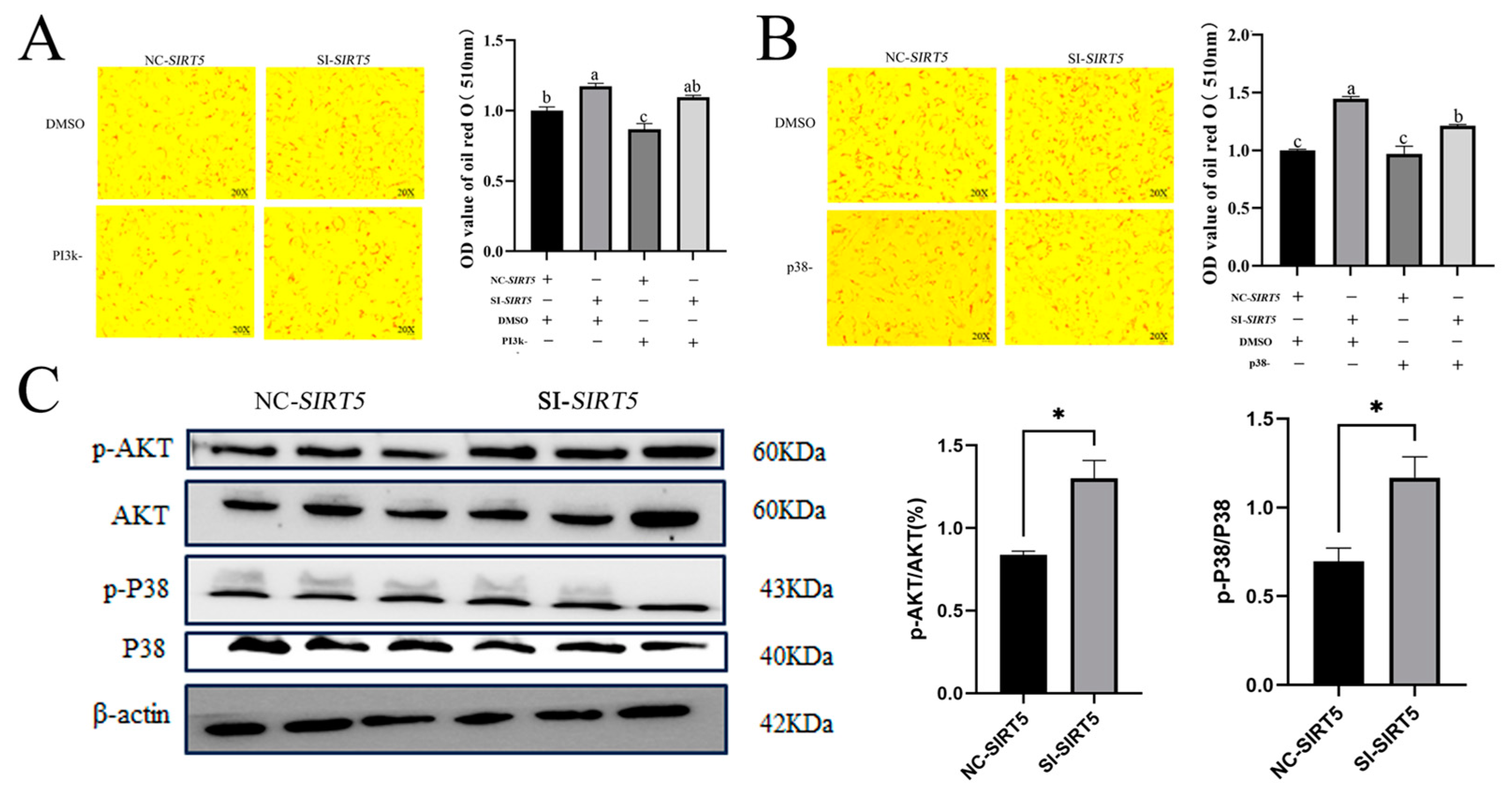
3.7. The SIRT5 Gene Promotes Lipid Deposition in Goat Intramuscular Precursor Adipocytes via the PI3k-Akt and p38 Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moloney, A.P.; Mooney, M.T.; Kerry, J.P.; Troy, D.J. Producing tender and flavoursome beef with enhanced nutritional characteristics. Proc. Nutr. Soc. 2001, 60, 221–229. [Google Scholar] [PubMed]
- Baik, M.; Kang, H.J.; Park, S.J.; Na, S.W.; Piao, M.; Kim, S.Y.; Fassah, D.M.; Moon, Y.S. Triennial growth and development symposium: Molecular mechanisms related to bovine intramuscular fat deposition in the longissimus muscle. J. Anim. Sci. 2017, 95, 2284–2303. [Google Scholar]
- Nishimura, T.; Ojima, K.; Liu, A.; Hattori, A.; Takahashi, K. Structural changes in the intramuscular connective tissue during development of bovine semitendinosus muscle. Tissue Cell 1996, 28, 527–536. [Google Scholar] [PubMed]
- Pande, S.; Raisuddin, S. Molecular and cellular regulatory roles of sirtuin protein. Crit. Rev. Food Sci. Nutr. 2023, 63, 9895–9913. [Google Scholar]
- D’Onofrio, N.; Servillo, L.; Balestrieri, M.L. SIRT1 and SIRT6 Signaling Pathways in Cardiovascular Disease Protection. Antioxid. Redox Signal 2018, 28, 711–732. [Google Scholar] [PubMed]
- Frye, R.A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999, 260, 273–279. [Google Scholar]
- Nishida, Y.; Rardin, M.J.; Carrico, C.; He, W.; Sahu, A.K.; Gut, P.; Najjar, R.; Fitch, M.; Hellerstein, M.; Gibson, B.W.; et al. SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol. Cell 2015, 59, 321–332. [Google Scholar]
- Yu, J.; Sadhukhan, S.; Noriega, L.G.; Moullan, N.; He, B.; Weiss, R.S.; Lin, H.; Schoonjans, K.; Auwerx, J. Metabolic characterization of a Sirt5 deficient mouse model. Sci. Rep. 2013, 3, 2806. [Google Scholar]
- Matsushita, N.; Yonashiro, R.; Ogata, Y.; Sugiura, A.; Nagashima, S.; Fukuda, T.; Inatome, R.; Yanagi, S. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes. Cells 2011, 16, 190–202. [Google Scholar]
- Sadhukhan, S.; Liu, X.; Ryu, D.; Nelson, O.D.; Stupinski, J.A.; Li, Z.; Chen, W.; Zhang, S.; Weiss, R.S.; Locasale, J.W.; et al. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc. Natl. Acad. Sci. USA 2016, 113, 4320–4325. [Google Scholar]
- Heinonen, T.; Ciarlo, E.; Théroude, C.; Pelekanou, A.; Herderschee, J.; Le Roy, D.; Roger, T. Sirtuin 5 Deficiency Does Not Compromise Innate Immune Responses to Bacterial Infections. Front. Immunol. 2018, 9, 2675. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Shi, W.; Tao, J.; Li, H.; Lin, X.; Yang, S.; Hua, P. SIRT5 and post-translational protein modifications: A potential therapeutic target for myocardial ischemia-reperfusion injury with regard to mitochondrial dynamics and oxidative metabolism. Eur. J. Pharmacol. 2018, 818, 410–418. [Google Scholar] [CrossRef]
- Buler, M.; Aatsinki, S.-M.; Izzi, V.; Uusimaa, J.; Hakkola, J. SIRT5 is under the control of PGC-1α and AMPK and is involved in regulation of mitochondrial energy metabolism. FASEB J. 2014, 28, 3225–3237. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.-P.; Xi, Q.-Y.; Sun, J.-J.; Cheng, X.; Zhu, Y.-L.; Ye, D.-Z.; Chen, T.; Wei, L.-M.; Ye, R.-S.; Jiang, Q.-Y.; et al. In low protein diets, microRNA-19b regulates urea synthesis by targeting SIRT5. Sci. Rep. 2016, 6, 33291. [Google Scholar] [CrossRef]
- Carrico, C.; Meyer, J.G.; He, W.; Gibson, B.W.; Verdin, E. The Mitochondrial Acylome Emerges: Proteomics, Regulation by Sirtuins, and Metabolic and Disease Implications. Cell Metab. 2018, 27, 497–512. [Google Scholar] [CrossRef]
- Kumar, S.; Lombard, D.B. Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 311–334. [Google Scholar] [CrossRef]
- Guedouari, H.; Daigle, T.; Scorrano, L.; Hebert-Chatelain, E. Sirtuin 5 protects mitochondria from fragmentation and degradation during starvation. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Shuai, L.; Zhang, L.-N.; Li, B.-H.; Tang, C.-L.; Wu, L.-Y.; Li, J.; Li, J.-Y. SIRT5 Regulates Brown Adipocyte Differentiation and Browning of Subcutaneous White Adipose Tissue. Diabetes 2019, 68, 1449–1461. [Google Scholar] [CrossRef]
- Hirschey, M.D.; Zhao, Y. Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Mol. Cell Proteom. 2015, 14, 2308–2315. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Kazak, L.; Spiegelman, B.M. New Advances in Adaptive Thermogenesis: UCP1 and Beyond. Cell Metab. 2019, 29, 27–37. [Google Scholar] [CrossRef]
- Hong, J.; Mei, C.; Raza, S.H.A.; Khan, R.; Cheng, G.; Zan, L. SIRT5 inhibits bovine preadipocyte differentiation and lipid deposition by activating AMPK and repressing MAPK signal pathways. Genomics 2020, 112, 1065–1076. [Google Scholar] [PubMed]
- Hong, J.; Mei, C.; Abbas Raza, S.H.; Khan, R.; Cheng, G.; Zan, L. SIRT6 cooperates with SIRT5 to regulate bovine preadipocyte differentiation and lipid metabolism via the AMPKα signaling pathway. Arch. Biochem. Biophys. 2020, 681, 108260. [Google Scholar] [CrossRef]
- Molinari, F.; Feraco, A.; Mirabilii, S.; Saladini, S.; Sansone, L.; Vernucci, E.; Tomaselli, G.; Marzolla, V.; Rotili, D.; Russo, M.A.; et al. SIRT5 Inhibition Induces Brown Fat-Like Phenotype in 3T3-L1 Preadipocytes. Cells 2021, 10, 1126. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Q.; Lin, Y.; Wang, Y.; Shi, H.; Huang, L.; Zhao, W.; Xiang, H.; Zhu, J. MCD Inhibits Lipid Deposition in Goat Intramuscular Preadipocytes. Genes. 2023, 14, 440. [Google Scholar] [PubMed]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar]
- Du, Y.; Hu, H.; Hua, C.; Du, K.; Wei, T. Tissue distribution, subcellular localization, and enzymatic activity analysis of human SIRT5 isoforms. Biochem. Biophys. Res. Commun. 2018, 503, 763–769. [Google Scholar] [CrossRef]
- Colak, G.; Pougovkina, O.; Dai, L.; Tan, M.; Te Brinke, H.; Huang, H.; Cheng, Z.; Park, J.; Wan, X.; Liu, X.; et al. Proteomic and Biochemical Studies of Lysine Malonylation Suggest Its Malonic Aciduria-associated Regulatory Role in Mitochondrial Function and Fatty Acid Oxidation. Mol. Cell Proteom. 2015, 14, 3056–3071. [Google Scholar] [CrossRef]
- Wang, G.; Meyer, J.G.; Cai, W.; Softic, S.; Li, M.E.; Verdin, E.; Newgard, C.; Schilling, B.; Kahn, C.R. Regulation of UCP1 and Mitochondrial Metabolism in Brown Adipose Tissue by Reversible Succinylation. Mol. Cell 2019, 74, 844–857. [Google Scholar] [CrossRef]
- Ogura, M.; Nakamura, Y.; Tanaka, D.; Zhuang, X.; Fujita, Y.; Obara, A.; Hamasaki, A.; Hosokawa, M.; Inagaki, N. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem. Biophys. Res. Commun. 2010, 393, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, W.; Wang, Y.; Li, H.; Zhang, C.; Wang, Y.; Lin, Y.; Shi, H.; Xiang, H.; Huang, L.; et al. Expression Variation of CPT1A Induces Lipid Reconstruction in Goat Intramuscular Precursor Adipocytes. Int. J. Mol. Sci. 2023, 24, 13415. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Xi, L.; Liu, Y.; Liu, R.; Wu, Z.; Jian, Z. SIRT5 promotes cell proliferation and invasion in hepatocellular carcinoma by targeting E2F1. Mol. Med. Rep. 2018, 17, 342–349. [Google Scholar] [CrossRef]
- Teng, P.; Cui, K.; Yao, S.; Fei, B.; Ling, F.; Li, C.; Huang, Z. SIRT5-mediated ME2 desuccinylation promotes cancer growth by enhancing mitochondrial respiration. Cell Death Differ. 2024, 31, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, C.; Tian, Y.; Yao, Y.; Mao, J.; Wang, H.; Li, Z.; Xu, Y.; Ye, M.; Wang, L. SIRT5 Promotes Hepatocellular Carcinoma Progression by Regulating Mitochondrial Apoptosis. J. Cancer 2019, 10, 3871–3882. [Google Scholar] [CrossRef]
- Gu, W.; Qian, Q.; Xu, Y.; Xu, X.; Zhang, L.; He, S.; Li, D. SIRT5 regulates autophagy and apoptosis in gastric cancer cells. J. Int. Med. Res. 2021, 49, 300060520986355. [Google Scholar] [CrossRef]
- Wu, Q.-J.; Zhang, T.-N.; Chen, H.-H.; Yu, X.-F.; Lv, J.-L.; Liu, Y.-Y.; Liu, Y.-S.; Zheng, G.; Zhao, J.-Q.; Wei, Y.-F.; et al. The sirtuin family in health and disease. Signal Transduct. Target. Ther. 2022, 7, 402. [Google Scholar]
- Katsyuba, E.; Romani, M.; Hofer, D.; Auwerx, J. NAD+ homeostasis in health and disease. Nat. Metab. 2020, 2, 9–31. [Google Scholar] [CrossRef]
- Chen, L.; Wang, J.; Ren, Y.; Ma, Y.; Liu, J.; Jiang, H.; Liu, C. Artesunate improves glucose and lipid metabolism in db/db mice by regulating the metabolic profile and the MAPK/PI3K/Akt signalling pathway. Phytomedicine 2024, 126, 155382. [Google Scholar] [PubMed]
- Xiao, H.; Sun, X.; Lin, Z.; Yang, Y.; Zhang, M.; Xu, Z.; Liu, P.; Liu, Z.; Huang, H. Gentiopicroside targets PAQR3 to activate the PI3K/AKT signaling pathway and ameliorate disordered glucose and lipid metabolism. Acta Pharm. Sin. B 2022, 12, 2887–2904. [Google Scholar] [PubMed]
- Santoleri, D.; Titchenell, P.M. Resolving the Paradox of Hepatic Insulin Resistance. Cell Mol. Gastroenterol. Hepatol. 2019, 7, 447–456. [Google Scholar] [PubMed]
- Yoshida, T.; Delafontaine, P. Mechanisms of IGF-1-Mediated Regulation of Skeletal Muscle Hypertrophy and Atrophy. Cells 2020, 9, 1970. [Google Scholar] [CrossRef]
- Cyr, N.E.; Steger, J.S.; Toorie, A.M.; Yang, J.Z.; Stuart, R.; Nillni, E.A. Central Sirt1 regulates body weight and energy expenditure along with the POMC-derived peptide α-MSH and the processing enzyme CPE production in diet-induced obese male rats. Endocrinology 2015, 156, 961–974. [Google Scholar]
- Wara, A.K.; Wang, S.; Wu, C.; Fang, F.; Haemmig, S.; Weber, B.N.; Aydogan, C.O.; Tesmenitsky, Y.; Aliakbarian, H.; Hawse, J.R.; et al. KLF10 Deficiency in CD4+ T Cells Triggers Obesity, Insulin Resistance, and Fatty Liver. Cell Rep. 2020, 33, 108550. [Google Scholar]
- Cahuzac, K.M.; Lubin, A.; Bosch, K.; Stokes, N.; Shoenfeld, S.M.; Zhou, R.; Lemon, H.; Asara, J.; Parsons, R.E. AKT activation because of PTEN loss upregulates xCT via GSK3β/NRF2, leading to inhibition of ferroptosis in PTEN-mutant tumor cells. Cell Rep. 2023, 42, 112536. [Google Scholar]
- He, Q.; Gao, L.; Zhang, F.; Yao, W.; Wu, J.; Song, N.; Luo, J.; Zhang, Y. The FoxO1-ATGL axis alters milk lipolysis homeostasis through PI3K/AKT signaling pathway in dairy goat mammary epithelial cells. J. Anim. Sci. 2023, 101. [Google Scholar]
- Seger, R.; Seger, D.; Reszka, A.A.; Munar, E.S.; Eldar-Finkelman, H.; Dobrowolska, G.; Jensen, A.M.; Campbell, J.S.; Fischer, E.H.; Krebs, E.G. Overexpression of mitogen-activated protein kinase kinase (MAPKK) and its mutants in NIH 3T3 cells. Evidence that MAPKK involvement in cellular proliferation is regulated by phosphorylation of serine residues in its kinase subdomains VII and VIII. J. Biol. Chem. 1994, 269, 25699–25709. [Google Scholar] [CrossRef]
- Park, H.-B.; Baek, K.-H. E3 ligases and deubiquitinating enzymes regulating the MAPK signaling pathway in cancers. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188736. [Google Scholar]
- Cameron, S.J.; Abe, J.-I.; Malik, S.; Che, W.; Yang, J. Differential role of MEK5alpha and MEK5beta in BMK1/ERK5 activation. J. Biol. Chem. 2004, 279, 1506–1512. [Google Scholar] [CrossRef] [PubMed]
- Wible, D.J.; Parikh, Z.; Cho, E.J.; Chen, M.-D.; Jeter, C.R.; Mukhopadhyay, S.; Dalby, K.N.; Varadarajan, S.; Bratton, S.B. Unexpected inhibition of the lipid kinase PIKfyve reveals an epistatic role for p38 MAPKs in endolysosomal fission and volume control. Cell Death Dis. 2024, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, W.; Li, H.; Xiang, H.; Zhang, C.; Du, Z.; Huang, L.; Zhu, J. MiR-196a Promotes Lipid Deposition in Goat Intramuscular Preadipocytes by Targeting MAP3K1 and Activating PI3K-Akt Pathway. Cells 2024, 13, 1459. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Yao, W.; Yang, C.; Zhang, W.; Wang, Y.; Lin, Y.; Du, Z.; Zhang, C.; Huang, L.; Zhang, M.; et al. SIRT5 Regulates Lipid Deposition in Goat Preadipocytes via PI3K-Akt and MAPK Signaling Pathways. Animals 2025, 15, 1072. https://doi.org/10.3390/ani15071072
Li H, Yao W, Yang C, Zhang W, Wang Y, Lin Y, Du Z, Zhang C, Huang L, Zhang M, et al. SIRT5 Regulates Lipid Deposition in Goat Preadipocytes via PI3K-Akt and MAPK Signaling Pathways. Animals. 2025; 15(7):1072. https://doi.org/10.3390/ani15071072
Chicago/Turabian StyleLi, Haiyang, Wenli Yao, Changheng Yang, Wenyang Zhang, Yong Wang, Yaqiu Lin, Zhanyu Du, Changhui Zhang, Lian Huang, Ming Zhang, and et al. 2025. "SIRT5 Regulates Lipid Deposition in Goat Preadipocytes via PI3K-Akt and MAPK Signaling Pathways" Animals 15, no. 7: 1072. https://doi.org/10.3390/ani15071072
APA StyleLi, H., Yao, W., Yang, C., Zhang, W., Wang, Y., Lin, Y., Du, Z., Zhang, C., Huang, L., Zhang, M., Fan, H., Zhu, J., & Xiang, H. (2025). SIRT5 Regulates Lipid Deposition in Goat Preadipocytes via PI3K-Akt and MAPK Signaling Pathways. Animals, 15(7), 1072. https://doi.org/10.3390/ani15071072





