Mechanism of circRNA_4083 Circularization and Its Role in Regulating Cell Viability
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Culture
2.2. Total RNA Extraction and Quality Assessment
2.3. Validation of circRNA_4083
2.4. RNase Digestion
2.5. qRT-PCR Analysis
2.6. Subcellular Localization
2.7. Plasmid Construction
2.8. Cell Transfection
2.9. Cell Viability Assay
2.10. Apoptosis Assay
2.11. Cell Cycle Analysis
2.12. Bioinformatics Analysis
2.13. Statistical Analysis
3. Results
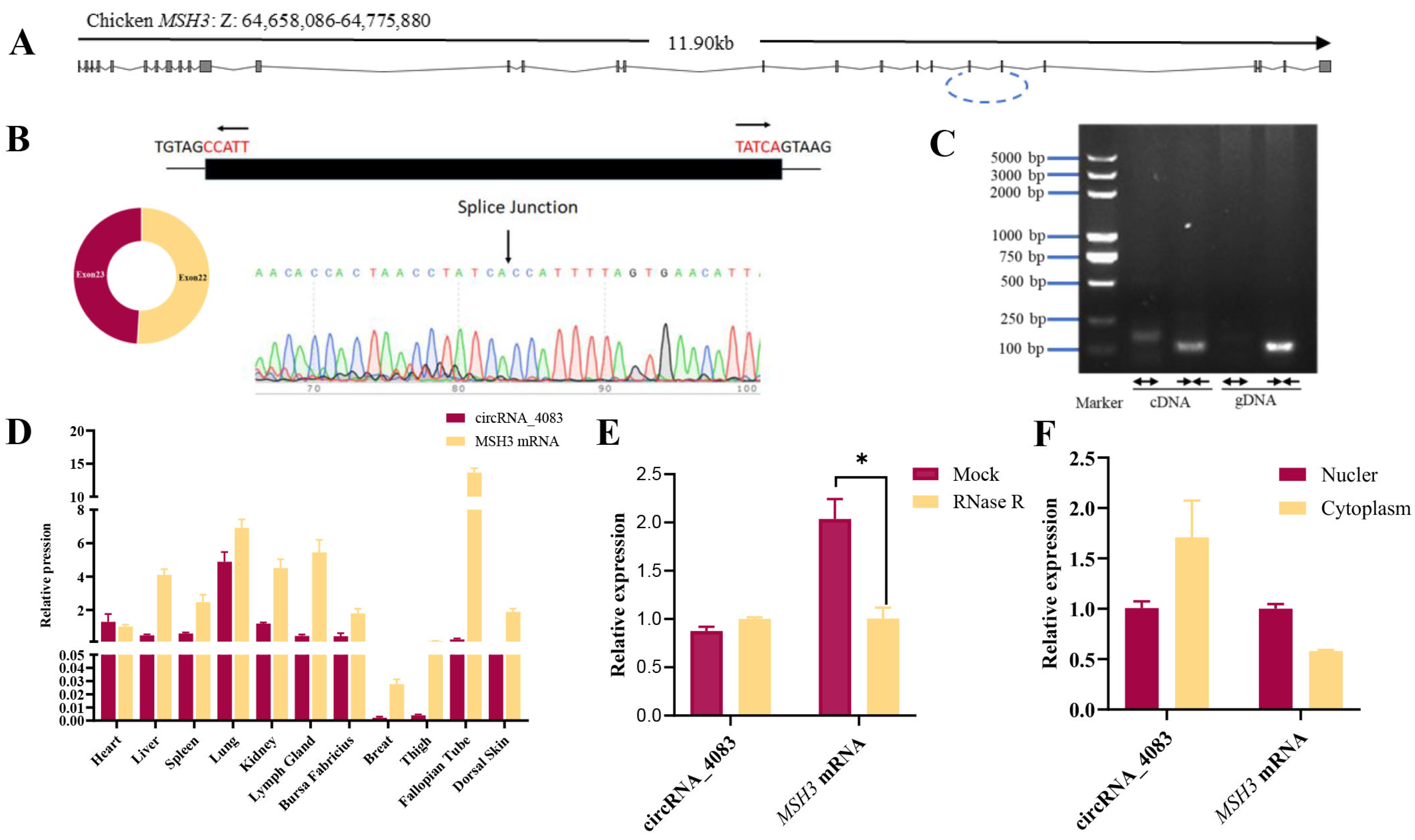
3.1. Characterization of Chicken-Derived circRNA_4083
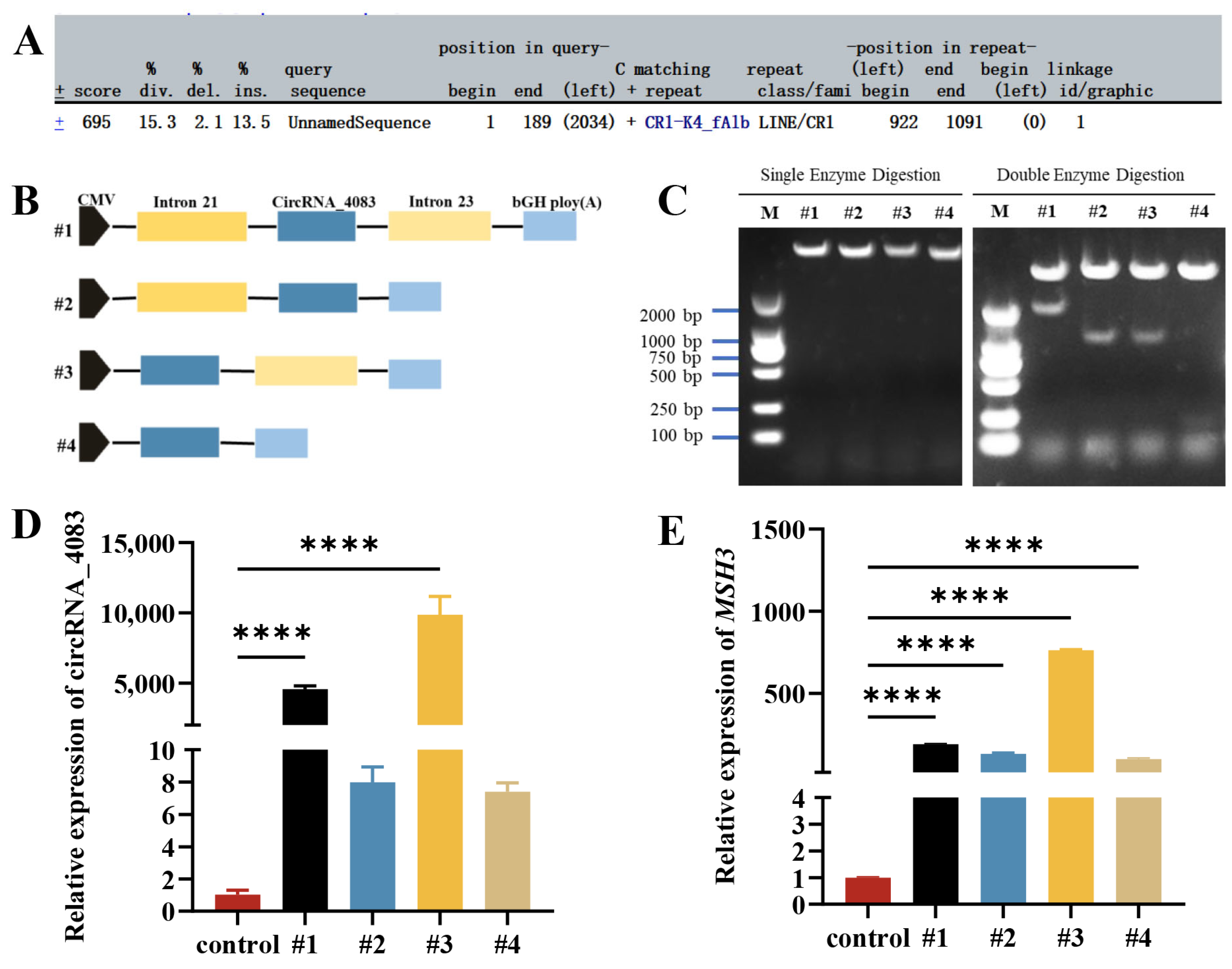
3.2. Circularization Mechanism of circRNA_4083
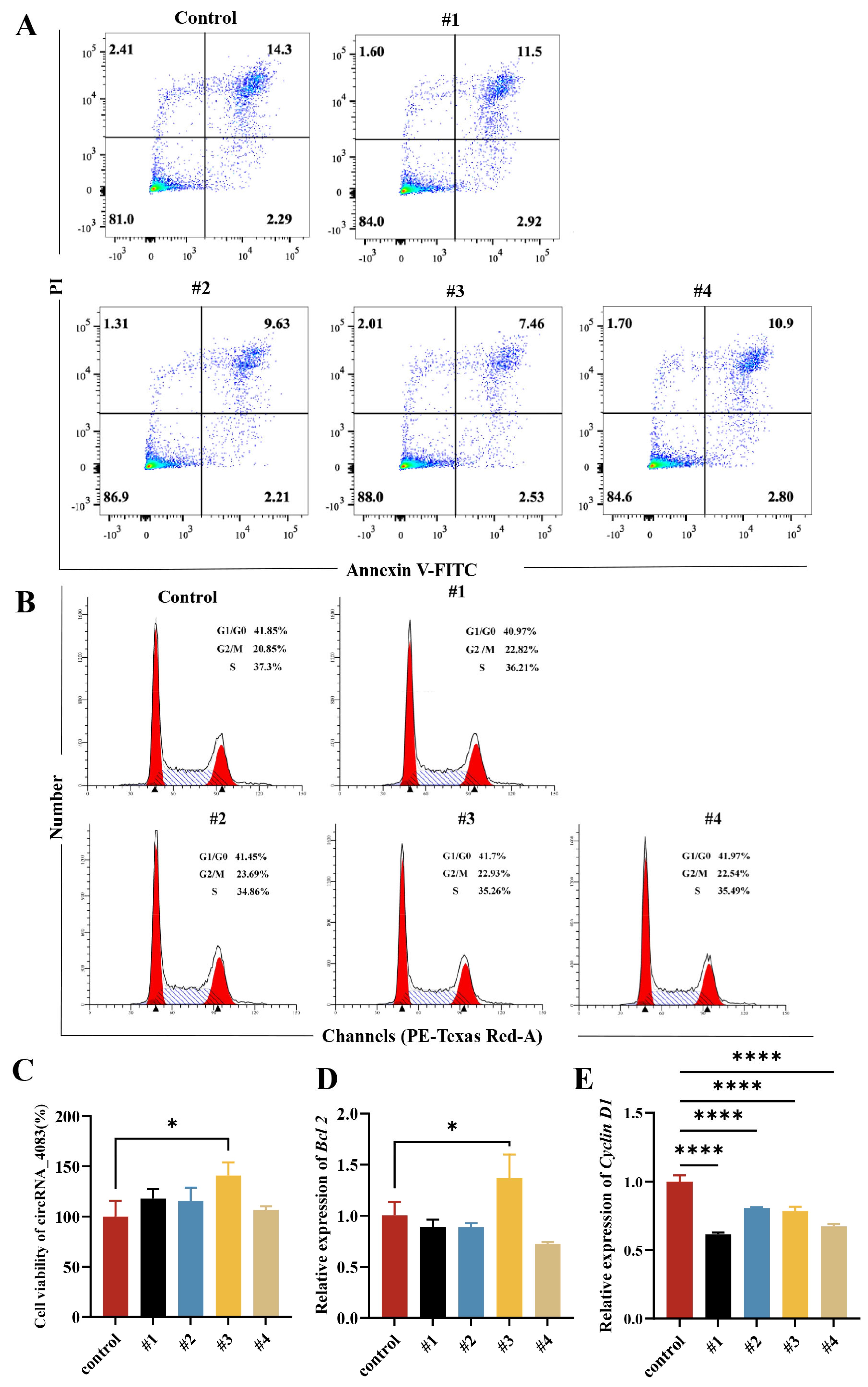
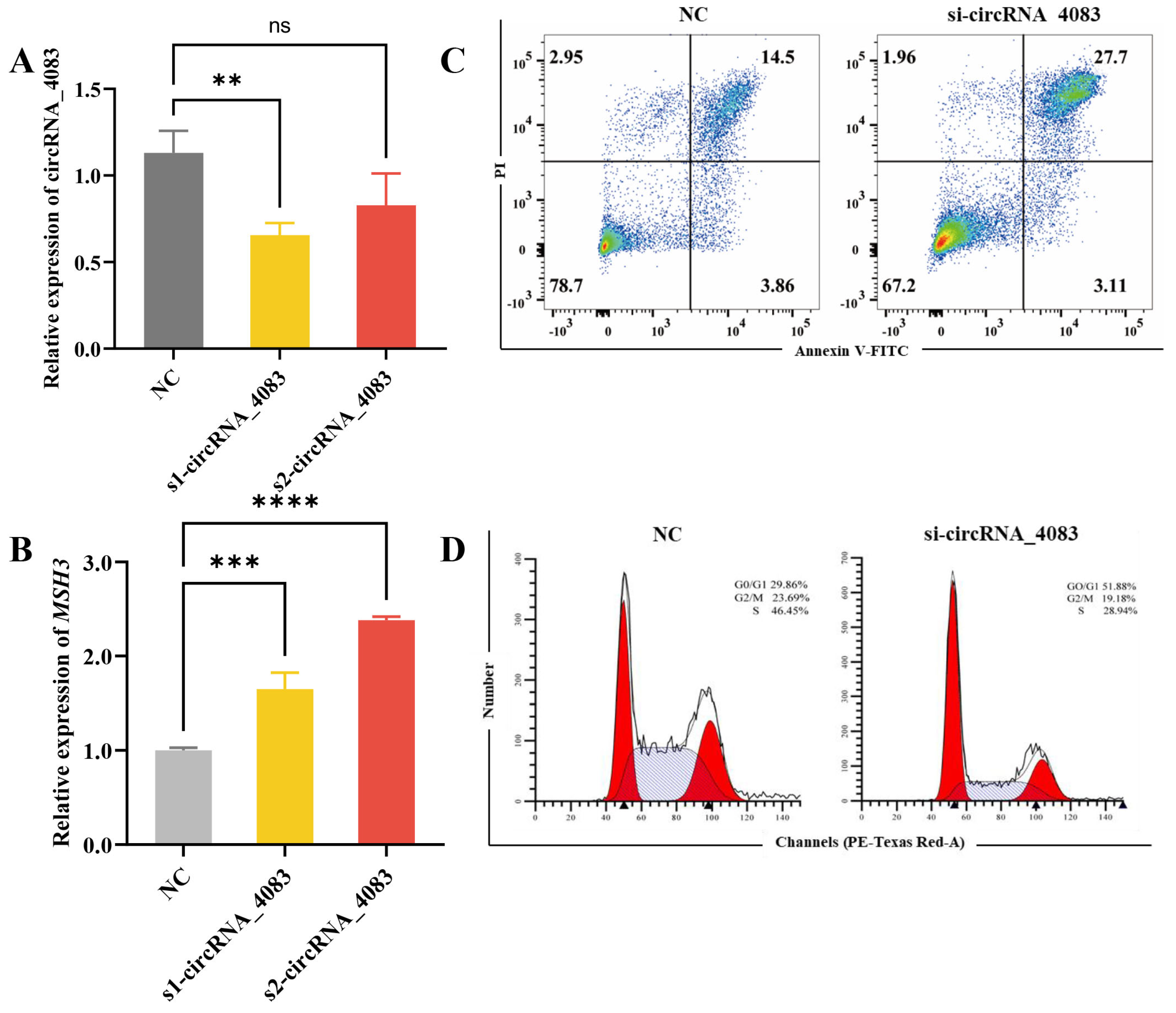
3.3. circRNA_4083 Suppresses Apoptosis and Promotes Cell Proliferation
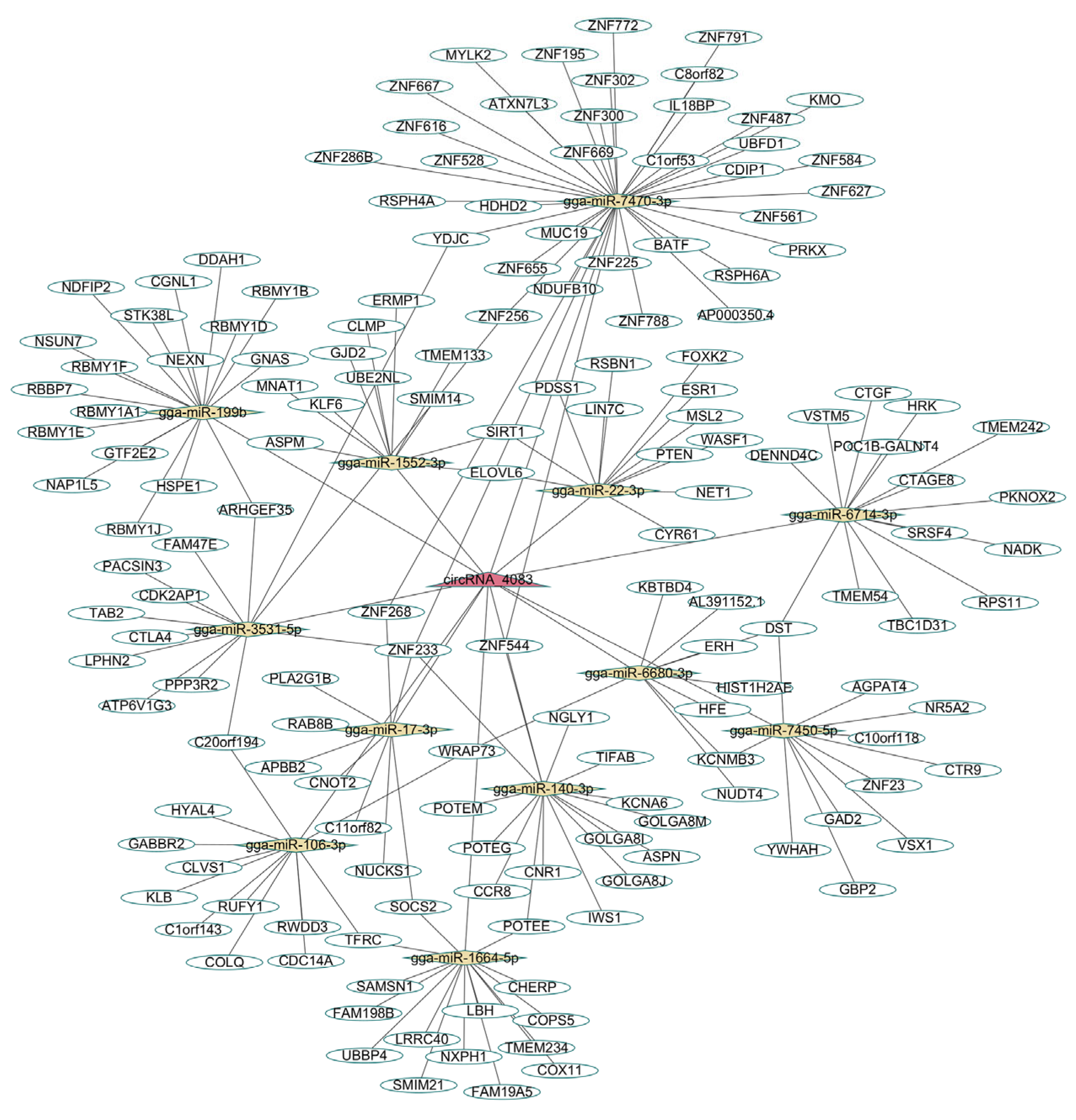
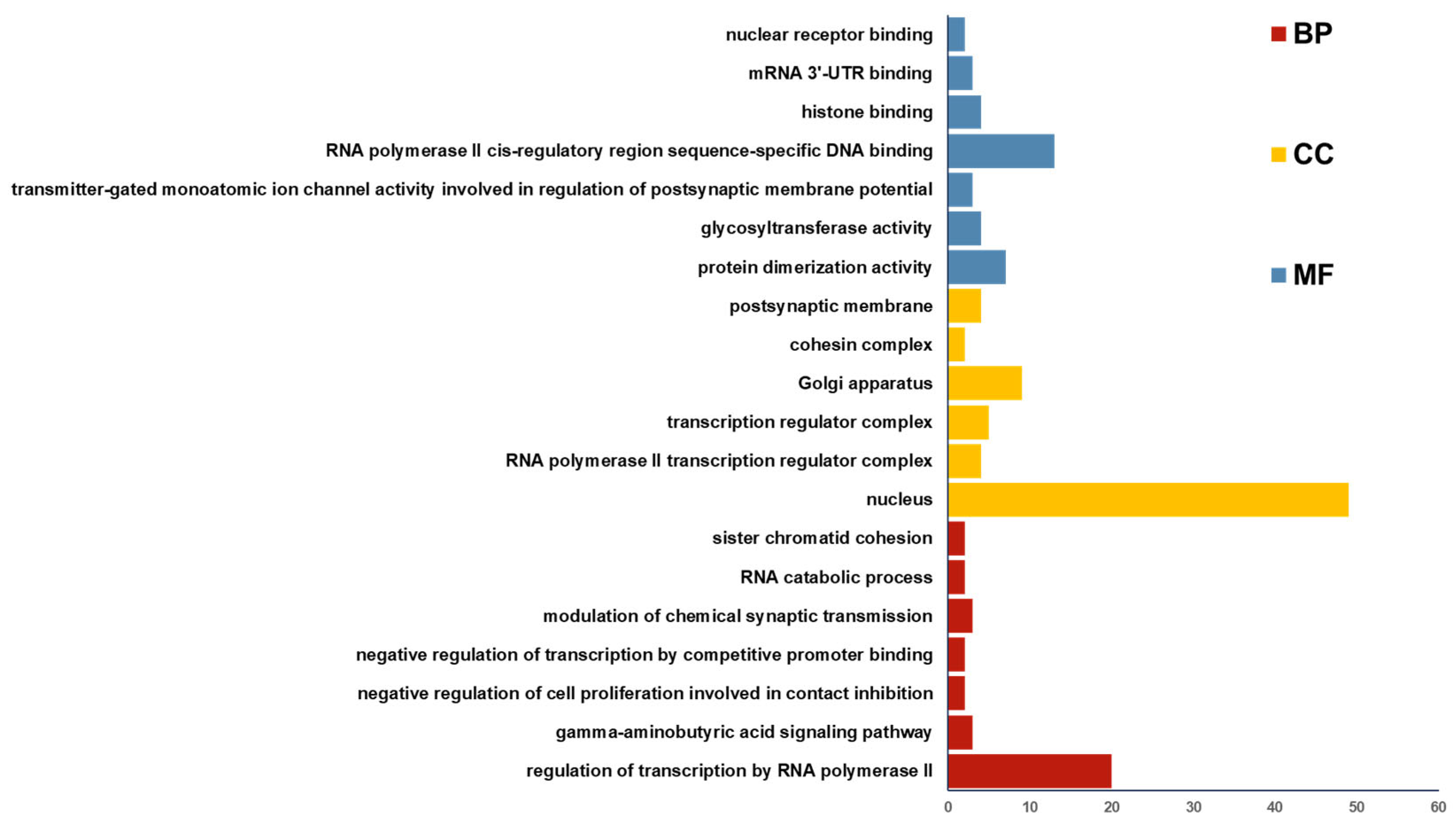
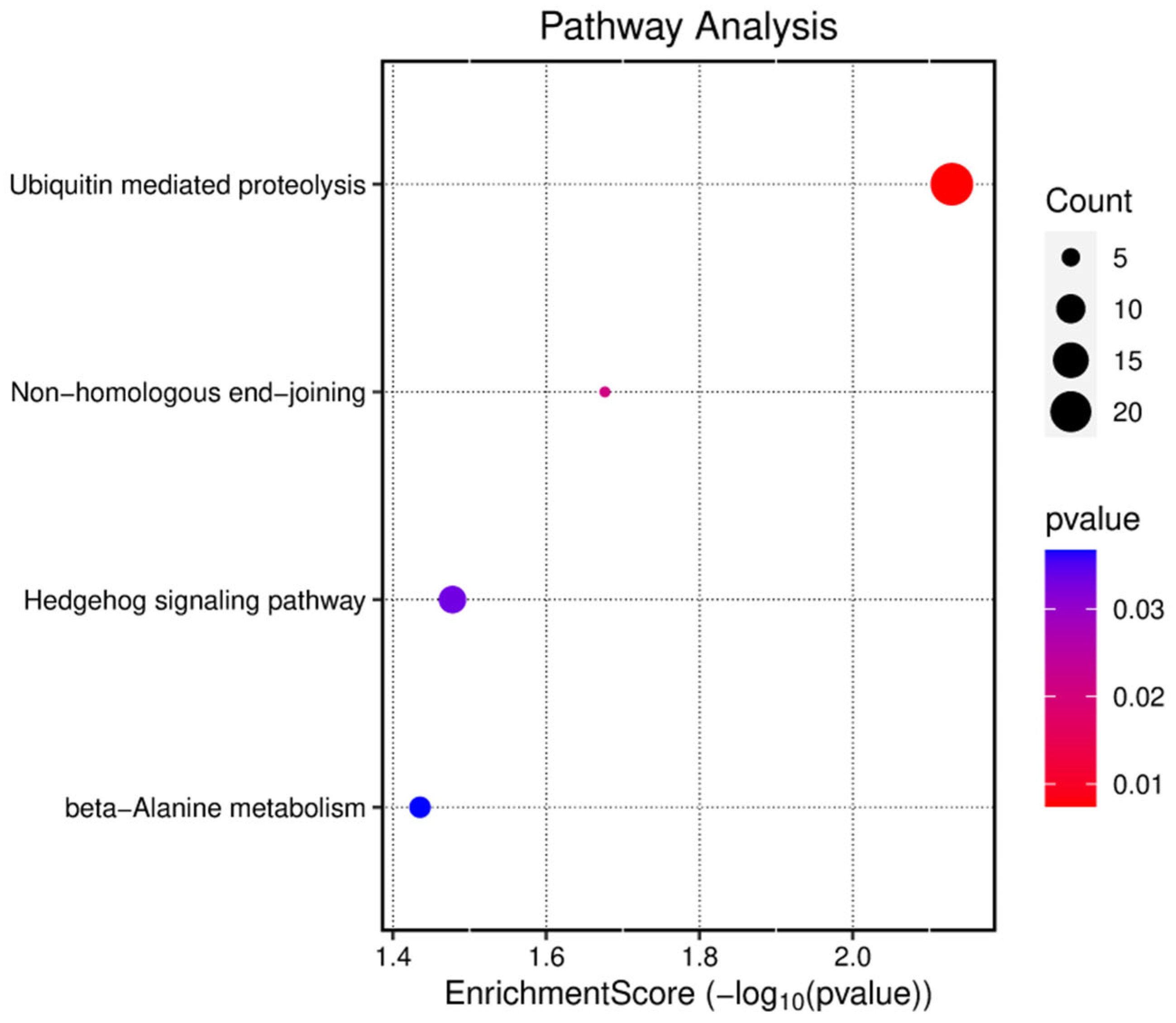
3.4. circRNA_4083 May Indirectly Regulate DNA Repair and Genomic Stability
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALV-J | Avian leukosis virus subgroup J |
| MSH3 | MutS Homolog 3 |
| DF-1 | Douglas Foster-1 |
References
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Dong, Y.; He, D.; Peng, Z.; Peng, W.; Shi, W.; Wang, J.; Li, B.; Zhang, C.; Duan, C. Circular RNAs in cancer: An emerging key player. J. Hematol. Oncol. 2017, 10, 2. [Google Scholar] [CrossRef]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995, 268, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, Y.; Li, P.; Zhu, D. RE: Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2019, 111, 435. [Google Scholar] [CrossRef]
- Han, D.; Li, J.; Wang, H.; Su, X.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.; et al. Circular RNA MTO1 acts as the sponge of miR-9 to suppress hepatocellular carcinoma progression. Hepatology 2017, 66, 1151–1164. [Google Scholar] [CrossRef]
- Wang, S.; Tang, D.; Wang, W.; Yang, Y.; Wu, X.; Wang, L.; Wang, D. circLMTK2 acts as a sponge of miR-150-5p and promotes proliferation and metastasis in gastric cancer. Mol. Cancer 2019, 18, 162. [Google Scholar] [CrossRef]
- Liu, C.; Zhu, X.; Niu, X.; Chen, L.; Ge, C. Elevated hsa_circRNA_101015, hsa_circRNA_101211, and hsa_circRNA_103470 in the Human Blood: Novel Biomarkers to Early Diagnose Acute Pancreatitis. BioMed Res. Int. 2020, 2020, 2419163. [Google Scholar] [CrossRef]
- Qiu, L.L.; Chang, G.B.; Li, Z.T.; Bi, Y.L.; Liu, X.P.; Chen, G.H. Comprehensive Transcriptome Analysis Reveals Competing Endogenous RNA Networks During Avian Leukosis Virus, Subgroup J-Induced Tumorigenesis in Chickens. Front. Physiol. 2018, 9, 996. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Erie, D.A. DNA MISMATCH REPAIR. Annu. Rev. Biochem. 2005, 74, 681–710. [Google Scholar] [CrossRef] [PubMed]
- Jiricny, J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346. [Google Scholar] [CrossRef]
- Young, S.J.; West, S.C. Coordinated roles of SLX4 and MutSβ in DNA repair and the maintenance of genome stability. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 157–177. [Google Scholar] [CrossRef]
- Edelmann, W.; Umar, A.; Yang, K.; Heyer, J.; Kucherlapati, M.; Lia, M.; Kneitz, B.; Avdievich, E.; Fan, K.H.; Wong, E.; et al. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res. 2000, 60, 803–807. [Google Scholar]
- van Oers, J.M.M.; Edwards, Y.; Chahwan, R.; Zhang, W.; Smith, C.; Pechuan, X.; Schaetzlein, S.; Jin, B.; Wang, Y.; Bergman, A.; et al. The MutSβ complex is a modulator of p53-driven tumorigenesis through its functions in both DNA double-strand break repair and mismatch repair. Oncogene 2014, 33, 3939–3946. [Google Scholar] [CrossRef]
- Tseng-Rogenski, S.; Koi, M.; Carethers, J. Inflammation-driven MSH3 Nuclear-to-Cytosol Shift Demonstrates MSH3 is Protein-bound in the Cytoplasm, and Influenced by Protein Modification and Structural Polymorphism. Gastroenterology 2016, 150, S963. [Google Scholar] [CrossRef]
- Suzuki, S.; Koi, M.; Carethers, J.M. Deletion of the mismatch repair protein MSH3 causes aneuploidy due to failure of initiation of Homologous Recombination (HR) repair of DNA double Strand Breaks (DSB). Gastroenterology 2020, 158, S532. [Google Scholar] [CrossRef]
- Koi, M.; Haraguchi, A.; Carethers, J.M. Genetic instability caused by loss of MSH3 in colorectal cancers. Gastroenterology 2024, 166, S174. [Google Scholar] [CrossRef]
- Aldous, S.G.; Smith, E.J.; Landles, C.; Osborne, G.F.; Cañibano-Pico, M.; Nita, I.M.; Phillips, J.; Zhang, Y.; Jin, B.; Hirst, M.B.; et al. A CAG repeat threshold for therapeutics targeting somatic instability in Huntington’s disease. Brain 2024, 147, 1784–1798. [Google Scholar] [CrossRef]
- Keogh, N.; Chan, K.Y.; Li, G.-M.; Lahue, R.S. MutSβ abundance and Msh3 ATP hydrolysis activity are important drivers of CTG•CAG repeat expansions. Nucleic Acids Res. 2017, 45, 10068–10078. [Google Scholar] [CrossRef]
- Nan, Z.; Xinjia, W.; Yu, L.; Yiwei, L.; Chengcheng, S.; Yumeng, S.; Ningye, M.; Yisheng, J. Mechanisms and therapeutic implications of gene expression regulation by circRNA-protein interactions in cancer. Commun. Biol. 2025, 8, 77. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Tang, J.; Zhang, Q.; Wang, C.; Qi, J.; Wei, Y.; Xu, J.; Yang, K.; Zhou, Z.; Wu, H.; et al. CircHIPK2 Contributes Cell Growth in Intestinal Epithelial of Colitis and Colorectal Cancer through Promoting TAZ Translation. Adv. Sci. 2024, 11, e2401588. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, J.; Xu, Z.; Yang, J.; Xu, Y.; Liu, Y.; Li, B.; Xie, J.; Li, J. Circular RNA hsa_circ_0000848 Promotes Trophoblast Cell Migration and Invasion and Inhibits Cell Apoptosis by Sponging hsa-miR-6768-5p. Front. Cell Dev. Biol. 2020, 8, 278. [Google Scholar] [CrossRef]
- Ji, Y.; Ni, C.; Shen, Y.; Xu, Z.; Tang, L.; Yu, F.; Zhu, L.; Lu, H.; Zhang, C.; Yang, S.; et al. ESRP1-mediated biogenesis of circPTPN12 inhibits hepatocellular carcinoma progression by PDLIM2/ NF-κB pathway. Mol. Cancer 2024, 23, 143. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Jamali, E. CircITCH: A Circular RNA With Eminent Roles in the Carcinogenesis. Front. Oncol. 2021, 11, 774979. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Wu, S.; Zhou, Z.; Ding, X.; Shi, R.; Thorne, R.F.; Zhang, X.D.; Hu, W.; Wu, M. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab. 2019, 30, 157–173.e7. [Google Scholar] [CrossRef]
- Feng, P.F.; Zhu, L.X.; Sheng, N.; Li, X.S.; Liu, P.G.; Chen, X.F. CircXRN2 accelerates colorectal cancer progression through regulating miR-149-5p/MACC1 axis and EMT. Sci. Rep. 2024, 14, 2448. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, X.; He, H.; Zhao, J.; Wei, Y.; Chen, Y.; Han, S.; Zhu, Y.; Zhang, Y.; Zhu, Q.; et al. Evolutionary conserved circular MEF2A RNAs regulate myogenic differentiation and skeletal muscle development. PLoS Genet. 2023, 19, e1010923. [Google Scholar] [CrossRef]
- Fan, A.; Zhang, Y.; Li, Y.; Meng, W.; Wu, F.; Pan, W.; Ma, Z.; Chen, W. Primary Cilia Formation Mediated by Hsa_Circ_0005185/OTUB1/RAB8A Complex Inhibits Prostate Cancer Progression by Suppressing Hedgehog Signaling Pathway. Adv. Sci. 2025, 12, e2411675. [Google Scholar] [CrossRef]
- Fei, D.; Wang, F.; Wang, Y.; Chen, J.; Chen, S.; Fan, L.; Yang, L.; Ren, Q.; Duangmano, S.; Du, F.; et al. Circular RNA ACVR2A promotes the progression of hepatocellular carcinoma through mir-511-5p targeting PI3K-Akt signaling pathway. Mol. Cancer 2024, 23, 159. [Google Scholar] [CrossRef] [PubMed]
- Riobo-Del Galdo, N.A.; Lara Montero, A.; Wertheimer, E.V. Role of Hedgehog Signaling in Breast Cancer: Pathogenesis and Therapeutics. Cells 2019, 8, 375. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wang, Y.; He, L.; Zhang, J.; Zhu, X.; Liu, N.; Wang, J.; Lu, T.; He, L.; Tian, Y.; et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol. Cancer 2021, 20, 132. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, X.; Yang, J.; Sun, Q.; Liu, Y.; Li, N.; Zhang, Z.; Xu, H. Circ-GLI1 promotes metastasis in melanoma through interacting with p70S6K2 to activate Hedgehog/GLI1 and Wnt/β-catenin pathways and upregulate Cyr61. Cell Death Dis. 2020, 11, 596. [Google Scholar] [CrossRef]
| Gene | Primer Sequence (5′-3′) |
|---|---|
| CircRNA_4083 | F:CTACTGGGAGAACAGGATCA |
| R:TTATCTTTCACAACTGGTCTGC | |
| MSH3 | F:CTGTATCTAAAGCGATTGGTC |
| R:GCTGATTATCTTTCACAACTGG | |
| GAPDH | F:GAACATCATCCCAGCGTCCA |
| R:CGGCAGGTCAGGTCAACAAC |
| Plasmid | Primer Sequence (5′-3′) |
|---|---|
| #1-intron21 | F:agcgtttaaacttaagcttggtaccGCACTGGAATAGGCTCCCCG |
| R:actaaaatggctctacaccaAAACAGGGAAAATAGAAAGAGG | |
| #1-4083 | F:tggtgtagagccattttagtGAACATTATCACACTGTATCTAAAGC |
| R:caatacttactgtgataggtTAGTGGTGTTTGGAACATACT | |
| #1-intron23 | F:accatgaatgacactatttgAGATACCCTTTGAGCTTCAAATCA |
| R:aacgggccctctagactcgaGTCTCCTGCTGCAACACCAATATCTG | |
| #2-4083 | F:tggtgtagagccattttagtGAACATTATCACACTGTATCTAAAGC |
| R:aacgggccctctagactcgagtgTGATAGGTTAGTGGTGTTTGGAACATACTG | |
| #3-4083 | F:agcgtttaaacttaagcttgGTACCCCATTTTAGTGAACATTATCACACTGTATCTAAAGC |
| R:caatacttactgtgataggtTAGTGGTGTTTGGAACATACT | |
| #4 | F:agcgtttaaacttaagcttggtaccCCATTTTAGTGAACATTATCACACTGTATCTAAAGC |
| R:aacgggccctctagactcgagtgTGATAGGTTAGTGGTGTTTGGAACATACTG |
| Name of siRNA | Primer Sequence (5′-3′) |
|---|---|
| s1-circRNA_4083 | CACTAACCTATCACCATTT |
| s2-circRNA_4083 | AACCTATCACCATTTTAGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Yang, T.; Wang, H.; Bai, H.; Chang, G.; Qiu, L. Mechanism of circRNA_4083 Circularization and Its Role in Regulating Cell Viability. Animals 2025, 15, 1527. https://doi.org/10.3390/ani15111527
Li W, Yang T, Wang H, Bai H, Chang G, Qiu L. Mechanism of circRNA_4083 Circularization and Its Role in Regulating Cell Viability. Animals. 2025; 15(11):1527. https://doi.org/10.3390/ani15111527
Chicago/Turabian StyleLi, Wenhao, Ting Yang, Haojie Wang, Hao Bai, Guobin Chang, and Lingling Qiu. 2025. "Mechanism of circRNA_4083 Circularization and Its Role in Regulating Cell Viability" Animals 15, no. 11: 1527. https://doi.org/10.3390/ani15111527
APA StyleLi, W., Yang, T., Wang, H., Bai, H., Chang, G., & Qiu, L. (2025). Mechanism of circRNA_4083 Circularization and Its Role in Regulating Cell Viability. Animals, 15(11), 1527. https://doi.org/10.3390/ani15111527





