Association Analysis of SNPs in GRHL2 and RORA Genes with Lambing Number in Small-Tailed Han Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Animal Preparation and Sample Collection
2.3. Blood DNA Extraction
2.4. PCR Amplification and Sanger Sequencing
2.5. Genotyping
2.6. Data Analysis
2.7. Using STRING Database to Predict Protein–Protein Interaction Network
3. Results
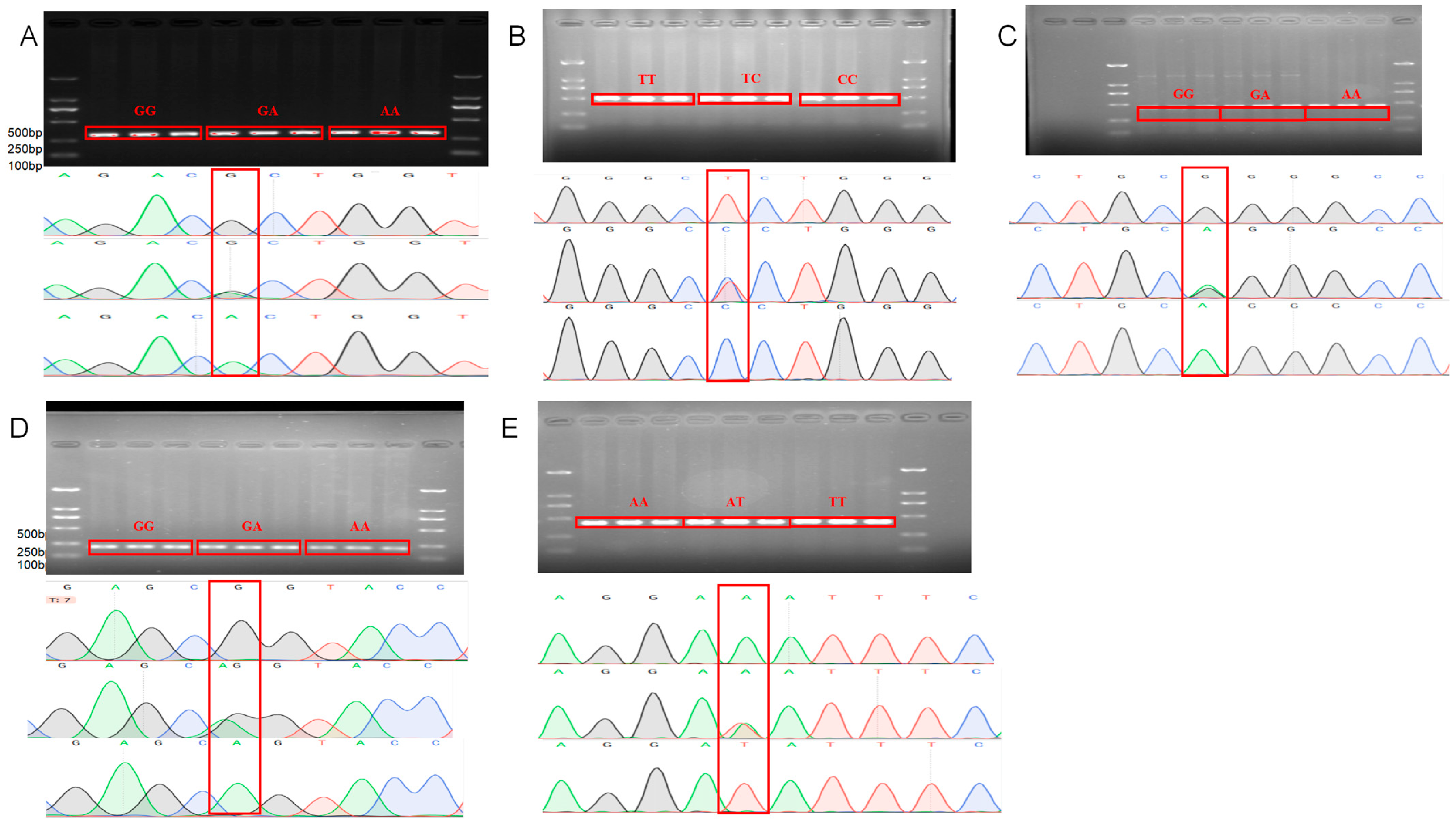
3.1. Identification of GRHL2 SNPs and RORA SNP Mutation Sites
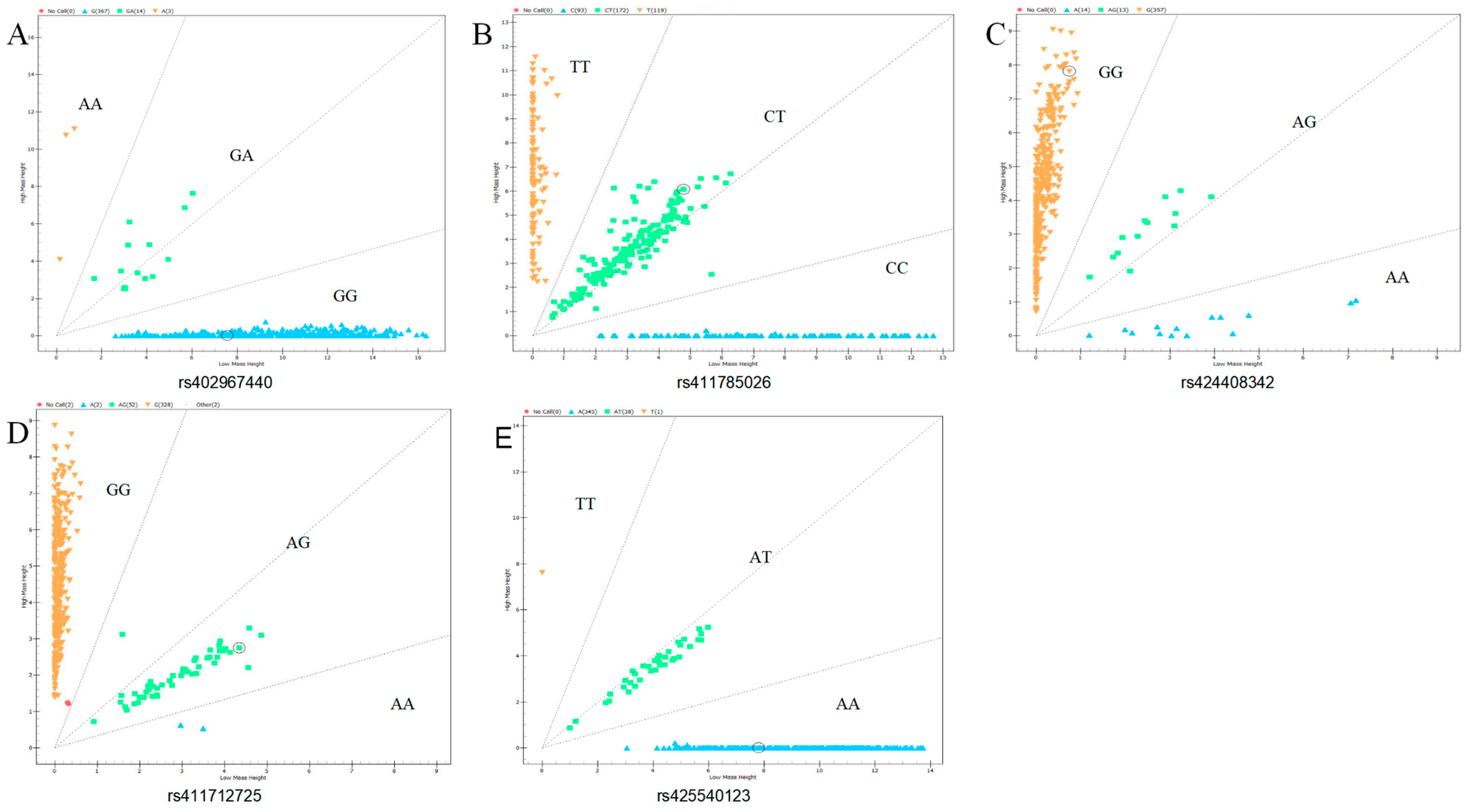
3.2. Genotyping and Population Genetic Analysis of Candidate SNPs in GRHL2 and RORA
3.3. Association Analysis of GRHL2 and RORA Candidate Loci with Lambing Number in Small-Tailed Han Sheep
| Gene | SNPs | Genotypes | 1st Parity Lambing Number | 2nd Parity Lambing Number | 3rd Parity Lambing Number | Average Lambing Number |
|---|---|---|---|---|---|---|
| GRHL2 | rs402967440 | GG (367) | 1.98 ± 0.05 | 1.68 ± 0.07 a | 0.68 ± 0.07 a | 1.45 ± 0.05 a |
| GA (14) | 2.36 ± 0.34 | 2.50 ± 0.39 b | 2.14 ± 0.53 b | 2.33 ± 0.34 b | ||
| AA (3) | 2.00 ± 0.58 | 2.00 ± 0.58 ab | 1.33 ± 0.67 ab | 1.78 ± 0.22 ab | ||
| GRHL2 | rs411785026 | TT (119) | 2.09 ± 0.09 | 1.76 ± 0.13 | 0.70 ± 0.11 | 1.52 ± 0.08 |
| CT (172) | 1.88 ± 0.08 | 1.71 ± 0.10 | 0.84 ± 0.11 | 1.49 ± 0.07 | ||
| CC (93) | 2.09 ± 0.11 | 1.67 ± 0.15 | 0.61 ± 0.13 | 1.46 ± 0.10 | ||
| GRHL2 | rs424408342 | GG (357) | 1.99 ± 0.05 | 1.76 ± 0.07 | 0.77 ± 0.07 | 1.51 ± 0.05 |
| GA (13) | 1.77 ± 0.28 | 1.15 ± 0.34 | 0.31 ± 0.24 | 1.08 ± 0.17 | ||
| AA (14) | 2.36 ± 0.20 | 1.21 ± 0.35 | 0.36 ± 0.07 | 1.31 ± 0.17 | ||
| GRHL2 | rs411712725 | GG (328) | 1.98 ± 0.06 a | 1.76 ± 0.07 a | 0.78 ± 0.08 | 1.51 ± 0.05 |
| GA (52) | 2.06 ± 0.15 a | 1.55 ± 0.19 a | 1.39 ± 0.18 | 1.39 ± 0.13 | ||
| AA (2) | 2.67 ± 0.33 a | 0.00 b | 0.00 | 0.89 ± 0.11 | ||
| RORA | rs425540123 | TT (1) | 3.00 ± 0.00 | 2.00 ± 0.00 | 0.00 | 1.67 ± 0.00 |
| AT (38) | 1.89 ± 0.17 | 1.66 ± 0.23 | 0.89 ± 0.23 | 1.48 ± 0.16 | ||
| AA (345) | 2.01 ± 0.05 | 1.72 ± 0.07 | 0.73 ± 0.07 | 1.49 ± 0.05 |
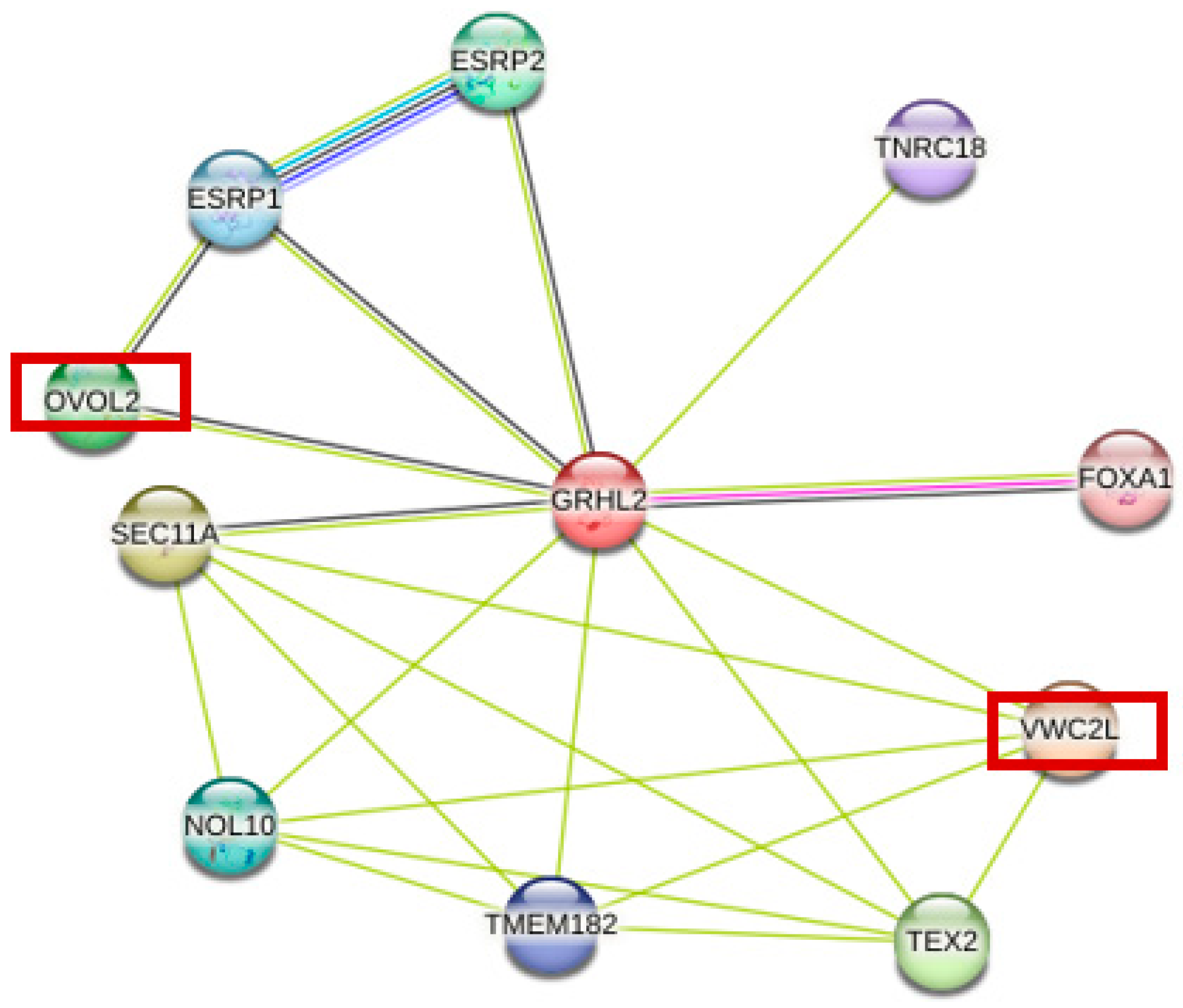
3.4. GRHL2-Related Protein Interaction Network Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdoli, R.; Zamani, P.; Mirhoseini, S.Z.; Ghavi Hossein-Zadeh, N.; Nadri, S. A review on prolificacy genes in sheep. Reprod. Domest. Anim. 2016, 51, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Nenova, R.; Dimitrova, I.; Stancheva, N.; Bozhilova, M.; Tzonev, T.; Minkova, T. Genetic markers associated to improving prolificacy of sheep. A review. Bulg. J. Agric. Sci. 2023, 29, 371–377. [Google Scholar]
- Baloch, F.S.; Altaf, M.T.; Liaqat, W.; Bedir, M.; Nadeem, M.A.; Cömertpay, G.; Çoban, N.; Habyarimana, E.; Barutçular, C.; Cerit, I.; et al. Recent advancements in the breeding of sorghum crop: Current status and future strategies for marker-assisted breeding. Front. Genet. 2023, 14, 1150616. [Google Scholar] [CrossRef]
- Tuersuntuoheti, M.; Zhang, J.; Zhou, W.; Zhang, C.L.; Liu, C.; Chang, Q.; Liu, S. Exploring the growth trait molecular markers in two sheep breeds based on genome-wide association analysis. PLoS ONE 2023, 18, e0283383. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
- Liu, K.; Liu, Y.; Chu, M. Detection of polymorphisms in six genes and their association analysis with litter size in sheep. Anim. Biotechnol. 2024, 35, 2309954. [Google Scholar] [CrossRef]
- Chu, M.X.; Liu, Z.H.; Jiao, C.L.; He, Y.Q.; Fang, L.; Ye, S.C.; Chen, G.H.; Wang, J.Y. Mutations in BMPR-IB and BMP-15 genes are associated with litter size in small tailed han sheep (Ovis aries). J. Anim. Sci. 2007, 85, 598–603. [Google Scholar] [CrossRef]
- Nagdy, H.; Mahmoud, K.G.M.; Kandiel, M.M.M.; Helmy, N.A.; Ibrahim, S.S.; Nawito, M.F.; Othman, O.E. PCR-RFLP of bone morphogenetic protein 15 (BMP15/FecX) gene as a candidate for prolificacy in sheep. Int. J. Vet. Sci. Med. 2018, 6, S68–s72. [Google Scholar] [CrossRef]
- Wang, F.; Chu, M.; Pan, L.; Wang, X.; He, X.; Zhang, R.; Tao, L.; La, Y.; Ma, L.; Di, R. Polymorphism detection of GDF9 gene and its association with litter size in luzhong mutton sheep (Ovis aries). Animals 2021, 11, 571. [Google Scholar] [CrossRef]
- Tsartsianidou, V.; Pavlidis, A.; Tosiou, E.; Arsenos, G.; Banos, G.; Triantafyllidis, A. Novel genomic markers and genes related to reproduction in prolific chios dairy sheep: A genome-wide association study. Animal 2023, 17, 100723. [Google Scholar] [CrossRef]
- Sundararajan, V.; Pang, Q.Y.; Choolani, M.; Huang, R.Y. Spotlight on the granules (Grainyhead-like proteins)—From an evolutionary conserved controller of epithelial trait to pioneering the chromatin landscape. Front. Mol. Biosci. 2020, 7, 213. [Google Scholar] [CrossRef] [PubMed]
- Boivin, F.J.; Schmidt-Ott, K.M. Functional roles of grainyhead-like transcription factors in renal development and disease. Pediatr. Nephrol. 2020, 35, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Werth, M.; Walentin, K.; Aue, A.; Schönheit, J.; Wuebken, A.; Pode-Shakked, N.; Vilianovitch, L.; Erdmann, B.; Dekel, B.; Bader, M.; et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 2010, 137, 3835–3845. [Google Scholar] [CrossRef]
- Hinze, C.; Ruffert, J.; Walentin, K.; Himmerkus, N.; Nikpey, E.; Tenstad, O.; Wiig, H.; Mutig, K.; Yurtdas, Z.Y.; Klein, J.D.; et al. GRHL2 is required for collecting duct epithelial barrier function and renal osmoregulation. J. Am. Soc. Nephrol. 2018, 29, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, S.; Darido, C.; Georgy, S.R.; Wilanowski, T.; Srivastava, S.; Ellett, F.; Pase, L.; Han, Y.; Meng, A.; Heath, J.K.; et al. Midbrain-hindbrain boundary patterning and morphogenesis are regulated by diverse grainy head-like 2-dependent pathways. Development 2012, 139, 525–536. [Google Scholar] [CrossRef]
- de Vries, M.E.; Carpinelli, M.R.; Fuller, J.N.; Sutton, Y.; Partridge, D.D.; Auden, A.; Anderson, P.J.; Jane, S.M.; Dworkin, S. Grainyhead-like 2 interacts with noggin to regulate tissue fusion in mouse. Development 2024, 151, dev202420. [Google Scholar] [CrossRef]
- de Vries, M.; Carpinelli, M.; Rutland, E.; Hatzipantelis, A.; Partridge, D.; Auden, A.; Anderson, P.J.; De Groef, B.; Wu, H.; Osterwalder, M.; et al. Interrogating the grainyhead-like 2 (GRHL2) genomic locus identifies an enhancer element that regulates palatogenesis in mouse. Dev. Biol. 2020, 459, 194–203. [Google Scholar] [CrossRef]
- Zhou, B.; Zhang, X.; Li, T.; Xie, R.; Zhou, J.; Luo, Y.; Yang, C. CircZDHHC20 represses the proliferation, migration and invasion in trophoblast cells by mir-144/GRHL2 axis. Cancer Cell Int. 2020, 20, 19. [Google Scholar] [CrossRef]
- Werner, S.; Frey, S.; Riethdorf, S.; Schulze, C.; Alawi, M.; Kling, L.; Vafaizadeh, V.; Sauter, G.; Terracciano, L.; Schumacher, U.; et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J. Biol. Chem. 2013, 288, 22993–23008. [Google Scholar] [CrossRef]
- Hu, F.; He, Z.; Sun, C.; Rong, D. Knockdown of GRHL2 inhibited proliferation and induced apoptosis of colorectal cancer by suppressing the PI3K/Akt pathway. Gene 2019, 700, 96–104. [Google Scholar] [CrossRef]
- Xiang, J.; Fu, X.; Ran, W.; Wang, Z. GRHL2 reduces invasion and migration through inhibition of TGFβ-induced EMT in gastric cancer. Oncogenesis 2017, 6, e284. [Google Scholar] [CrossRef] [PubMed]
- Walentin, K.; Hinze, C.; Werth, M.; Haase, N.; Varma, S.; Morell, R.; Aue, A.; Pötschke, E.; Warburton, D.; Qiu, A.; et al. A GRHL2-dependent gene network controls trophoblast branching morphogenesis. Development 2015, 142, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Helzer, K.T.; Szatkowski Ozers, M.; Meyer, M.B.; Benkusky, N.A.; Solodin, N.; Reese, R.M.; Warren, C.L.; Pike, J.W.; Alarid, E.T. The phosphorylated estrogen receptor α (ER) cistrome identifies a subset of active enhancers enriched for direct ER-DNA binding and the transcription factor GRHL2. Mol. Cell Biol. 2019, 39, e00417-18. [Google Scholar] [CrossRef] [PubMed]
- Reese, R.M.; Helzer, K.T.; Allen, K.O.; Zheng, C.; Solodin, N.; Alarid, E.T. GRHL2 enhances phosphorylated estrogen receptor (ER) chromatin binding and regulates ER-mediated transcriptional activation and repression. Mol. Cell Biol. 2022, 42, e0019122. [Google Scholar] [CrossRef]
- Carpinelli, M.R.; de Vries, M.E.; Auden, A.; Butt, T.; Deng, Z.; Partridge, D.D.; Miles, L.B.; Georgy, S.R.; Haigh, J.J.; Darido, C.; et al. Inactivation of Zeb1 in GRHL2-deficient mouse embryos rescues mid-gestation viability and secondary palate closure. Dis. Model. Mech. 2020, 13, dmm042218. [Google Scholar] [CrossRef]
- Nikolopoulou, E.; Hirst, C.S.; Galea, G.; Venturini, C.; Moulding, D.; Marshall, A.R.; Rolo, A.; De Castro, S.C.P.; Copp, A.J.; Greene, N.D.E. Spinal neural tube closure depends on regulation of surface ectoderm identity and biomechanics by GRHL2. Nat. Commun. 2019, 10, 2487. [Google Scholar] [CrossRef]
- Liu, D.; Wei, B.; Liang, L.; Sheng, Y.; Sun, S.; Sun, X.; Li, M.; Li, H.; Yang, C.; Peng, Y.; et al. The circadian clock component RORA increases immunosurveillance in melanoma by inhibiting PD-L1 expression. Cancer Res. 2024, 84, 2265–2281. [Google Scholar] [CrossRef]
- Xiao, L.; Wang, M.; Zhang, W.; Song, Y.; Zeng, J.; Li, H.; Yu, H.; Li, L.; Gao, P.; Yao, P. Maternal diabetes-mediated RORA suppression contributes to gastrointestinal symptoms in autism-like mouse offspring. BMC Neurosci. 2022, 23, 8. [Google Scholar] [CrossRef]
- Billon, C.; Sitaula, S.; Burris, T.P. Metabolic characterization of a novel RORα knockout mouse model without ataxia. Front. Endocrinol. 2017, 8, 141. [Google Scholar] [CrossRef]
- Haim-Vilmovsky, L.; Henriksson, J.; Walker, J.A.; Miao, Z.; Natan, E.; Kar, G.; Clare, S.; Barlow, J.L.; Charidemou, E.; Mamanova, L.; et al. Mapping RORA expression in resting and activated CD4+ T cells. PLoS ONE 2021, 16, e0251233. [Google Scholar] [CrossRef]
- Brązert, M.; Kranc, W.; Nawrocki, M.J.; Sujka-Kordowska, P.; Konwerska, A.; Jankowski, M.; Kocherova, I.; Celichowski, P.; Jeseta, M.; Ożegowska, K.; et al. New markers for regulation of transcription and macromolecule metabolic process in porcine oocytes during in vitro maturation. Mol. Med. Rep. 2020, 21, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Gelaye, B.; Denis, M.; Tadesse, M.G.; Luque Fernandez, M.A.; Enquobahrie, D.A.; Ananth, C.V.; Sanchez, S.E.; Williams, M.A. Circadian clock-related genetic risk scores and risk of placental abruption. Placenta 2015, 36, 1480–1486. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, X.; Qi, T.; Hui, Y.; Yan, H.; Qu, L.; Lan, X.; Pan, C. Whole-genome sequencing to identify candidate genes for litter size and to uncover the variant function in goats (Capra hircus). Genomics 2021, 113, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Sarachana, T.; Xu, M.; Wu, R.C.; Hu, V.W. Sex hormones in autism: Androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PLoS ONE 2011, 6, e17116. [Google Scholar] [CrossRef]
- Molinari, E.; Bar, H.; Pyle, A.M.; Patrizio, P. Transcriptome analysis of human cumulus cells reveals hypoxia as the main determinant of follicular senescence. Mol. Hum. Reprod. 2016, 22, 866–876. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, H.; Mao, C.; Jiang, F.; Lu, X.; Han, X.; Hao, K.; Lan, X.; Zhang, Q.; Pan, C. Detection of the 23-bp nucleotide sequence mutation in retinoid acid receptor related orphan receptor alpha (RORA) gene and its effect on sheep litter size. Anim. Biotechnol. 2022, 33, 70–78. [Google Scholar] [CrossRef]
- Pan, Z.; Li, S.; Liu, Q.; Wang, Z.; Zhou, Z.; Di, R.; Miao, B.; Hu, W.; Wang, X.; Hu, X.; et al. Whole-genome sequences of 89 chinese sheep suggest role of rxfp2 in the development of unique horn phenotype as response to semi-feralization. Gigascience 2018, 7, giy019. [Google Scholar] [CrossRef]
- Zhou, M.; Pan, Z.; Cao, X.; Guo, X.; He, X.; Sun, Q.; Di, R.; Hu, W.; Wang, X.; Zhang, X.; et al. Single nucleotide polymorphisms in the hira gene affect litter size in small tail han sheep. Animals 2018, 8, 71. [Google Scholar] [CrossRef]
- Xia, Q.; Wang, X.; Pan, Z.; Zhang, R.; Wei, C.; Chu, M.; Di, R. Genetic diversity and phylogenetic relationship of nine sheep populations based on microsatellite markers. Arch. Anim. Breed. 2021, 64, 7–16. [Google Scholar] [CrossRef]
- Liskova, P.; Dudakova, L.; Evans, C.J.; Rojas Lopez, K.E.; Pontikos, N.; Athanasiou, D.; Jama, H.; Sach, J.; Skalicka, P.; Stranecky, V.; et al. Ectopic GRHL2 expression due to non-coding mutations promotes cell state transition and causes posterior polymorphous corneal dystrophy 4. Am. J. Hum. Genet. 2018, 102, 447–459. [Google Scholar] [CrossRef]
- Matsushita, T.; Sakai, M.; Yoshida, H.; Morita, S.; Hieda, Y.; Sakai, T. GRHL2 regulation of SPINT1 expression controls salivary gland development. Biochem. Biophys. Res. Commun. 2018, 504, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Lv, X.; Zhang, L. GRHL2 acts as an anti-oncogene in bladder cancer by regulating ZEB1 in epithelial-mesenchymal transition (EMT) process. Onco Targets Ther. 2020, 13, 2511–2522. [Google Scholar] [CrossRef] [PubMed]
- Chung, V.Y.; Tan, T.Z.; Tan, M.; Wong, M.K.; Kuay, K.T.; Yang, Z.; Ye, J.; Muller, J.; Koh, C.M.; Guccione, E.; et al. GRHL2-mir-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep. 2016, 6, 19943. [Google Scholar] [CrossRef] [PubMed]
- Faddaoui, A.; Sheta, R.; Bachvarova, M.; Plante, M.; Gregoire, J.; Renaud, M.C.; Sebastianelli, A.; Gobeil, S.; Morin, C.; Ghani, K.; et al. Suppression of the grainyhead transcription factor 2 gene (GRHL2) inhibits the proliferation, migration, invasion and mediates cell cycle arrest of ovarian cancer cells. Cell Cycle 2017, 16, 693–706. [Google Scholar] [CrossRef]
- Zhang, Y.; Chan, H.L.; Garcia-Martinez, L.; Karl, D.L.; Weich, N.; Slingerland, J.M.; Verdun, R.E.; Morey, L. Estrogen induces dynamic ERα and RING1B recruitment to control gene and enhancer activities in luminal breast cancer. Sci. Adv. 2020, 6, eaaz7249. [Google Scholar] [CrossRef]
- Du, X.; He, X.; Liu, Q.; Liu, Q.; Di, R.; Chu, M. Identification of photoperiod-induced specific mirnas in the adrenal glands of sunite sheep (Ovis aries). Front. Vet. Sci. 2022, 9, 888207. [Google Scholar] [CrossRef]
- Li, B.; Wang, W.; Huang, Y.; Han, L.; Li, J.; Zheng, N.; Wu, Z.; Zhang, X.; Li, X.; Deng, L.; et al. Lithium treatment promotes the activation of primordial follicles through PI3K/Akt signaling†. Biol. Reprod. 2022, 107, 1059–1071. [Google Scholar] [CrossRef]
- Kalous, J.; Aleshkina, D.; Anger, M. A role of PI3K/Akt signaling in oocyte maturation and early embryo development. Cells 2023, 12, 1830. [Google Scholar] [CrossRef]
- Maidarti, M.; Anderson, R.A.; Telfer, E.E. Crosstalk between pten/PI3K/Akt signalling and DNA damage in the oocyte: Implications for primordial follicle activation, oocyte quality and ageing. Cells 2020, 9, 200. [Google Scholar] [CrossRef]
- Barberino, R.S.; Macedo, T.J.S.; Lins, T.; Menezes, V.G.; Silva, R.L.S.; Monte, A.P.O.; Palheta, R.C., Jr.; Smitz, J.E.J.; Matos, M.H.T. Immunolocalization of melatonin receptor type 1 in the sheep ovary and involvement of the PI3K/Akt/foxo3a signaling pathway in the effects of melatonin on survival and in vitro activation of primordial follicles. Mol. Reprod. Dev. 2022, 89, 485–497. [Google Scholar] [CrossRef]
- López-Cardona, A.P.; Pérez-Cerezales, S.; Fernández-González, R.; Laguna-Barraza, R.; Pericuesta, E.; Agirregoitia, N.; Gutiérrez-Adán, A.; Agirregoitia, E. CB1 cannabinoid receptor drives oocyte maturation and embryo development via PI3K/Akt and mapk pathways. Faseb. J. 2017, 31, 3372–3382. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liang, W.; Luo, Y.; Wang, J.; Liu, X.; Li, S.; Hao, Z. Transforming growth factor-β1 mediates the smad4/bmf pathway to regulate ovarian granulosa cell apoptosis in small tail han sheep. Theriogenology 2024, 214, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Juengel, J.L.; Bibby, A.H.; Reader, K.L.; Lun, S.; Quirke, L.D.; Haydon, L.J.; McNatty, K.P. The role of transforming growth factor-beta (tgf-beta) during ovarian follicular development in sheep. Reprod. Biol. Endocrinol. 2004, 2, 78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhu, Q.; Xie, Z.; Chen, Y.; Qiao, Y.; Li, L.; Jing, N. The zinc finger transcription factor Ovol2 acts downstream of the bone morphogenetic protein pathway to regulate the cell fate decision between neuroectoderm and mesendoderm. J. Biol. Chem. 2013, 288, 6166–6177. [Google Scholar] [CrossRef]
- Bondestam, J.; Kaivo-oja, N.; Kallio, J.; Groome, N.; Hydén-Granskog, C.; Fujii, M.; Moustakas, A.; Jalanko, A.; ten Dijke, P.; Ritvos, O. Engagement of activin and bone morphogenetic protein signaling pathway smad proteins in the induction of inhibin B production in ovarian granulosa cells. Mol. Cell Endocrinol. 2002, 195, 79–88. [Google Scholar] [CrossRef]
- Findlay, J.K.; Drummond, A.E.; Dyson, M.; Baillie, A.J.; Robertson, D.M.; Ethier, J.F. Production and actions of inhibin and activin during folliculogenesis in the rat. Mol. Cell Endocrinol. 2001, 180, 139–144. [Google Scholar] [CrossRef]
- Liu, J.; Xu, L.; Ding, X.; Ma, Y. Genome-wide association analysis of reproductive traits in chinese holstein cattle. Genes 2023, 15, 12. [Google Scholar] [CrossRef]
| Primer Name | Primer Sequence (5′-3′) | Length (bp) |
|---|---|---|
| GRHL2-SNP (rs402967440) | F-GATGTTGCGACTGTTTGGG | 19 |
| R-CAGAACTCCTGGGCTTTTGT | 20 | |
| GRHL2-SNP (rs411785026) | F-CTTGGACAAAGGAAGCGGC | 19 |
| R-GGACGACAACATCATTGAGCACT | 23 | |
| GRHL2-SNP (rs424408342) | F-CGGAGAAAAGGTGGACAATAAAT | 23 |
| R-TGAAAGAATGGCTGAGGGGAC | 21 | |
| GRHL2-SNP (rs411712725) | F-AATCGTCCCCTACTCTTCCTCA | 22 |
| R-TTGGCACCCCGCATACTTTA | 20 | |
| RORA-SNP (rs425540123) | F-GCATCACTCATAACCTTGACCTT | 23 |
| R-GGCATCATTTTGACATTCTCCT | 22 |
| Gene | Primer Name | Primer Sequence (5′-3′) | Length (bp) |
|---|---|---|---|
| GRHL2 rs402967440 (g.75350274G > A) | Forward | ACGTTGGATGTGCTGGATGGGTGTGAGAAG | 30 |
| Reverse | ACGTTGGATGACAAGCGTTCTCTCTCCATC | 30 | |
| Extension | CTGGCACTTCTGAAACCAGT | 20 | |
| GRHL2 rs411785026 (g.75351375T > C) | Forward | ACGTTGGATGAATGGACTGAAAGGGAGAGC | 30 |
| Reverse | ACGTTGGATGTATTGTCCACCTTTTCTCCG | 30 | |
| Extension | CCAAAGAGTCCTGGTGGGCT | 20 | |
| GRHL2 rs424408342 (g.75351555G > A) | Forward | ACGTTGGATGGTCGTGTAACAGCAGTCGTC | 30 |
| Reverse | ACGTTGGATGGAAATCTGAGCCCCATTCTG | 30 | |
| Extension | GGTGAGGTGGGCCTGCG | 17 | |
| GRHL2 rs411712725 (g.75371268G > A) | Forward | ACGTTGGATGCCTACTCTTCCTCATGACAG | 30 |
| Reverse | ACGTTGGATGAAAGAAGAAGGGATGAAGCG | 30 | |
| Extension | GGGCGGGCCCCAGGGCGAGCG | 21 | |
| RORA rs425540123 (g.46741101A > T) | Forward | ACGTTGGATGTCGAGCTACCCTTTGATCAG | 30 |
| Reverse | ACGTTGGATGTCTGCAATTCTGGCAGCTAC | 30 | |
| Extension | AGAAAAGGCAAAGAAGGAT | 19 |
| Gene | Loci | Genotype Frequency (Genotype Count) | Allele Frequency | PIC | HE | NE | Chi-Square Test (p-Value) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| GRHL2 | rs402967440 | GG | GA | AA | G | A | ||||
| 0.96 (367) | 0.04 (14) | 0.01 (3) | 0.97 | 0.03 | 0.05 | 0.05 | 1.05 | 0.00 | ||
| rs411785026 | TT | CT | CC | T | C | |||||
| 0.31 (119) | 0.45 (172) | 0.24 (93) | 0.53 | 0.47 | 0.37 | 0.50 | 1.99 | 0.05 | ||
| rs424408342 | GG | GA | AA | G | A | |||||
| 0.93 (357) | 0.03 (13) | 0.04 (14) | 0.95 | 0.05 | 0.10 | 0.10 | 1.11 | 0.00 | ||
| rs411712725 | GG | GA | AA | G | A | |||||
| 0.86 (328) | 0.14 (52) | 0.01 (2) | 0.93 | 0.07 | 0.13 | 0.14 | 1.16 | 0.97 | ||
| RORA | rs425540123 | AA | AT | TT | A | T | ||||
| 0.90 (345) | 0.10 (38) | 0.00 (1) | 0.95 | 0.05 | 0.09 | 0.10 | 1.11 | 0.97 | ||
| Description | Score | |
|---|---|---|
| VWC2L | Von Willebrand factor C domain containing 2 like | 0.654 |
| SEC11A | Signal peptidase complex catalytic subunit SEC11 | 0.609 |
| TEX2 | Testis expressed 2 | 0.573 |
| OVOL2 | Ovo like zinc finger 2 | 0.571 |
| ESRP2 | Epithelial splicing regulatory protein 2 | 0.557 |
| NOL10 | Nucleolar protein 10 | 0.551 |
| ESRP1 | Epithelial splicing regulatory protein 1 | 0.541 |
| TMEM182 | Transmembrane protein 182 | 0.515 |
| TNRC18 | Trinucleotide repeat containing 18 | 0.513 |
| FOXA1 | Forkhead box A1 | 0.507 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, X.; Liu, K.; Wang, X.; Di, R.; He, X.; Liu, Y.; Chu, M. Association Analysis of SNPs in GRHL2 and RORA Genes with Lambing Number in Small-Tailed Han Sheep. Animals 2025, 15, 1432. https://doi.org/10.3390/ani15101432
Pu X, Liu K, Wang X, Di R, He X, Liu Y, Chu M. Association Analysis of SNPs in GRHL2 and RORA Genes with Lambing Number in Small-Tailed Han Sheep. Animals. 2025; 15(10):1432. https://doi.org/10.3390/ani15101432
Chicago/Turabian StylePu, Xiufen, Kai Liu, Xiangyu Wang, Ran Di, Xiaoyun He, Yufang Liu, and Mingxing Chu. 2025. "Association Analysis of SNPs in GRHL2 and RORA Genes with Lambing Number in Small-Tailed Han Sheep" Animals 15, no. 10: 1432. https://doi.org/10.3390/ani15101432
APA StylePu, X., Liu, K., Wang, X., Di, R., He, X., Liu, Y., & Chu, M. (2025). Association Analysis of SNPs in GRHL2 and RORA Genes with Lambing Number in Small-Tailed Han Sheep. Animals, 15(10), 1432. https://doi.org/10.3390/ani15101432







