An Analysis of a Transposable Element Expression Atlas during 27 Developmental Stages in Porcine Skeletal Muscle: Unveiling Molecular Insights into Pork Production Traits
Abstract
Simple Summary
Abstract
1. Background
2. Materials and Methods
2.1. RNA-Seq Data Analysis
2.2. TE Annotation and Quantification
2.3. Multivariate Analysis of Detectably Expressed TEs
2.4. Weighted Gene Co-Expression Analysis
2.5. Module Function Analysis
2.6. TE-Mediated Gene Regulatory Network Construction
3. Results
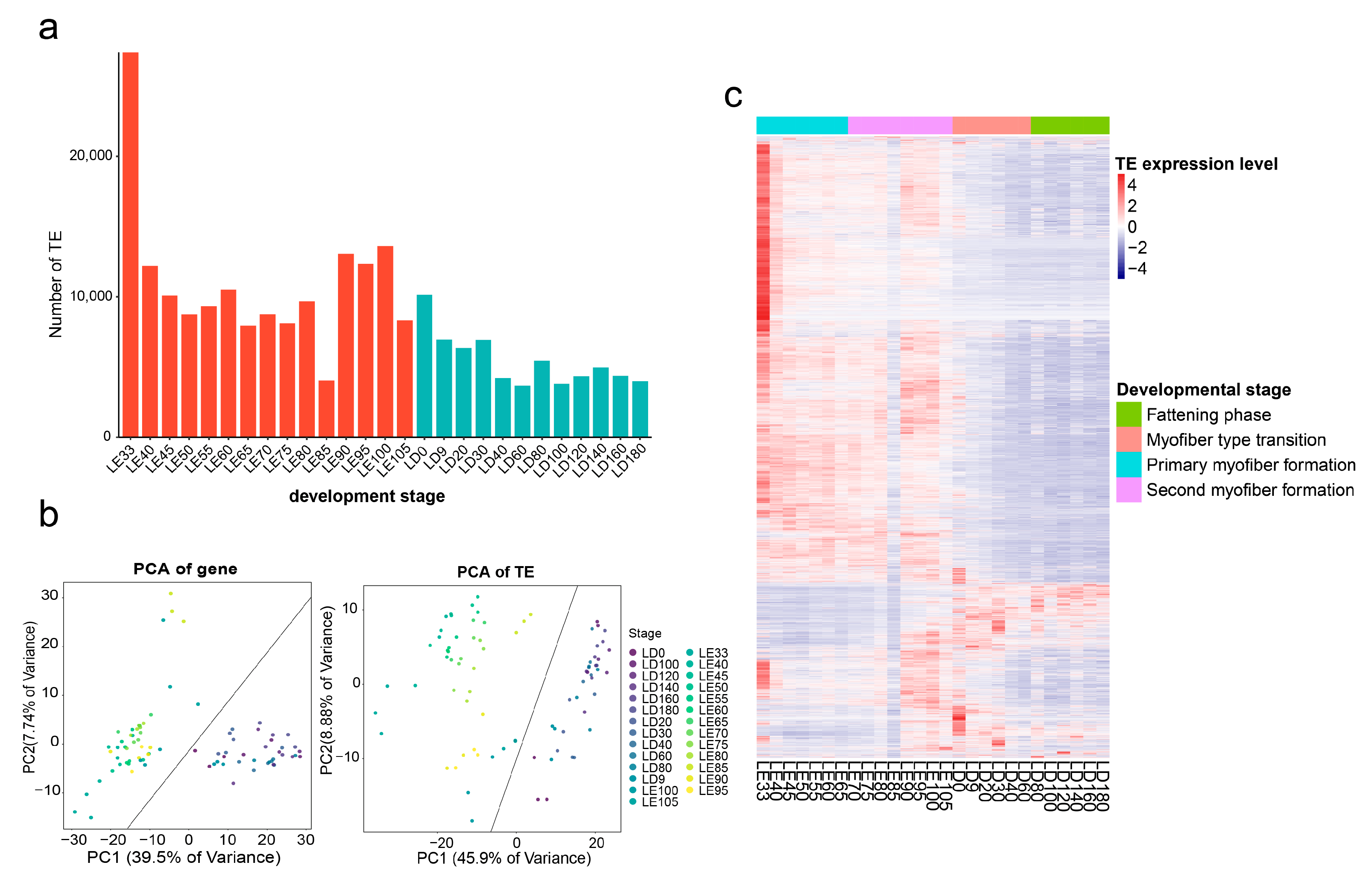
3.1. The Construction of a Dynamic Expression Atlas of TEs
3.2. Genomic Distribution and Epigenetic Features of TE Expression
3.3. Self-Expressed TEs Regulate Skeletal Muscle Development
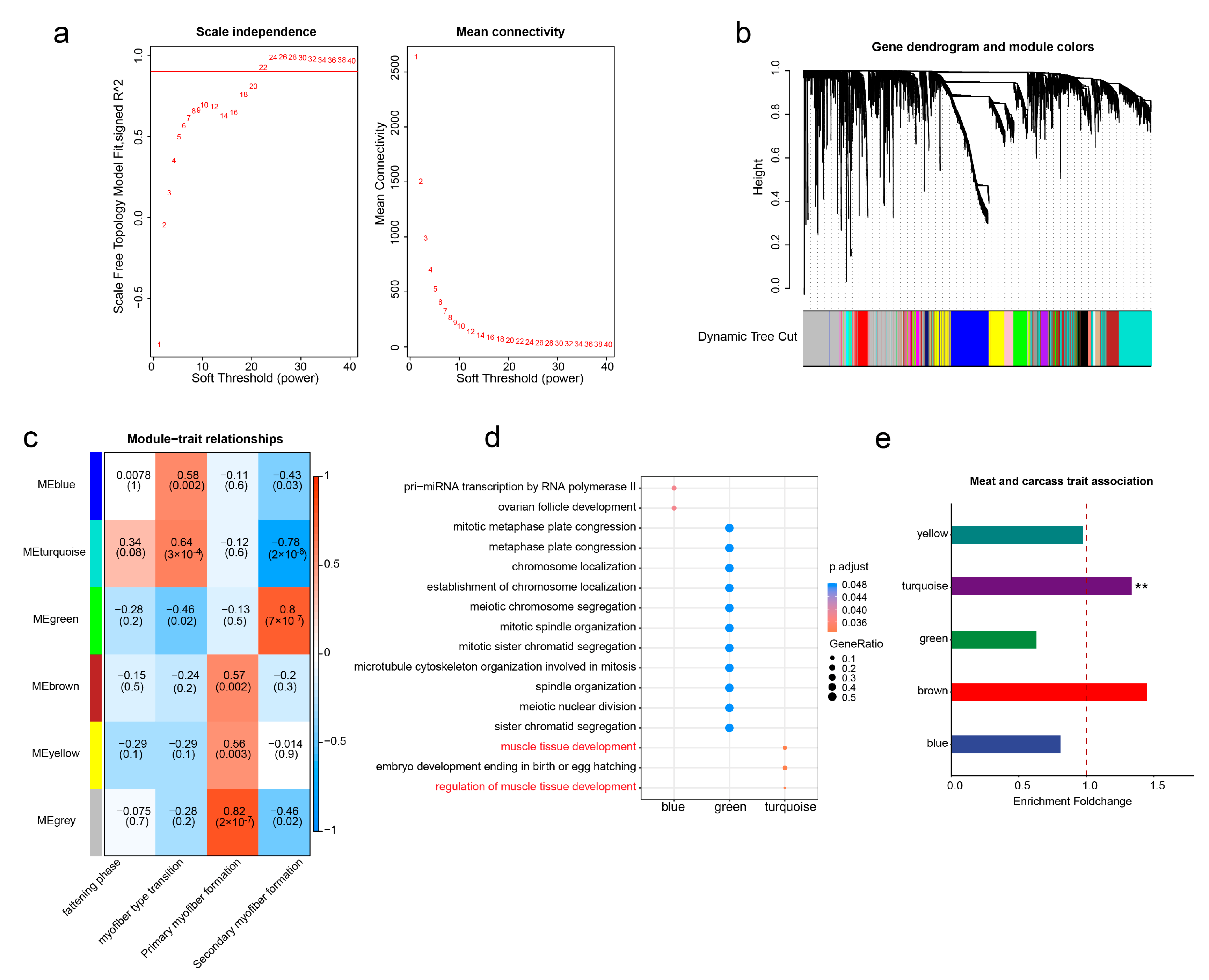
3.4. WGCNA Identification of the Key Modules Regulating Muscle Development
3.5. Construction of TE-Mediated Gene Regulatory Networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Young, L.D. Survival, body weights, feed efficiency, and carcass traits of 7/8 White Composite and 1/8 Duroc, 1/8 Meishan, 1/8 Fengjing, or 1/8 Minzhu pigs. J. Anim. Sci. 1998, 76, 1550–1558. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ai, H.; Fang, X.; Yang, B.; Huang, Z.; Chen, H.; Mao, L.; Zhang, F.; Zhang, L.; Cui, L.; He, W.; et al. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet. 2015, 47, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Yu, Y.; Feng, W.; Du, H.; Yu, J.; Kang, H.; Zheng, X.; Wang, Z.; Liu, G.E.; Ernst, C.W.; et al. Evidence of evolutionary history and selective sweeps in the genome of Meishan pig reveals its genetic and phenotypic characterization. Gigascience 2018, 7, giy058. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, L.; Tian, S.; Lin, Y.; Tang, Q.; Zhou, X.; Li, D.; Yeung, C.; Che, T.; Jin, L.; et al. Comprehensive variation discovery and recovery of missing sequence in the pig genome using multiple de novo assemblies. Genome Res. 2017, 27, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Nakajima, I.; Arakawa, A.; Mikawa, S.; Matsumoto, T.; Uenishi, H.; Nakamura, Y.; Taniguchi, M. Differences in gene expression profiles for subcutaneous adipose, liver, and skeletal muscle tissues between Meishan and Landrace pigs with different backfat thicknesses. PLoS ONE 2018, 13, e204135. [Google Scholar] [CrossRef]
- Zhou, S.; Ding, R.; Meng, F.; Wang, X.; Zhuang, Z.; Quan, J.; Geng, Q.; Wu, J.; Zheng, E.; Wu, Z.; et al. A meta-analysis of genome-wide association studies for average daily gain and lean meat percentage in two Duroc pig populations. BMC Genom. 2021, 22, 12. [Google Scholar] [CrossRef]
- Gao, G.; Gao, N.; Li, S.; Kuang, W.; Zhu, L.; Jiang, W.; Yu, W.; Guo, J.; Li, Z.; Yang, C.; et al. Genome-Wide Association Study of Meat Quality Traits in a Three-Way Crossbred Commercial Pig Population. Front. Genet. 2021, 12, 614087. [Google Scholar] [CrossRef]
- Hamilton, D.N.; Ellis, M.; Miller, K.D.; McKeith, F.K.; Parrett, D.F. The effect of the Halothane and Rendement Napole genes on carcass and meat quality characteristics of pigs. J. Anim. Sci. 2000, 78, 2862–2867. [Google Scholar] [CrossRef]
- Van Laere, A.S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M.; et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef]
- Ma, J.; Yang, J.; Zhou, L.; Ren, J.; Liu, X.; Zhang, H.; Yang, B.; Zhang, Z.; Ma, H.; Xie, X.; et al. A splice mutation in the PHKG1 gene causes high glycogen content and low meat quality in pig skeletal muscle. PLoS Genet. 2014, 10, e1004710. [Google Scholar] [CrossRef]
- Kim, K.S.; Larsen, N.; Short, T.; Plastow, G.; Rothschild, M.F. A missense variant of the porcine melanocortin-4 receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mamm. Genome 2000, 11, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Murani, E.; Muraniova, M.; Ponsuksili, S.; Schellander, K.; Wimmers, K. Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity. BMC Dev. Biol. 2007, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Beermann, D.H.; Cassens, R.G. Indirect fluorescence of primary and secondary myofibers in developing porcine muscle. J. Histochem. Cytochem. 1977, 25, 439–442. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, M.; Yan, E.; Wang, Y.; Ma, C.; Zhang, P.; Yin, J. Dietary Malic Acid Supplementation Induces Skeletal Muscle Fiber-Type Transition of Weaned Piglets and Further Improves Meat Quality of Finishing Pigs. Front. Nutr. 2021, 8, 825495. [Google Scholar] [CrossRef]
- Scheffler, T.L.; Scheffler, J.M.; Park, S.; Kasten, S.C.; Wu, Y.; McMillan, R.P.; Hulver, M.W.; Frisard, M.I.; Gerrard, D.E. Fiber hypertrophy and increased oxidative capacity can occur simultaneously in pig glycolytic skeletal muscle. Am. J. Physiol. Cell Physiol. 2014, 306, C354–C363. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Yan, J.; Chen, M.; Zhu, M.; Tang, Y.; Liu, S.; Tang, Z. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 2021, 49, 1313–1329. [Google Scholar] [CrossRef] [PubMed]
- Corbett, R.J.; Ford, L.M.; Raney, N.E.; Grabowski, J.M.; Ernst, C.W. Pig fetal skeletal muscle development is associated with genome-wide DNA hypomethylation and corresponding alterations in transcript and microRNA expression. Genome 2023, 66, 68–79. [Google Scholar] [CrossRef]
- Cai, S.; Hu, B.; Wang, X.; Liu, T.; Lin, Z.; Tong, X.; Xu, R.; Chen, M.; Duo, T.; Zhu, Q.; et al. Integrative single-cell RNA-seq and ATAC-seq analysis of myogenic differentiation in pig. BMC Biol. 2023, 21, 19. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, J.; Fan, X.; Chen, J.; Wang, Z.; Liu, X.; Yi, G.; Liu, Y.; Niu, Y.; Zhang, L.; et al. The genome variation and developmental transcriptome maps reveal genetic differentiation of skeletal muscle in pigs. PLoS Genet. 2021, 17, e1009910. [Google Scholar] [CrossRef]
- Tan, Y.; Gan, M.; Shen, L.; Li, L.; Fan, Y.; Chen, Y.; Chen, L.; Niu, L.; Zhao, Y.; Jiang, A.; et al. Profiling and Functional Analysis of Long Noncoding RNAs and mRNAs during Porcine Skeletal Muscle Development. Int. J. Mol. Sci. 2021, 22, 503. [Google Scholar] [CrossRef]
- Salavati, M.; Woolley, S.A.; Cortes, A.Y.; Halstead, M.M.; Stenhouse, C.; Johnsson, M.; Ashworth, C.J.; Archibald, A.L.; Donadeu, F.X.; Hassan, M.A.; et al. Profiling of open chromatin in developing pig (Sus scrofa) muscle to identify regulatory regions. G3 2022, 12, jkab424. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Hou, X.; Liu, X.; Wang, L.; Gao, H.; Zhao, F.; Shi, L.; Shi, L.; Yan, H.; Deng, T.; et al. The landscape of chromatin accessibility in skeletal muscle during embryonic development in pigs. J. Anim. Sci. Biotechnol. 2021, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Zhang, J.; Wang, Y.; Zhu, X.; Hu, S.; Zeng, J.; Liang, F.; Tang, Q.; Chen, Y.; Chen, L.; et al. Reorganization of chromatin architecture during prenatal development of porcine skeletal muscle. DNA Res. 2021, 28, dsab003. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Wang, S.; Wang, S.; Zeng, J.; Hong, L.; Li, Z.; Yang, J.; Cai, G.; Zheng, E.; Wu, Z.; et al. Genome-Wide Analysis of H3K27me3 in Porcine Embryonic Muscle Development. Front. Cell Dev. Biol. 2021, 9, 739321. [Google Scholar] [CrossRef] [PubMed]
- Sultana, T.; Zamborlini, A.; Cristofari, G.; Lesage, P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 2017, 18, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Campos-Sanchez, R.; Cremona, M.A.; Pini, A.; Chiaromonte, F.; Makova, K.D. Integration and Fixation Preferences of Human and Mouse Endogenous Retroviruses Uncovered with Functional Data Analysis. PLoS Comput. Biol. 2016, 12, e1004956. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.; Kural, D.; Stromberg, M.P.; Walker, J.A.; Konkel, M.K.; Stutz, A.M.; Urban, A.E.; Grubert, F.; Lam, H.Y.; Lee, W.P.; et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011, 7, e1002236. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Bao, Z.; Zhang, X.; Eddy, S.R.; Wessler, S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature 2004, 431, 569–573. [Google Scholar] [CrossRef]
- Chuong, E.B.; Rumi, M.A.; Soares, M.J.; Baker, J.C. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013, 45, 325–329. [Google Scholar] [CrossRef]
- Jacques, P.E.; Jeyakani, J.; Bourque, G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 2013, 9, e1003504. [Google Scholar] [CrossRef]
- Xing, J.; Wang, H.; Belancio, V.P.; Cordaux, R.; Deininger, P.L.; Batzer, M.A. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc. Natl. Acad. Sci. USA 2006, 103, 17608–17613. [Google Scholar] [CrossRef]
- Kapusta, A.; Kronenberg, Z.; Lynch, V.J.; Zhuo, X.; Ramsay, L.; Bourque, G.; Yandell, M.; Feschotte, C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013, 9, e1003470. [Google Scholar] [CrossRef] [PubMed]
- Bendall, M.L.; de Mulder, M.; Iniguez, L.P.; Lecanda-Sanchez, A.; Perez-Losada, M.; Ostrowski, M.A.; Jones, R.B.; Mulder, L.; Reyes-Teran, G.; Crandall, K.A.; et al. Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput. Biol. 2019, 15, e1006453. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Tam, O.H.; Paniagua, E.; Hammell, M. TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 2015, 31, 3593–3599. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Babarinde, I.A.; Sun, L.; Xu, S.; Chen, R.; Shi, J.; Wei, Y.; Li, Y.; Ma, G.; Zhuang, Q.; et al. Identifying transposable element expression dynamics and heterogeneity during development at the single-cell level with a processing pipeline scTE. Nat. Commun. 2021, 12, 1456. [Google Scholar] [CrossRef]
- Rodriguez-Quiroz, R.; Valdebenito-Maturana, B. SoloTE for improved analysis of transposable elements in single-cell RNA-Seq data using locus-specific expression. Commun. Biol. 2022, 5, 1063. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.R.; Ardeljan, D.; Pacyna, C.N.; Payer, L.M.; Burns, K.H. SQuIRE reveals locus-specific regulation of interspersed repeat expression. Nucleic Acids Res. 2019, 47, e27. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, M.; Fan, X.; Yao, Y.; Yan, J.; Tang, Y.; Liu, S.; Li, K.; Tang, Z. Developmental atlas of the RNA editome in Sus scrofa skeletal muscle. DNA Res. 2019, 26, 261–272. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Lee, B.T.; Barber, G.P.; Benet-Pages, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, C.M.; et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022, 50, D1115–D1122. [Google Scholar] [CrossRef] [PubMed]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 4, 4.10.1–4.10.14. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, M.; Wu, T.; Zhan, L.; Li, L.; Chen, M.; Xie, W.; Xie, Z.; Hu, E.; Xu, S.; et al. Exploring Epigenomic Datasets by ChIPseeker. Curr. Protoc. 2022, 2, e585. [Google Scholar] [CrossRef]
- Zhao, Y.; Hou, Y.; Xu, Y.; Luan, Y.; Zhou, H.; Qi, X.; Hu, M.; Wang, D.; Wang, Z.; Fu, Y.; et al. A compendium and comparative epigenomics analysis of cis-regulatory elements in the pig genome. Nat. Commun. 2021, 12, 2217. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Hu, Z.L.; Park, C.A.; Wu, X.L.; Reecy, J.M. Animal QTLdb: An improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013, 41, D871–D879. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef] [PubMed]
- Lanciano, S.; Cristofari, G. Measuring and interpreting transposable element expression. Nat. Rev. Genet. 2020, 21, 721–736. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.C.; Rovira, Q.; Wells, J.; Feschotte, C.; Vaquerizas, J.M. Zebrafish transposable elements show extensive diversification in age, genomic distribution, and developmental expression. Genome Res. 2022, 32, 1408–1423. [Google Scholar] [CrossRef] [PubMed]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liao, Y.; Zhang, H.; Jiang, Y.; Peng, Z.; Ren, R.; Li, X.; Wang, H. Tcf12 is required to sustain myogenic genes synergism with MyoD by remodelling the chromatin landscape. Commun. Biol. 2022, 5, 1201. [Google Scholar] [CrossRef]
- Zhu, X.; Huang, Q.; Luo, J.; Kong, D.; Zhang, Y. Mini-review: Gene regulatory network benefits from three-dimensional chromatin conformation and structural biology. Comput. Struct. Biotechnol. J. 2023, 21, 1728–1737. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Yu, Z.; Qiu, X.; Jiang, R.; Li, W. Uncovering the gene regulatory network of type 2 diabetes through multi-omic data integration. J. Transl. Med. 2022, 20, 604. [Google Scholar] [CrossRef]
- Gao, X.; Chandra, T.; Gratton, M.O.; Quelo, I.; Prud’Homme, J.; Stifani, S.; St-Arnaud, R. HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J. Cell Biol. 2001, 154, 1161–1171. [Google Scholar] [CrossRef]
- Morgan, N.V.; Brueton, L.A.; Cox, P.; Greally, M.T.; Tolmie, J.; Pasha, S.; Aligianis, I.A.; van Bokhoven, H.; Marton, T.; Al-Gazali, L.; et al. Mutations in the embryonal subunit of the acetylcholine receptor (CHRNG) cause lethal and Escobar variants of multiple pterygium syndrome. Am. J. Hum. Genet. 2006, 79, 390–395. [Google Scholar] [CrossRef]
- Munoz, M.; Garcia-Casco, J.M.; Caraballo, C.; Fernandez-Barroso, M.A.; Sanchez-Esquiliche, F.; Gomez, F.; Rodriguez, M.; Silio, L. Identification of Candidate Genes and Regulatory Factors Underlying Intramuscular Fat Content Through Longissimus Dorsi Transcriptome Analyses in Heavy Iberian Pigs. Front. Genet. 2018, 9, 608. [Google Scholar] [CrossRef]
- Muller, J.S.; Baumeister, S.K.; Schara, U.; Cossins, J.; Krause, S.; von der Hagen, M.; Huebner, A.; Webster, R.; Beeson, D.; Lochmuller, H.; et al. CHRND mutation causes a congenital myasthenic syndrome by impairing co-clustering of the acetylcholine receptor with rapsyn. Brain 2006, 129, 2784–2793. [Google Scholar] [CrossRef]
- Yanay, N.; Elbaz, M.; Konikov-Rozenman, J.; Elgavish, S.; Nevo, Y.; Fellig, Y.; Rabie, M.; Mitrani-Rosenbaum, S.; Nevo, Y. Pax7, Pax3 and Mamstr genes are involved in skeletal muscle impaired regeneration of dy2J/dy2J mouse model of Lama2-CMD. Hum. Mol. Genet. 2019, 28, 3369–3390. [Google Scholar] [CrossRef]
- Katoku-Kikyo, N.; Paatela, E.; Houtz, D.L.; Lee, B.; Munson, D.; Wang, X.; Hussein, M.; Bhatia, J.; Lim, S.; Yuan, C.; et al. Per1/Per2-Igf2 axis-mediated circadian regulation of myogenic differentiation. J. Cell Biol. 2021, 220, e202101057. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.; Bowen, L.; Yongzhou, B.; Zhen, W.; Choulin, C.; Yuanyuan, Z.; Shenghua, Q.; Tao, S.; Zhonglin, T.; Yuwen, L. Multi-omics analysis reveals critical cis-regulatory roles of transposable elements in livestock genomes. bioRxiv 2023. [Google Scholar] [CrossRef]
- Walsh, C.P.; Chaillet, J.R.; Bestor, T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998, 20, 116–117. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef]
- Goodier, J.L. Restricting retrotransposons: A review. Mob. DNA 2016, 7, 16. [Google Scholar] [CrossRef]
- He, J.; Fu, X.; Zhang, M.; He, F.; Li, W.; Abdul, M.M.; Zhou, J.; Sun, L.; Chang, C.; Li, Y.; et al. Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nat. Commun. 2019, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Cossins, J.; Vernon, A.E.; Zhang, Y.; Philpott, A.; Jones, P.H. Hes6 regulates myogenic differentiation. Development 2002, 129, 2195–2207. [Google Scholar] [CrossRef]
- Valdebenito-Maturana, B.; Torres, F.; Carrasco, M.; Tapia, J.C. Differential regulation of transposable elements (TEs) during the murine submandibular gland development. Mob. DNA 2021, 12, 23. [Google Scholar] [CrossRef]
- van Berkum, N.L.; Lieberman-Aiden, E.; Williams, L.; Imakaev, M.; Gnirke, A.; Mirny, L.A.; Dekker, J.; Lander, E.S. Hi-C: A method to study the three-dimensional architecture of genomes. J. Vis. Exp. 2010, 39, 1869. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Lei, B.; Liu, Y. An Analysis of a Transposable Element Expression Atlas during 27 Developmental Stages in Porcine Skeletal Muscle: Unveiling Molecular Insights into Pork Production Traits. Animals 2023, 13, 3581. https://doi.org/10.3390/ani13223581
Wang C, Lei B, Liu Y. An Analysis of a Transposable Element Expression Atlas during 27 Developmental Stages in Porcine Skeletal Muscle: Unveiling Molecular Insights into Pork Production Traits. Animals. 2023; 13(22):3581. https://doi.org/10.3390/ani13223581
Chicago/Turabian StyleWang, Chao, Bowen Lei, and Yuwen Liu. 2023. "An Analysis of a Transposable Element Expression Atlas during 27 Developmental Stages in Porcine Skeletal Muscle: Unveiling Molecular Insights into Pork Production Traits" Animals 13, no. 22: 3581. https://doi.org/10.3390/ani13223581
APA StyleWang, C., Lei, B., & Liu, Y. (2023). An Analysis of a Transposable Element Expression Atlas during 27 Developmental Stages in Porcine Skeletal Muscle: Unveiling Molecular Insights into Pork Production Traits. Animals, 13(22), 3581. https://doi.org/10.3390/ani13223581



