Excavation of Genes Related to the Mining of Growth, Development, and Meat Quality of Two Crossbred Sheep Populations Based on Comparative Transcriptomes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation, Collection, and Measures of Productive Performance
2.2. Masson Staining
2.3. Total RNA Preparation, cDNA Library, and Sequencing
2.4. Transcriptome Assembly and DEG Identification
2.5. Gene Behavior Network and Co-Expression Analysis Targeting DEGs
2.6. Quantitative Real-Time PCR Assays for Target Genes
2.7. Statistical Analysis
3. Results
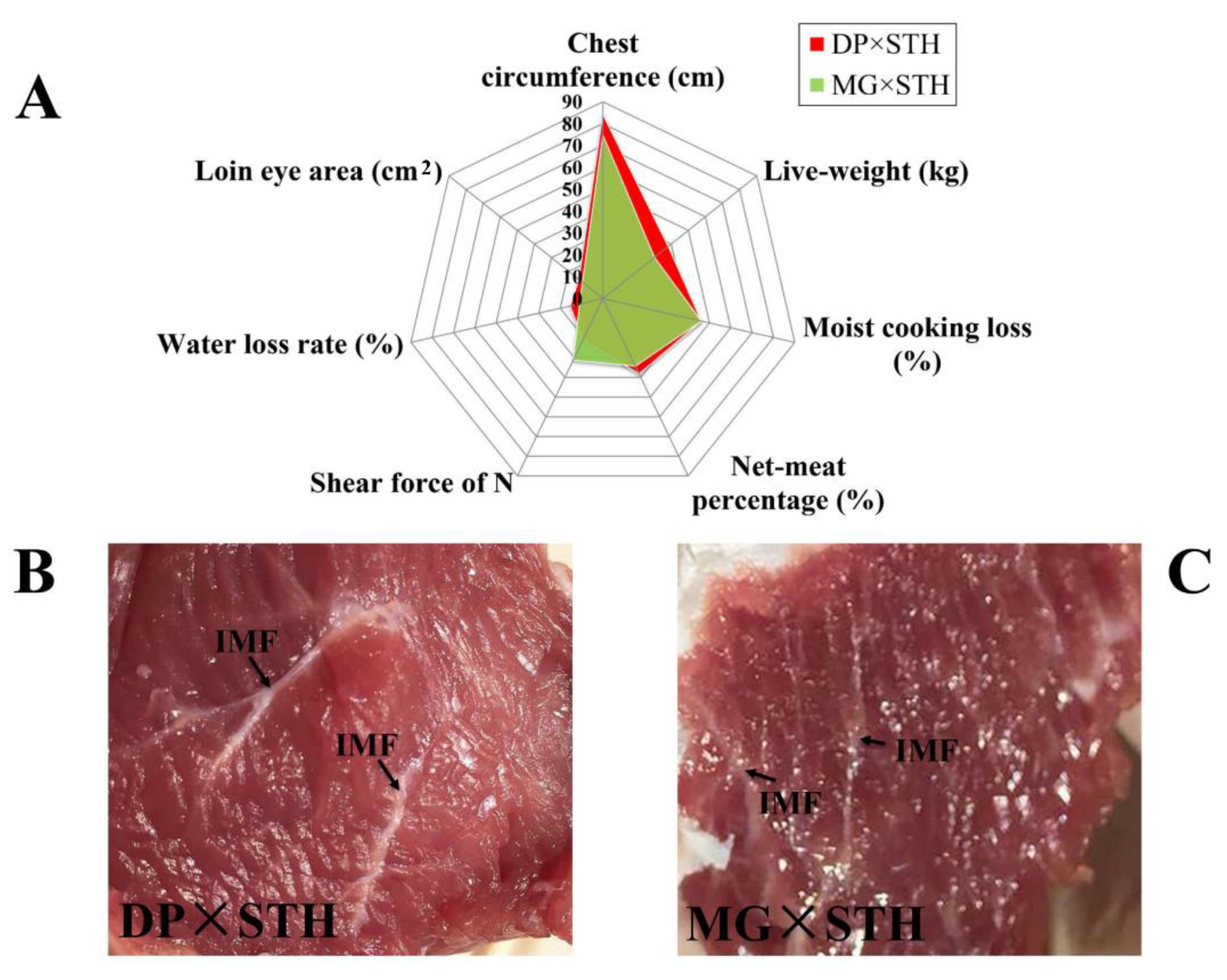
3.1. Productive Performance and Meat Quality
3.2. Masson Staining of the Longissimus dorsi
3.3. Transcriptome Analysis of Longissimus dorsi Tissues
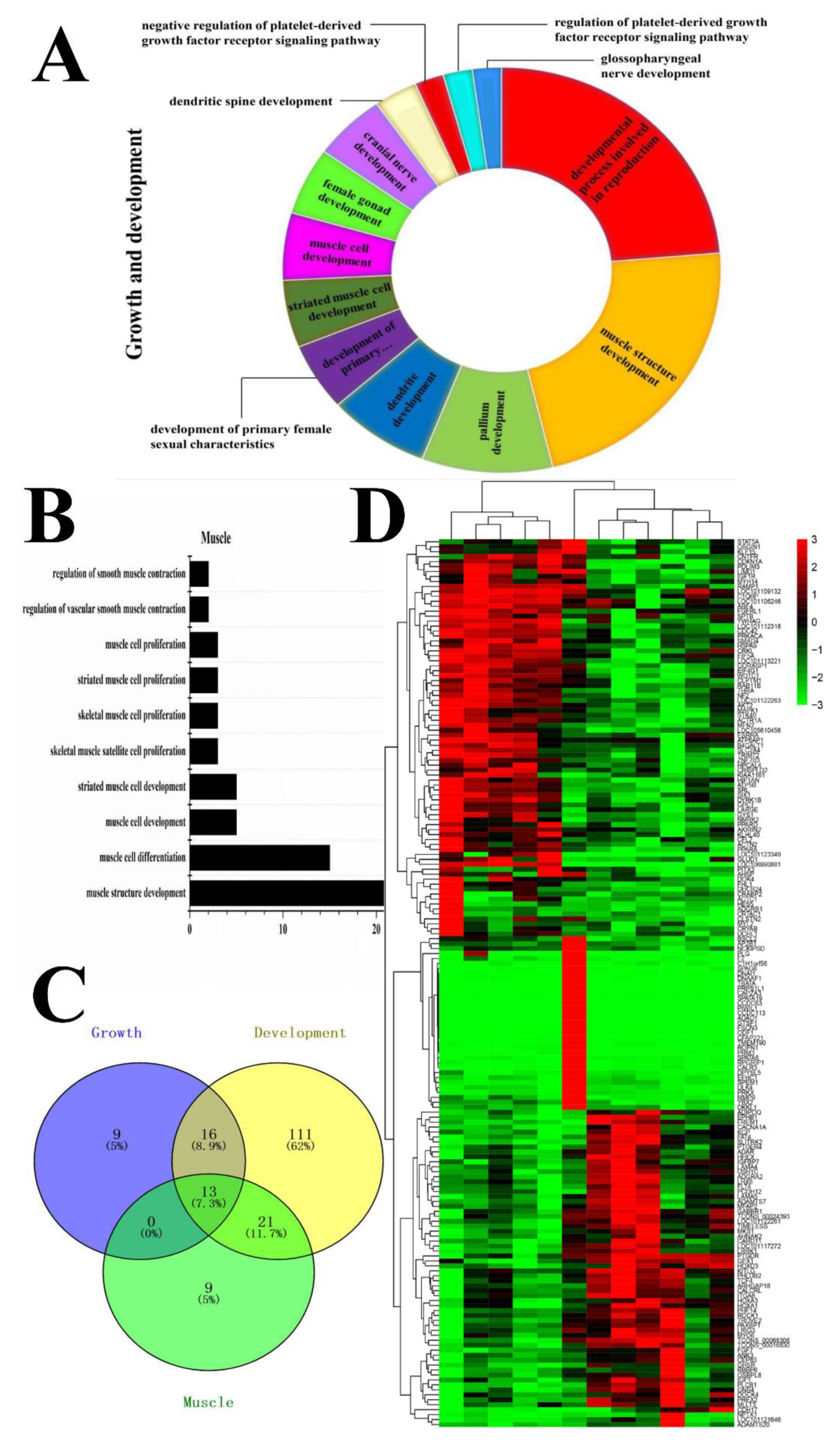
3.4. Functional Analysis of Candidate DEGs Related to Growth, Development, and Meat Quality
3.5. Validation of the Target Genes Related to Growth, Development and Meat Quality in the Longissimus dorsi
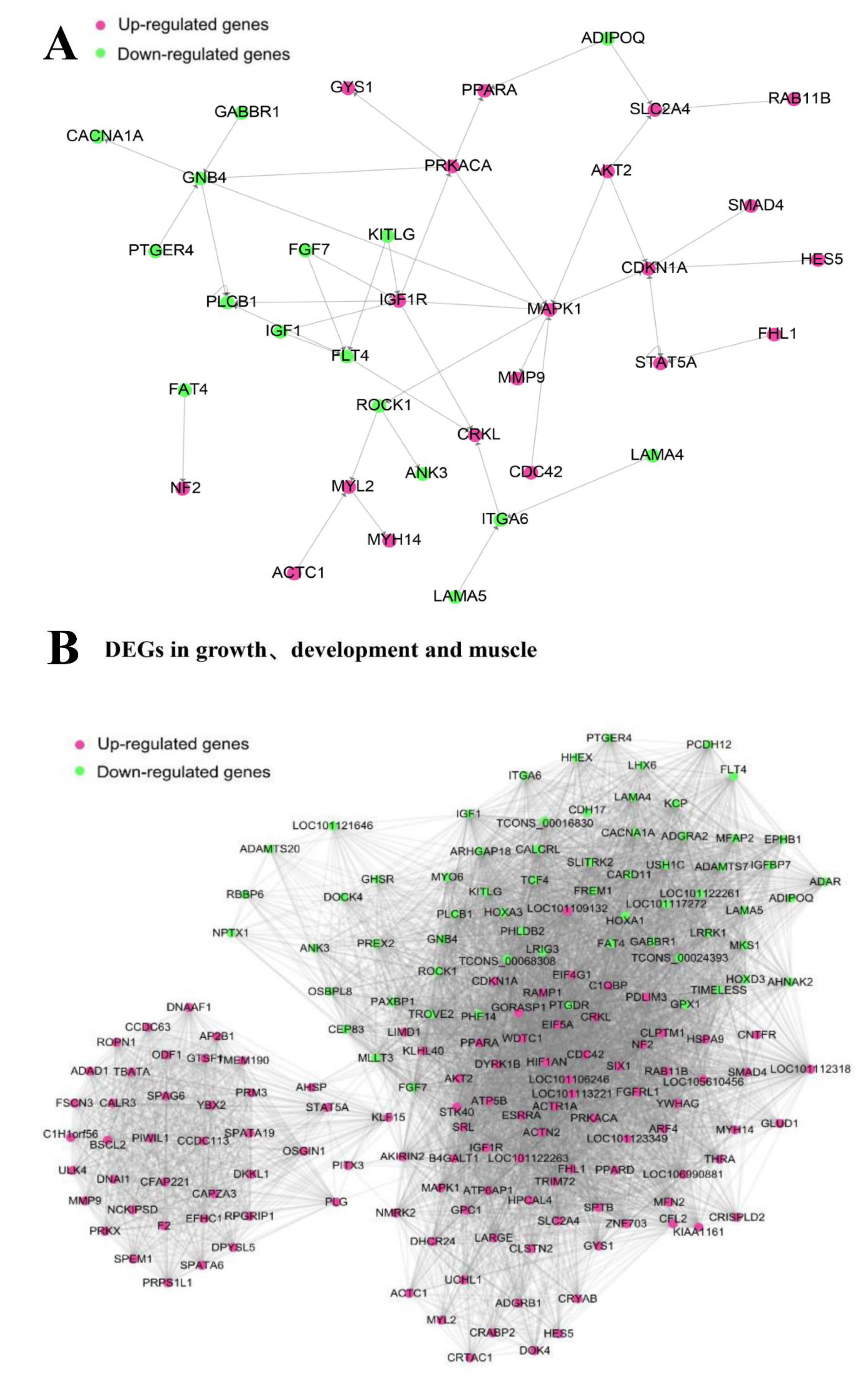
3.6. Gene-Act-Network and Co-Expression Analysis of Targeted DEGs in Sheep Growth, Development, and Meat Quality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Tang, J.S. Study on Screening Polytocous Candidate Genes in Sheep Based Transcriptome Sequencing and Proteomics. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2019. [Google Scholar]
- Miao, X.; Luo, Q.; Qin, X. Genome-wide analysis reveals the differential regulations of mRNAs and miRNAs in Dorset and Small Tail Han sheep muscles. Gene 2015, 562, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Guo, J.; Li, F.; Niu, C. Evaluation of crossbreeding of Australian superfine merinos with gansu alpine finewool sheep to improve wool characteristics. PLoS ONE 2016, 11, e0166374. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2010, 10, 57–63. [Google Scholar]
- Kumar, H.; Iskender, A.U.; Srikanth, K.; Kim, H.; Zhunushov, A.T.; Chooq, H.; Jang, G.W.; Lim, Y.; Song, K.D.; Park, J.E. Transcriptome of chicken liver tissues reveals the candidate genes and pathways responsible for adaptation into two different climatic conditions. Animals 2019, 9, 1076. [Google Scholar] [CrossRef]
- Esteve-Codina, A.; Kofler, R.; Palmieri, N.; Bussotti, G.; Notredame, C.; Pérez-Enciso, M. Exploring the gonad transcriptome of two extreme male pigs with RNA-seq. BMC Genom. 2011, 12, 552. [Google Scholar] [CrossRef]
- McCabe, M.; Waters, S.; Morris, D.; Kenny, D.; Lynn, D.; Creevey, C. RNA-seq analysis of differential gene expression in liver from lactating dairy cows divergent in negative energy balance. BMC Genom. 2012, 13, 193. [Google Scholar] [CrossRef]
- Chao, T.; Wang, G.; Wang, J.; Liu, Z.; Ji, Z.; Hou, L.; Zhang, C. Identification and Classification of New Transcripts in Dorper and Small-Tailed Han Sheep Skeletal Muscle Transcriptomes. PLoS ONE 2016, 11, e0159638. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q.; Qin, X.; Guo, Y.; Zhao, H. Genome-wide mRNA-seq profiling reveals predominant down-regulation of lipid metabolic processes in adipose tissues of Small Tail Han than Dorset sheep. Biochem. Biophys. Res. Commun. 2015, 467, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Feng, X.P.; Wang, H.L.; Meng, C.H.; Zhang, J.; Qian, Y.; Zhong, J.F.; Cao, S.X. Transcriptome analysis reveals corresponding genes and key pathways involved in heat stress in Hu sheep. Cell Stress Chaperones 2019, 24, 1045–1054. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, G.; Wang, J.; Ji, Z.; Dong, F.; Chao, T. Analysis of Differential Gene Expression and Novel Transcript Units of Ovine Muscle Transcriptomes. PLoS ONE 2014, 9, e89817. [Google Scholar] [CrossRef] [PubMed]
- Govoni, K.; Amaar, Y.; Krämer, A.; Winter, E.; Baylink, D.; Mohan, S. Regulation of insulin-like growth factor binding protein-5, four and a half lim-2, and a disintegrin and metalloprotease-9 expression in osteoblasts. Growth Horm. IGF Res. 2006, 16, 49–56. [Google Scholar] [CrossRef][Green Version]
- Rudnicki, M.A.; Jaenisch, R. The MyoD family of transcription factors and skeletal myogenesis. BioEssays 1995, 17, 203–209. [Google Scholar] [CrossRef]
- Sun, L.; Bai, M.; Xiang, L.; Zhang, G.; Ma, W.; Jiang, H. Comparative transcriptome profiling of longissimus muscle tissues from Qianhua Mutton Merino and Small Tail Han sheep. Sci. Rep. 2016, 6, 33586. [Google Scholar] [CrossRef] [PubMed]
- Przysucha, T.; Grodzki, H.; Gołebiewski, M.; Slósarz, J.; Piotrowski, T. Evaluation of the performance of Scottish Highland beef cattle in Poland. Med. Weter. 2013, 69, 252–254. [Google Scholar]
- Chulayo, A.; Muchenje, V. Effect of pre-slaughter conditions on physico-chemical characteristics of mutton from three sheep breeds slaughtered at a smallholder rural abattoir. S. Afr. J. Anim. Sci. 2014, 43, 64. [Google Scholar] [CrossRef]
- Beccavin, C.; Chevalier, B.; Cogburn, L.A.; Simon, J.; Duclos, M.J. Insulin-like growth factors and body growth in chickens divergently selected for high or low growth rate. J. Endocrinol. 2001, 168, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Schreurs, N.; Johnson, P.; Corner-Thomas, R.; Agnew, M.; Silcock, P.; Eyres, G.; Maclennan, G.; Realini, C. Carcass characteristics and meat quality of commercial lambs reared in different forage systems. Livest. Sci. 2020, 232, 103908. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Ghosh, S.; Chan, C.-K.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Loehr, J.; Carey, J.; Ylönen, H.; Suhonen, J. Coat darkness is associated with social dominance and mating behaviour in a mountain sheep hybrid lineage. Anim. Behav. 2008, 76, 1545–1553. [Google Scholar] [CrossRef]
- Lahnalampi, M.; Heinäniemi, M.; Sinkkonen, L.; Wabitsch, M.; Carlberg, C. Time-Resolved Expression Profiling of the Nuclear Receptor Superfamily in Human Adipogenesis. PLoS ONE 2010, 5, e12991. [Google Scholar] [CrossRef] [PubMed]
- Acunzo, J.; Katsogiannou, M.; Rocchi, P. Small heat shock proteins HSP27 (HspB1), αB-crystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. Int. J. Biochem. Cell Biol. 2012, 44, 1622–1631. [Google Scholar] [CrossRef]
- Gonçalves, D.A.; Silveira, W.A.; Manfredi, L.H.; Graça, F.A.; Armani, A.; Bertaggia, E.; O´neill, B.T.; Lautherbach, N.; Machado, J.; Nogara, L.; et al. Insulin/IGF1 signalling mediates the effects of β2-adrenergic agonist on muscle proteostasis and growth. J. Cachexia Sarcopenia Muscle 2019, 10, 455–475. [Google Scholar] [CrossRef] [PubMed]
- Macías-Gómez, N.M.; Hernández-Terrones, M.C.; Ramírez-Guerrero, A.A.; Leal-Ugarte, E.; Gutiérrez-Angulo, M.; Peregrina-Sandoval, J. ADIPOQ rs2241766 SNP as protective marker against DIBC development in Mexican population. PLoS ONE 2019, 14, e0214080. [Google Scholar] [CrossRef]
- Perrot, G.; Colin-Pierre, C.; Ramont, L.; Proult, I.; Garbar, C.; Bardey, V.; Brézillon, S. Decreased expression of GPC1 in human skin keratinocytes and epidermis during ageing. Exp. Gerontol. 2019, 126, 110693. [Google Scholar] [CrossRef]
- Amann, R.; Wyder, S.; Slavotinek, A.M.; Trueb, B. The FgfrL1 receptor is required for development of slow muscle fibers. Dev. Biol. 2014, 394, 228–241. [Google Scholar] [CrossRef]
- Heanue, T.A.; Reshef, R.; Davis, R.J.; Mardon, G.; Oliver, G.; Tomarev, S.; Lassar, A.B.; Tabin, C.J. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999, 13, 3231–3243. [Google Scholar] [CrossRef]
- Liu, R.; Kang, X.; Xu, L.; Yuan, W.; Dong, Y. The Study on the Function of MYL2 Gene in Muscle Growth; Chinese Academy of Agricultural Sciences (CAAS), Scientech Documentation and Information Centre: Beijing, China, 2018. [Google Scholar]
- Lee, S. Oxidative Stress in Muscle Progenitor Cell Function; Emory University: Atlanta, GA, USA, 2008. [Google Scholar]
- Joo, S.; Kauffman, R.; Kim, B.; Park, G. The relationship of sarcoplasmic and myofibrillar protein solubility to colour and water-holding capacity in porcine longissimus muscle. Meat Sci. 1999, 52, 291–297. [Google Scholar] [CrossRef]
- Cross, H.R.; Smith, G.C.; Carpenter, Z.L. Palatability of individual muscles from ovine leg steaks as related to chemical and histological traits. J. Food Sci. 2010, 37, 282–285. [Google Scholar] [CrossRef]
- Coles, C.A.; Wadeson, J.; Knight, M.I.; Café, L.M.; Johns, W.H.; White, J.D.; Greenwood, P.L.; McDonagh, M.B. A disintegrin and metalloprotease-12 is type I myofiber specific in and cattle. J. Anim. Sci. 2014, 92, 1473–1483. [Google Scholar] [CrossRef]
- Ju, J.; Mittal, G. Relationships of physical properties of fat-substitutes, cooking methods and fat levels with quality of ground beef patties. J. Food Process. Preserv. 2000, 24, 125–142. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, J.; Wang, X.; Song, Y.; Liu, T.; Cheng, S.; Zhang, Q. Excavation of Genes Related to the Mining of Growth, Development, and Meat Quality of Two Crossbred Sheep Populations Based on Comparative Transcriptomes. Animals 2021, 11, 1492. https://doi.org/10.3390/ani11061492
Shi J, Wang X, Song Y, Liu T, Cheng S, Zhang Q. Excavation of Genes Related to the Mining of Growth, Development, and Meat Quality of Two Crossbred Sheep Populations Based on Comparative Transcriptomes. Animals. 2021; 11(6):1492. https://doi.org/10.3390/ani11061492
Chicago/Turabian StyleShi, Jinping, Xueying Wang, Yali Song, Ting Liu, Shuru Cheng, and Quanwei Zhang. 2021. "Excavation of Genes Related to the Mining of Growth, Development, and Meat Quality of Two Crossbred Sheep Populations Based on Comparative Transcriptomes" Animals 11, no. 6: 1492. https://doi.org/10.3390/ani11061492
APA StyleShi, J., Wang, X., Song, Y., Liu, T., Cheng, S., & Zhang, Q. (2021). Excavation of Genes Related to the Mining of Growth, Development, and Meat Quality of Two Crossbred Sheep Populations Based on Comparative Transcriptomes. Animals, 11(6), 1492. https://doi.org/10.3390/ani11061492





