Genome-Wide Analyses Identifies Known and New Markers Responsible of Chicken Plumage Color
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Samples, Genotyping and Quality Control
2.2. Genome-Wide Analyses
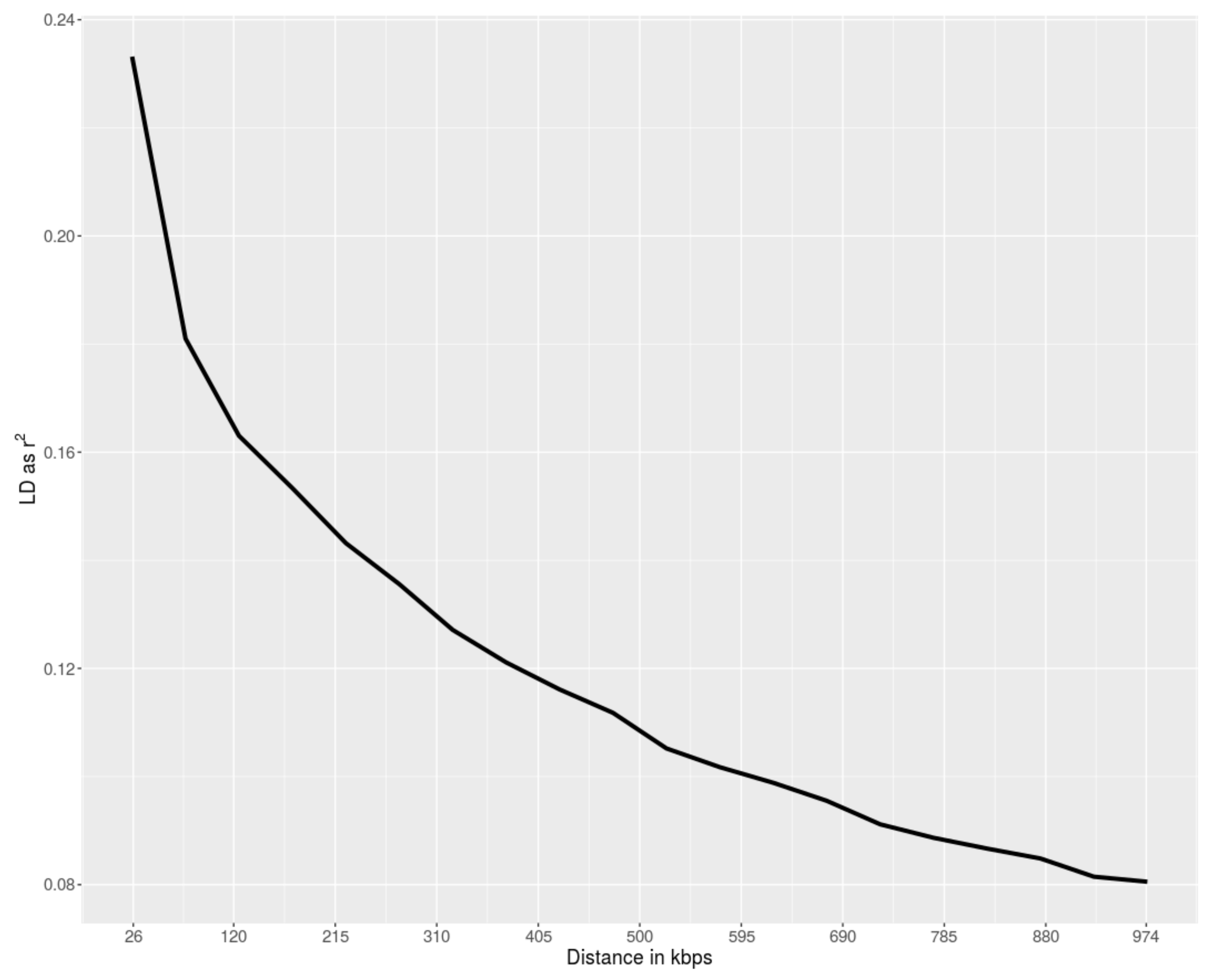
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Leroy, G.; Besbes, B.; Boettcher, P.; Hoffmann, I.; Capitan, A.; Baumung, R. Rare phenotypes in domestic animals: Unique resources for multiple applications. Anim. Genet. 2016, 47, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Kijas, J.W.; Serrano, M.; McCulloch, R.; Li, Y.; Salces Ortiz, J.; Calvo, J.H.; Pérez-Guzmán, M.D. The International Sheep Genomics Consortium. Genome wide association for a dominant pigmentation gene in sheep. J. Anim. Breed. Genet. 2013, 130, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Schiavo, G.; Bertolini, F.; Utzeri, V.J.; Ribani, A.; Geraci, C.; Santoro, L.; Ovilo, C.; Fernandez, A.I.; Gallo, M.; Fontanesi, L. Taking advantage from phenotype variability in a local animal genetic resource: Identification of genomic regions associated with the hairless phenotype in Casertana pigs. Anim. Genet. 2018, 49, 321–325. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Sottile, G.; Sutera, A.M.; Di Gerlando, R.; Tolone, M.; Moscarelli, A.; Sardina, M.T.; Portolano, B. Genome-wide association study reveals the locus responsible for microtia in Valle del Belice sheep breed. Anim. Genet. 2018, 49, 636–640. [Google Scholar] [CrossRef]
- Zanetti, E.; De Marchi, M.; Dalvit, C.; Cassandro, M. Genetic characterization of local Italian breeds of chickens undergoing in situ conservation. Poult. Sci. 2010, 89, 420–427. [Google Scholar] [CrossRef]
- De Marchi, M.; Dalvit, C.; Targhetta, C.; Cassandro, M. Assessing genetic variability in two ancient chicken breeds of Padova area. Ital. J. Anim. Sci. 2005, 4 (Suppl. 3), 151–153. [Google Scholar] [CrossRef]
- Viale, E.; Zanetti, E.; Özdemir, D.; Broccanello, C.; Dalmasso, A.; De Marchi, M.; Cassandro, M. Development and validation of a novel SNP panel for the genetic characterization of Italian chicken breeds by next-generation sequencing discovery and array genotyping. Poult. Sci. 2017, 96, 3858–3866. [Google Scholar] [CrossRef]
- Bai, H.; Sun, Y.; Liu, N.; Xue, F.; Li, Y.; Xu, S.; Ye, J.; Zhang, L.; Chen, Y.; Chen, J. Single SNP-and pathway-based genome-wide association studies for beak deformity in chickens using high-density 600K SNP arrays. BMC Genom. 2018, 19, 501. [Google Scholar] [CrossRef]
- Wang, Q.; Pi, J.; Shen, J.; Pan, A.; Qu, L. Genome-wide association study confirms that the chromosome Z harbours a region responsible for rumplessness in Hongshan chickens. Anim. Genet. 2018, 49, 326–328. [Google Scholar] [CrossRef]
- Yang, L.; Du, X.; Wei, S.; Gu, L.; Li, N.; Gong, Y.; Li, S. Genome-wide association analysis identifies potential regulatory genes for eumelanin pigmentation in chicken plumage. Anim. Genet. 2017, 48, 611–614. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, J.R.; Armengol, L.; Sole, X.; Guino, E.; Mercader, J.M.; Estivill, X.; Moreno, V. SNPASSOC: An R package to perform whole genome association studies. Bioinformatics 2007, 23, 654–655. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D. qqman: An R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv 2014, 2, 005165. [Google Scholar]
- VanLiere, J.M.; Rosenberg, N.A. Mathematical properties of the r2 measure of linkage disequilibrium. Theor. Popul. Biol. 2008, 74, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, S.; Ben Jemaa, S.; Sottile, G.; Casu, S.; Portolano, B.; Ciani, E.; Pilla, F. Combined approaches to identify genomic regions involved in phenotypic differentiation between low divergent breeds: Application in Sardinian sheep populations. J. Anim. Breed. Genet. 2019, 136, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wang, P.; Fu, W.; Dong, T.; Qi, C.; Liu, L.; Guo, G.; Li, C.; Ciu, X.; Zhang, S.; et al. Genome-wide association study for pigmentation traits in Chinese Holstein population. Anim. Genet. 2014, 45, 740–744. [Google Scholar] [CrossRef]
- Edea, Z.; Dadi, H.; Dessie, T.; Kim, I.H.; Kim, K.S. Association of MITF loci with coat color spotting patterns in Ethiopian cattle. Genes Genom. 2017, 39, 285–293. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Sottile, G.; Sardina, M.T.; Sutera, A.M.; Tolone, M.; Di Gerlando, R.; Portolano, B. A combined genome-wide approach identifies a new potential candidate marker associated with the coat color sidedness in cattle. Livest. Sci. 2019, 225, 91–95. [Google Scholar] [CrossRef]
- Muniz, M.M.M.; Caetano, A.R.; McManus, C.; Cavalcanti, L.C.G.; Façanha, D.A.E.; Leite, J.H.G.M.; Facò, O.; Paiva, S.R. Application of genomic data to assist a community- based breeding program: A preliminary study of coat color genetics in Morada Nova sheep. Livest. Sci. 2016, 190, 89–93. [Google Scholar] [CrossRef]
- Kumar, C.; Song, S.; Dewani, P.; Kumar, M.; Parkash, O.; Ma, Y.; Jiang, L. Population structure, genetic diversity and selection signatures within seven indigenous Pakistani goat populations. Anim. Genet. 2018, 49, 592–604. [Google Scholar] [CrossRef]
- Nazari-Ghadikolaei, A.; Mehrabani-Yeganeh, H.; Miarei-Aashtiani, S.R.; Staiger, E.A.; Rashidi, A.; Huson, H.J. Genome-wide association studies identify candidate genes for coat color and Mohair Traits in the Iranian Markhoz Goat. Front. Genet. 2018, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.M.; Coville, J.L.; Coquerelle, G.; Gourichon, D.; Oulmouden, A.; Tixier-Boichard, M. Complete association between a retroviral insertion in the tyrosinase gene and the recessive white mutation in chickens. BMC Genom. 2006, 7, 19. [Google Scholar]
- Liu, W.B.; Chen, S.R.; Zheng, J.X.; Qu, L.J.; Xu, G.Y.; Yang, N. Developmental phenotypic-genotypic associations of tyrosinase and melanocortin 1 receptor genes with changing profiles in chicken plumage pigmentation. Poult. Sci. 2010, 89, 1110–1114. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, F.; Cao, J.; Liu, X. Skin transcriptome profiles associated with skin color in chickens. PLoS ONE 2015, 10, e0127301. [Google Scholar] [CrossRef]
- Beleza, S.; Johnson, N.A.; Candille, S.I.; Absher, D.M.; Coram, M.A.; Lopes, J.; Campos, J.; Araùjo, I.I.; Anderson, T.M.; Vilhjàlmsson, B.J.; et al. Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet. 2013, 9, e1003372. [Google Scholar] [CrossRef]
- Lloyd-Jones, L.R.; Robinson, M.R.; Moser, G.; Zeng, J.; Beleza, S.; Barsh, G.S.; Tang, H.; Visscher, P.M. Inference on the genetic basis of eye and skin color in an admixed population via Bayesian linear mixed models. Genetics 2017, 206, 1113–1126. [Google Scholar] [CrossRef]
- Lona-Durazo, F.; Hernandez-Pacheco, N.; Fan, S.; Zhang, T.; Choi, J.; Kovacs, M.A.; Loftus, S.K.; Le, P.; Edwards, M.; Fortes-Lima, C.A.; et al. Meta-analysis of GWA studies provides new insights on the genetic architecture of skin pigmentation in recently admixed populations. BMC Genet. 2019, 20, 59. [Google Scholar] [CrossRef]
- Miao, Y.; Soudy, F.; Xu, Z.; Liao, M.; Zhao, S.; Li, X. Candidate Gene Identification of Feed Efficiency and Coat Color Traits in a C57BL/6J× Kunming F2 Mice Population Using Genome-Wide Association Study. BioMed Res. Int. 2017, 2017. [Google Scholar] [CrossRef]
- Pfeffer, S.R. Rab GTPases: Specifying and deciphering organelle identity and function. Trends Cell Biol. 2001, 11, 487–491. [Google Scholar] [CrossRef]
- Wasmeier, C.; Romao, M.; Plowright, L.; Bennett, D.C.; Raposo, G.; Seabra, M.C. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol. 2006, 175, 271–281. [Google Scholar] [CrossRef]
- Brooks, B.P.; Larson, D.M.; Chan, C.C.; Kjellstrom, S.; Smith, R.S.; Crawford, M.A.; Lamoreux, L.; Huizing, M.; Hess, R.; Jiao, X. Analysis of ocular hypopigmentation in Rab38cht/cht mice. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3905–3913. [Google Scholar] [CrossRef] [PubMed]
- Kumano, K.; Masuda, S.; Sata, M.; Saito, T.; Lee, S.Y.; Sakata-Yanagimoto, M.; Tomita, T.; Iwatsubo, T.; Natsugari, H.; Kurokawa, M.; et al. Both Notch1 and Notch2 contribute to the regulation of melanocyte homeostasis. Pigment. Cell Melanoma Res. 2008, 21, 70–78. [Google Scholar] [CrossRef]
- Park, M.N.; Choi, J.A.; Lee, K.T.; Lee, H.J.; Choi, B.H.; Kim, H.; Kim, T.H.; Cho, S.; Lee, T. Genome-wide association study of chicken plumage pigmentation. Asian-Aust. J. Anim. Sci. 2013, 26, 1523. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.M.; Nelson, R.M. Characterization of genetic diversity and gene mapping in two Swedish local chicken breeds. Front. Genet. 2015, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Shah, T.M.; Patel, N.V.; Patel, A.B.; Upadhyay, M.R.; Mohapatra, A.; Singh, K.M.; Deshpande, S.D.; Joshi, C.G. A genome-wide approach to screen for genetic variants in broilers (Gallus gallus) with divergent feed conversion ratio. Mol. Genet. Genom. 2016, 291, 1715–1725. [Google Scholar] [CrossRef]
- Lee, J.; Karnuah, A.B.; Rekaya, R.; Anthony, N.B.; Aggrey, S.E. Transcriptomic analysis to elucidate the molecular mechanisms that underlie feed efficiency in meat-type chickens. Mol. Genet. Genom. 2015, 290, 1673–1682. [Google Scholar] [CrossRef]
- Lyu, S.J.; Tian, Y.D.; Wang, S.H.; Han, R.L.; Mei, X.X.; Kang, X.T. A novel 2-bp indel within Krüppel-like factor 15 gene (KLF15) and its associations with chicken growth and carcass traits. Brit. Poult. Sci. 2014, 55, 427–434. [Google Scholar] [CrossRef]


| Nearest Gene | ||||||
| GGA | SNP | Position (bp) | p-Value | FST | Name | Distance (kb) |
| 1 | AX-75371751 | 184995531 | 5.45e-11 | 0.745 | MAML2 | 2.01 |
| 1 | AX-75373909 | 185836576 | 5.45e-11 | 0.773 | LOC107052349 | 2.90 |
| 1 | AX-75374539 | 186058014 | 5.45e-11 | 0.858 | CCDC67 | 29.43 |
| 1 | AX-75375587 | 186464423 | 5.45e-11 | 0.858 | FAT3 | Within |
| 1 | AX-75376255 | 186722445 | 5.45e-11 | 0.886 | ||
| 1 | AX-75376262 | 186735600 | 5.45e-11 | 0.886 | ||
| 1 | AX-75378645 | 187660456 | 9.01e-13 | 0.809 | NAALAD2 | Within |
| 1 | AX-75378836 | 187723578 | 9.01e-13 | 0.809 | FOLH1 | 2.90 |
| 1 | AX-75378888 | 187743605 | 9.01e-13 | 0.809 | FOLH1 | 22.92 |
| 1 | AX-75379333 | 187911192 | 9.01e-13 | 0.809 | NOX4 | 8.02 |
| 1 | AX-75379334 | 187911433 | 9.01e-13 | 0.809 | NOX4 | 8.26 |
| 1 | AX-75379450 | 187960805 | 9.01e-13 | 0.809 | TYR | Within |
| 1 | AX-77278759 | 188025840 | 9.01e-13 | 0.809 | GRM5 | Within |
| 1 | AX-75379693 | 188066880 | 9.01e-13 | 0.809 | ||
| 1 | AX-75379724 | 188079273 | 9.01e-13 | 0.809 | ||
| 1 | AX-75379753 | 188089989 | 9.01e-13 | 0.809 | ||
| 1 | AX-75379761 | 188093458 | 9.01e-13 | 0.809 | ||
| 1 | AX-75379775 | 188096972 | 9.01e-13 | 0.809 | ||
| 1 | AX-75379792 | 188102761 | 9.01e-13 | 0.809 | ||
| 1 | AX-75379800 | 188106002 | 9.01e-13 | 0.809 | ||
| 1 | AX-75379813 | 188112765 | 9.01e-13 | 0.809 | ||
| 1 | AX-75380172 | 188238879 | 9.01e-13 | 0.809 | ||
| 1 | AX-75380766 | 188476552 | 9.01e-13 | 0.809 | RAB38 | 88.89 |
| 1 | AX-75380808 | 188490865 | 9.01e-13 | 0.809 | RAB38 | 103.21 |
| 1 | AX-80852333 | 188493037 | 9.01e-13 | 0.809 | RAB38 | 105.38 |
| 1 | AX-75380927 | 188538625 | 9.01e-13 | 0.809 | TMEM135 | 61.51 |
| 1 | AX-75380931 | 188540546 | 9.01e-13 | 0.809 | TMEM135 | 59.59 |
| 3 | AX-76506116 | 55929533 | 5.45e-11 | 0.745 | HBS1L | 9.40 |
| 3 | AX-76506117 | 55930178 | 5.45e-11 | 0.745 | HBS1L | 9.40 |
| 8 | AX-77109355 | 4012906 | 2e-10 | 0.757 | CRIP1 | 7.01 |
| 8 | AX-77109358 | 4014014 | 2e-10 | 0.757 | CRIP1 | 8.12 |
| 8 | AX-77109696 | 4164384 | 2e-10 | 0.757 | SEC22B | Within |
| 8 | AX-77109700 | 4167984 | 2e-10 | 0.757 | ||
| 8 | AX-77109855 | 4230320 | 2e-10 | 0.757 | NOTCH2 | Within |
| 8 | AX-77109898 | 4249450 | 2e-10 | 0.757 | ||
| 12 | AX-75680106 | 10597665 | 2e-10 | 0.755 | KLF15 | 37.98 |
| 12 | AX-75680164 | 10627473 | 2e-10 | 0.755 | KLF15 | 8.17 |
| 12 | AX-75680170 | 10629579 | 2e-10 | 0.755 | KLF15 | 6.07 |
| 21 | AX-76239008 | 2640299 | 2e-10 | 0.757 | C21H1ORF159 | 0.53 |
| 21 | AX-76239099 | 2657895 | 2e-10 | 0.757 | C21H1ORF159 | 1.85 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mastrangelo, S.; Cendron, F.; Sottile, G.; Niero, G.; Portolano, B.; Biscarini, F.; Cassandro, M. Genome-Wide Analyses Identifies Known and New Markers Responsible of Chicken Plumage Color. Animals 2020, 10, 493. https://doi.org/10.3390/ani10030493
Mastrangelo S, Cendron F, Sottile G, Niero G, Portolano B, Biscarini F, Cassandro M. Genome-Wide Analyses Identifies Known and New Markers Responsible of Chicken Plumage Color. Animals. 2020; 10(3):493. https://doi.org/10.3390/ani10030493
Chicago/Turabian StyleMastrangelo, Salvatore, Filippo Cendron, Gianluca Sottile, Giovanni Niero, Baldassare Portolano, Filippo Biscarini, and Martino Cassandro. 2020. "Genome-Wide Analyses Identifies Known and New Markers Responsible of Chicken Plumage Color" Animals 10, no. 3: 493. https://doi.org/10.3390/ani10030493
APA StyleMastrangelo, S., Cendron, F., Sottile, G., Niero, G., Portolano, B., Biscarini, F., & Cassandro, M. (2020). Genome-Wide Analyses Identifies Known and New Markers Responsible of Chicken Plumage Color. Animals, 10(3), 493. https://doi.org/10.3390/ani10030493






