Outcome of Different Sequencing and Assembly Approaches on the Detection of Plasmids and Localization of Antimicrobial Resistance Genes in Commensal Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Bacterial Isolates
2.1.1. Antimicrobial Susceptibility Testing
2.1.2. PFGE Profiling and Plasmid Prediction
2.1.3. Gene Prediction with PCR
2.1.4. In Vitro Filter Mating Experiments
2.2. Genomic DNA Extraction for Whole-Genome Sequencing (WGS)
2.3. Whole-Genome Sequencing
2.4. De Novo Assembly Strategies and Genome Characterization
2.5. Accession Numbers
3. Results and Discussion
3.1. Impact of Different Long- and Short-Read Sequencing Approaches
3.2. Small Plasmids Are Difficult to Detect
3.3. Hybrid Assembly Allows a Deep Insight into the Plasmid Structure
3.4. Common Mistakes in Resistance Gene Detection and Phenotype Evaluation
3.5. Evaluation of the Phenotype
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karp, B.E.; Tate, H.; Plumblee, J.R.; Dessai, U.; Whichard, J.M.; Thacker, E.L.; Hale, K.R.; Wilson, W.; Friedman, C.R.; Griffin, P.M.; et al. National Antimicrobial Resistance Monitoring System: Two Decades of Advancing Public Health Through Integrated Surveillance of Antimicrobial Resistance. Foodborne Pathog. Dis. 2017, 14, 545–557. [Google Scholar] [CrossRef]
- Florez-Cuadrado, D.; Moreno, M.A.; Ugarte-Ruíz, M.; Domínguez, L. Antimicrobial Resistance in the Food Chain in the European Union. Adv. Food Nutr. Res. 2018, 86, 115–136. [Google Scholar] [PubMed]
- McArthur, A.G.; Tsang, K.K. Antimicrobial resistance surveillance in the genomic age. Ann. N. Y. Acad. Sci. 2017, 1388, 78–91. [Google Scholar] [CrossRef]
- Schrijver, R.; Stijntjes, M.; Rodríguez-Baño, J.; Tacconelli, E.; Rajendran, N.B.; Voss, A. Review of antimicrobial resistance surveillance programmes in livestock and meat in EU with focus on humans. Clin. Microbiol. Infect. 2018, 24, 577–590. [Google Scholar] [CrossRef]
- Varona, O.M.; Chaintarli, K.; Muller-Pebody, B.; Anjum, M.F.; Eckmanns, T.; Norström, M.; Boone, I.; Tenhagen, B.-A. Monitoring Antimicrobial Resistance and Drug Usage in the Human and Livestock Sector and Foodborne Antimicrobial Resistance in Six European Countries. Infect. Drug Resist. 2020, 13, 957–993. [Google Scholar] [CrossRef] [PubMed]
- Angers-Loustau, A.; Petrillo, M.; Bengtsson-Palme, J.; Berendonk, T.; Blais, B.; Chan, K.-G.; Coque, T.M.; Hammer, P.; Heß, S.; Kagkli, D.M.; et al. The challenges of designing a benchmark strategy for bioinformatics pipelines in the identification of antimicrobial resistance determinants using next generation sequencing technologies. F1000Research 2018, 7. [Google Scholar] [CrossRef]
- EFSA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, e05182. [Google Scholar]
- Abdelbary, M.M.; Senn, L.; Moulin, E.; Prod’Hom, G.; Croxatto, A.; Greub, G.; Blanc, D.S. Evaluating the use of whole-genome sequencing for outbreak investigations in the lack of closely related reference genome. Infect. Genet. Evol. 2018, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. Review on the Applications of Next Generation Sequencing Technologies as Applied to Food-Related Microbiome Studies. Front. Microbiol. 2017, 8, 1829. [Google Scholar] [CrossRef]
- Koren, S.; Phillippy, A.M. One chromosome, one contig: Complete microbial genomes from long-read sequencing and assembly. Curr. Opin. Microbiol. 2015, 23, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Aung, K.T.; Tay, M.Y.; Seow, K.L.G.; Ng, L.C.; Schlundt, J. Extended-spectrum β-lactamase-producing Proteus mirabilis with multidrug resistance isolated from raw chicken in Singapore: Genotypic and phenotypic analysis. J. Glob. Antimicrob. Resist. 2019, 19, 252–254. [Google Scholar] [CrossRef] [PubMed]
- McCombie, W.R.; McPherson, J.D.; Mardis, E.R. Next-Generation Sequencing Technologies. Cold Spring Harb. Perspect. Med. 2019, 9, a036798. [Google Scholar] [CrossRef] [PubMed]
- Besser, J.; Carleton, H.; Gerner-Smidt, P.; Lindsey, R.; Trees, E. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin. Microbiol. Infect. 2018, 24, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Satou, K.; Shiroma, A.; Teruya, K.; Shimoji, M.; Nakano, K.; Juan, A.; Tamotsu, H.; Terabayashi, Y.; Aoyama, M.; Teruya, M.; et al. Complete Genome Sequences of Eight Helicobacter pylori Strains with Different Virulence Factor Genotypes and Methylation Profiles, Isolated from Patients with Diverse Gastrointestinal Diseases on Okinawa Island, Japan, Determined Using PacBio Single-Molecule Real-Time Technology. Genome Announc. 2014, 2, e00286–e00314. [Google Scholar]
- Ellington, M.; Ekelund, O.; Aarestrup, F.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.; Hopkins, K.; et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef]
- De Lannoy, C.; de Ridder, D.; Risse, J. The long reads ahead: De novo genome assembly using the MinION. F1000Research 2017, 6, 1083. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H. Safety profile of the fluoroquinolones: Focus on levofloxacin. Drug Saf. 2010, 33, 353–369. [Google Scholar] [CrossRef]
- Ezelarab, H.A.A.; Abbas, S.H.; Hassan, H.A.; Abuo-Rahma, G.E.-D.A. Recent updates of fluoroquinolones as antibacterial agents. Arch. Pharm. 2018, 351, e1800141. [Google Scholar] [CrossRef]
- Robicsek, A.; Jacoby, G.A.; Hooper, D.C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006, 6, 629–640. [Google Scholar] [CrossRef]
- Hansen, L.H.; Johannesen, E.; Burmølle, M.; Sørensen, A.H.; Sørensen, S.J. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob. Agents Chemother. 2004, 48, 3332–3337. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef]
- Vinué, L.; Corcoran, M.A.; Hooper, D.C.; Jacoby, G.A. Mutations That Enhance the Ciprofloxacin Resistance of Escherichia coli with qnrA1. Antimicrob. Agents Chemother. 2015, 60, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Poeta, P.; Hébraud, M.; Capelo, J.L.; Igrejas, G. Mechanisms of quinolone action and resistance: Where do we stand? J. Med. Microbiol. 2017, 66, 551–559. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Commission implementing decision of 12 november 2013 on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria. Off. J. Eur. Union 2013, L 303, 26–39. [Google Scholar]
- Dortet, L.; Bonnin, R.A.; Pennisi, I.; Gauthier, L.; Jousset, A.B.; Dabos, L.; Furniss, R.C.D.; Mavridou, D.A.I.; Bogaerts, P.; Glupczynski, Y.; et al. Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: The MALDIxin test. J. Antimicrob. Chemother. 2018, 73, 3359–3367. [Google Scholar] [CrossRef] [PubMed]
- PulsNet. Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli Non-O157 (STEC), Salmonella Serotypes, Shigella Sonnei and Shigella Flexneri. 2017. Available online: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf (accessed on 26 June 2018).
- Rodríguez-Martínez, J.M.; Briales, A.; Velasco, C.; Conejo, M.C.; Martínez-Martínez, L.; Pascual, A. Mutational analysis of quinolone resistance in the plasmid-encoded pentapeptide repeat proteins QnrA, QnrB and QnrS. J. Antimicrob. Chemother. 2009, 63, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Guillard, T.; Moret, H.; Brasme, L.; Carlier, A.; Vernet-Garnier, V.; Cambau, E.; De Champs, C. Rapid detection of qnr and qepA plasmid-mediated quinolone resistance genes using real-time PCR. Diagn. Microbiol. Infect. Dis. 2011, 70, 253–259. [Google Scholar] [CrossRef]
- Guerra, B.; Soto, S.M.; Arguüelles, J.M.; Mendoza, M.C. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:-]. Antimicrob. Agents Chemother. 2001, 45, 1305–1308. [Google Scholar] [CrossRef]
- Sambrook, J.; Russel, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 3, p. 2344. [Google Scholar]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, J.; Wang, J.; Butaye, P.; Kelly, P.; Li, M.; Yang, F.; Gong, J.; Yassin, A.K.; Guo, W.; et al. Newly identified colistin resistance genes, mcr-4 and mcr-5, from upper and lower alimentary tract of pigs and poultry in China. PLoS ONE 2018, 13, e0193957. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Seemann, T. Abricate. 2014. Available online: https://github.com/tseemann/abricate (accessed on 4 September 2019).
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Ingle, D.J.; Valcanis, M.; Kuzevski, A.; Tauschek, M.; Inouye, M.; Stinear, T.; Levine, M.M.; Robins-Browne, R.M.; Holt, K.E. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 2016, 2, e000064. [Google Scholar]
- Schwengers, O.; Barth, P.; Falgenhauer, L.; Hain, T.; Chakraborty, T.; Goesmann, A. Platon: Identification and characterization of bacterial plasmid contigs in short-read draft assemblies exploiting protein sequence-based replicon distribution scores. Microb. Genom. 2020, 6, e000398. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Redondo-Salvo, S.; Vielva, L.; De La Cruz, F. MOBscan: Automated Annotation of MOB Relaxases. Methods Mol. Biol. 2020, 2075, 295–308. [Google Scholar] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.J.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Nouws, S.; Bogaerts, B.; Verhaegen, B.; Denayer, S.; Piérard, D.; Marchal, K.; Roosens, N.H.C.; Vanneste, K.; De Keersmaecker, S.C.J. Impact of DNA extraction on whole genome sequencing analysis for characterization and relatedness of Shiga toxin-producing Escherichia coli isolates. Sci. Rep. 2020, 10, 14649. [Google Scholar] [CrossRef]
- Taylor, T.L.; Volkening, J.D.; DeJesus, E.; Simmons, M.; Dimitrov, K.M.; Tillman, G.E.; Suarez, D.L.; Afonso, C.L. Rapid, multiplexed, whole genome and plasmid sequencing of foodborne pathogens using long-read nanopore technology. Sci. Rep. 2019, 9, 16350. [Google Scholar] [CrossRef]
- Oakeson, K.F.; Wagner, J.M.; Rohrwasser, A.; Atkinson-Dunn, R. Whole-Genome Sequencing and Bioinformatic Analysis of Isolates from Foodborne Illness Outbreaks of Campylobacter jejuni and Salmonella enterica. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef]
- De Maio, N.; Shaw, L.P.; Hubbard, A.; George, S.; Sanderson, N.D.; Swann, J.; Wick, R.; AbuOun, M.; Stubberfield, E.; Hoosdally, S.J.; et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 2019, 5, e000294. [Google Scholar] [CrossRef]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Kumar, K.R.; Cowley, M.J.; Davis, R.L. Next-Generation Sequencing and Emerging Technologies. Semin. Thromb. Hemost. 2019, 45, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [PubMed]
- González-Escalona, N.; Allard, M.A.; Brown, E.W.; Sharma, S.; Hoffmann, M. Nanopore sequencing for fast determination of plasmids, phages, virulence markers, and antimicrobial resistance genes in Shiga toxin-producing Escherichia coli. PLoS ONE 2019, 14, e0220494. [Google Scholar] [CrossRef]
- Forde, B.M.; Ben Zakour, N.L.; Stanton-Cook, M.; Phan, M.-D.; Totsika, M.; Peters, K.M.; Chan, K.G.; Schembri, M.A.; Upton, M.; Beatson, S.A. The complete genome sequence of Escherichia coli EC958: A high quality reference sequence for the globally disseminated multidrug resistant E. coli O25b:H4-ST131 clone. PLoS ONE 2014, 9, e104400. [Google Scholar]
- Dolejska, M.; Papagiannitsis, C.C. Plasmid-mediated resistance is going wild. Plasmid 2018, 99, 99–111. [Google Scholar] [CrossRef]
- Sethuvel, D.P.M.; Anandan, S.; Ragupathi, N.K.D.; Gajendiran, R.; Kuroda, M.; Shibayama, K.; Veeraraghavan, B. IncFII plasmid carrying antimicrobial resistance genes in Shigella flexneri: Vehicle for dissemination. J. Glob. Antimicrob. Resist. 2019, 16, 215–219. [Google Scholar] [CrossRef]
- Madec, J.Y.; Haenni, M. Antimicrobial resistance plasmid reservoir in food and food-producing animals. Plasmid 2018, 99, 72–81. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; A Wagenaar, J.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Laczny, C.C.; Galata, V.; Plum, A.; Posch, A.E.; Keller, A. Assessing the heterogeneity of in silico plasmid predictions based on whole-genome-sequenced clinical isolates. Brief. Bioinform. 2019, 20, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Villa, L.; Minoia, M.; Guardabassi, L.; Carattoli, A. Complete sequences of IncHI1 plasmids carrying blaCTX-M-1 and qnrS1 in equine Escherichia coli provide new insights into plasmid evolution. J. Antimicrob. Chemother. 2014, 69, 2388–2393. [Google Scholar] [CrossRef] [PubMed]
- Dupouy, V.; Abdelli, M.; Moyano, G.; Arpaillange, N.; Bibbal, D.; Cadiergues, M.-C.; Lopez-Pulin, D.; Sayah-Jeanne, S.; De Gunzburg, J.; Saint-Lu, N.; et al. Prevalence of Beta-Lactam and Quinolone/Fluoroquinolone Resistance in Enterobacteriaceae from Dogs in France and Spain-Characterization of ESBL/pAmpC Isolates, Genes, and Conjugative Plasmids. Front. Vet. Sci. 2019, 6, 279. [Google Scholar] [CrossRef] [PubMed]
- Liakopoulos, A.; Van Der Goot, J.; Bossers, A.; Betts, J.; Brouwer, M.S.M.; Kant, A.; Smith, H.; Ceccarelli, D.; Mevius, D. Genomic and functional characterisation of IncX3 plasmids encoding blaSHV-12 in Escherichia coli from human and animal origin. Sci. Rep. 2018, 8, 7674. [Google Scholar] [CrossRef]
- Walther-Rasmussen, J.; Hoiby, N. OXA-type carbapenemases. J. Antimicrob. Chemother. 2006, 57, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Long, S.W.; McDermott, P.F.; Olsen, R.J.; Olson, R.; Stevens, R.L.; Tyson, G.H.; Zhao, S.; Davis, J.J. Using Machine Learning To Predict Antimicrobial MICs and Associated Genomic Features for Nontyphoidal Salmonella. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Liu, Z.; Deng, D.; Lu, H.; Sun, J.; Lv, L.; Li, S.; Peng, G.; Ma, X.; Li, J.; Li, Z.; et al. Evaluation of Machine Learning Models for Predicting Antimicrobial Resistance of Actinobacillus pleuropneumoniae from Whole Genome Sequences. Front. Microbiol. 2020, 11, 48. [Google Scholar] [CrossRef]
- Collineau, L.; Boerlin, P.; Carson, C.A.; Chapman, B.; Fazil, A.; Hetman, B.; McEwen, S.A.; Parmley, E.J.; Reid-Smith, R.J.; Taboada, E.N.; et al. Integrating Whole-Genome Sequencing Data Into Quantitative Risk Assessment of Foodborne Antimicrobial Resistance: A Review of Opportunities and Challenges. Front. Microbiol. 2019, 10, 1107. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Satola, S.W.; Read, T.D. Genome-Based Prediction of Bacterial Antibiotic Resistance. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Pontes, D.S.; De Araujo, R.S.A.; Dantas, N.; Scotti, L.; Scotti, M.T.; De Moura, R.O.; Mendonca-Junior, F.J.B. Genetic Mechanisms of Antibiotic Resistance and the Role of Antibiotic Adjuvants. Curr. Top. Med. Chem. 2018, 18, 42–74. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Horinouchi, T.; Furusawa, C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 2014, 5, 5792. [Google Scholar] [CrossRef] [PubMed]
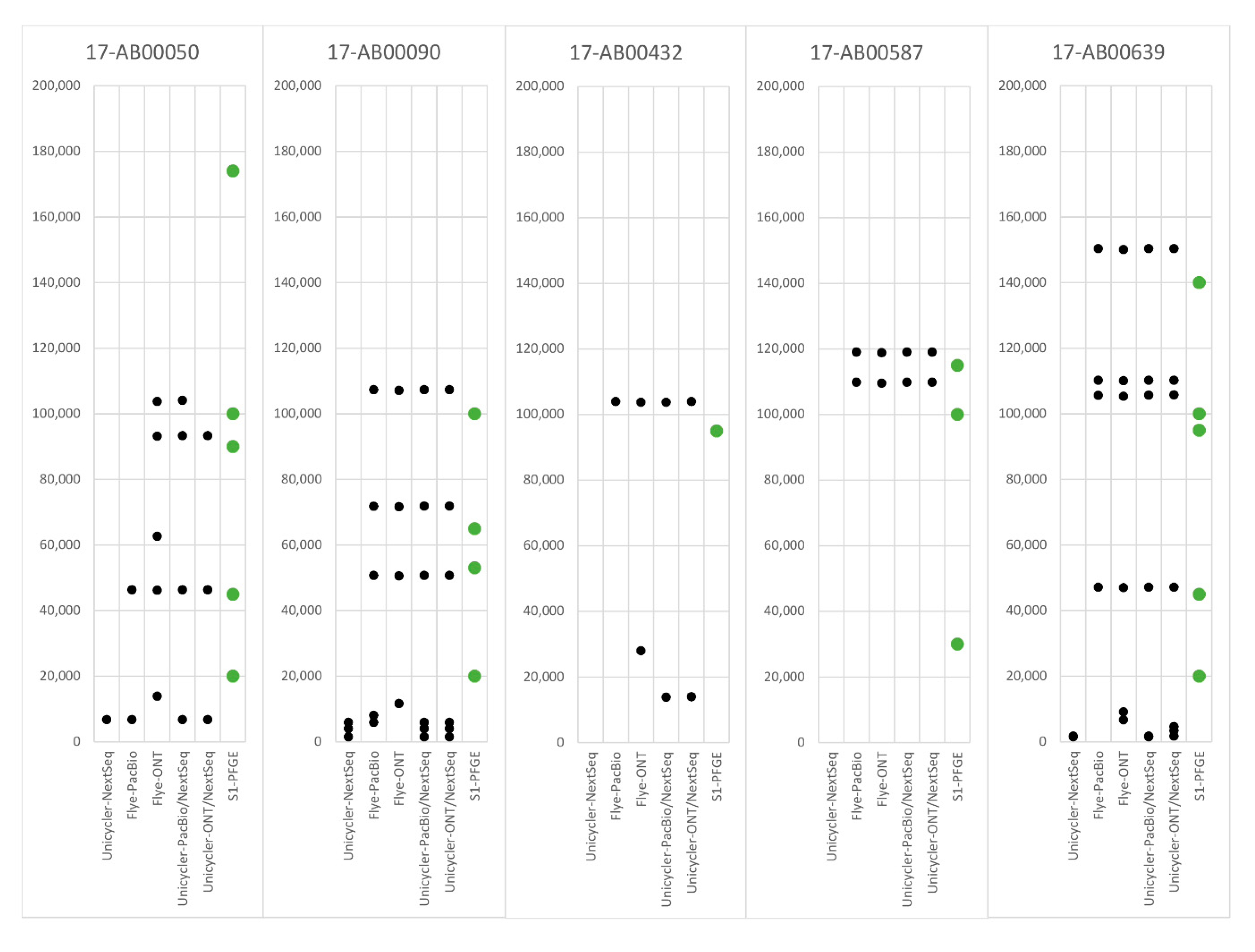
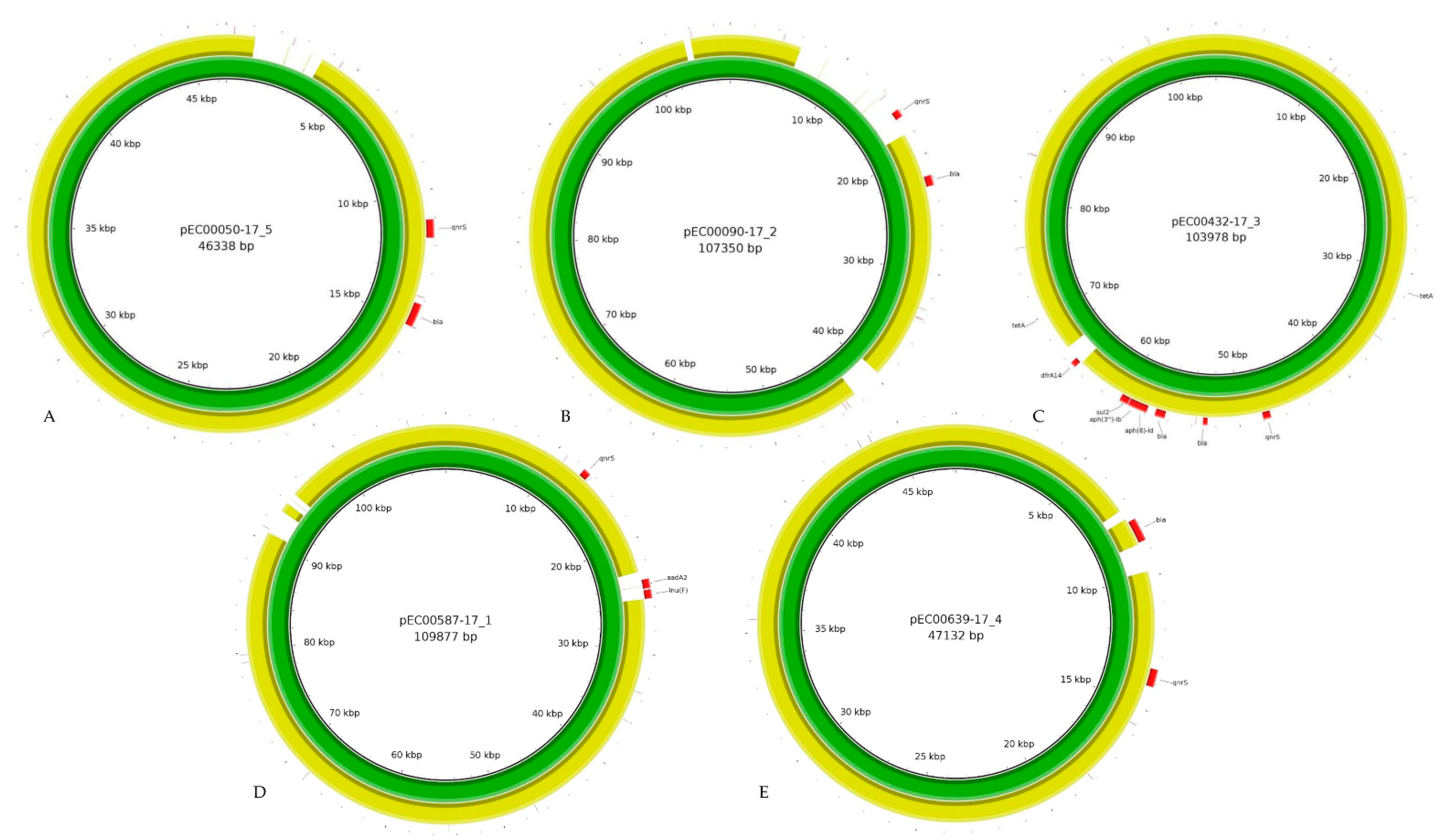
| Isolate ID | Matrix | Date of Isolation | AMR Profile | Sizes of Identified Plasmids + |
|---|---|---|---|---|
| 17-AB00050 | cecum, broiler | 22 November 2016 | AMP, CEF, CIP, FOT, GEN, SMX, TAZ | 174 kb; 100 kb; 90 kb; 45 kb *; <20.5 kb |
| 17-AB00090 | feces, turkey | 14 December 2016 | AMP, CIP, NAL, TET | 100 kb *; 65 kb; 53 kb; <20.5 kb |
| 17-AB00432 | cecum, calf | 21 February 2017 | AMP, CEF, CIP, FOT, NAL, TAZ, TET, TMP, SMX | 95 kb * |
| 17-AB00587 | meat, bovine | 23 March 2017 | AMP, CEF, CIP, FOT, TAZ | 115 kb; 100 kb *; 30 kb |
| 17-AB00639 | cecum, pig | 24 April 2017 | AMP, CEF, CIP, FOT, GEN, SMX, TAZ, TMP | 140 kb; 100 kb; 95 kb; 45 kb *; <20.5 kb |
| 17-AB00050 | 17-AB00090 | 17-AB00432 | 17-AB00587 | 17-AB00639 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Contigs (Plasmidal Content Relative to Total Contig Length) | Number of Circular Contigs/All Contigs < 10 kb | Longest Contig [bp] | Number of Contigs (Plasmidal Content Relative to Total Contig Length) | Number of Circular Contigs/Contigs < 10 kb | Longest Contig [bp] | Number of Contigs (Plasmidal Content Relative to Total Contig Length) | Number of Circular Contigs/Contigs < 10 kb | Longest Contig [bp] | Number of Contigs (Plasmidal Content Relative to Total Contig Length) | Number of Circular Contigs/Contigs < 10 kb | Longest Contig [bp] | Number of Contigs (Plasmidal Content Relative to Total Contig Length) | Number of Circular Contigs/Contigs < 10 kb | Longest Contig [bp] | |
| Unicycler-NextSeq | 466 (5.5%) | 2/2 | 341,211 | 186 (4.3%) | 3/3 | 502,527 | 159 (1.6%) | 0/0 | 314,433 | 201 (4.2%) | 0/0 | 485,939 | 183 (7.6%) | 2/2 | 340,946 |
| Flye-PacBio | 13 (4.6%) | 4/1 | 4094,393 | 6 (4.6%) | 6/2 | 5004,742 | 2 (2.1%) | 2/0 | 4,736,227 | 3 (4.4%) | 3/0 | 4,938,765 | 7 (7.9%) | 4/1 | 3,693,252 |
| Flye-ONT | 8 (5.3%) | 7/0 | 5,524,427 | 5 (4.6%) | 5/0 | 4,970,722 | 3 (2.7%) | 3/0 | 4,728,917 | 3 (4.4%) | 3/0 | 4,931,189 | 7 (8.3%) | 7/4 | 4,756,147 |
| Unicycler-PacBio/NextSeq | 27 (4.2%) | 6/2 | 4,341,057 | 7 (4.6%) | 7/3 | 5,004,751 | 3 (2.4%) | 3/0 | 4,736,229 | 3 (4.4%) | 3/0 | 4,938,758 | 13 (8.1%) | 7/2 | 4,763,387 |
| Unicycler-ONT/NextSeq | 22 (4.0%) | 6/2 | 5,533,851 | 7 (4.6%) | 7/3 | 5,004,751 | 3 (2.4%) | 3/0 | 4,736,229 | 3 (4.4%) | 3/0 | 4,938,758 | 9 (8.2%) | 9/2 | 4,763,387 |
| 17-AB00050 | 17-AB00090 | 17-AB00432 | 17-AB00587 | 17-AB00639 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Contig Size [bp] | Plasmid Marker | Other AMR Genes | Contig Size [bp] | lPlasmid Marker | Other AMR Genes | Contig Size [bp] | Plasmid Marker | Other AMR Genes | Contig Size [bp] | Plasmid Marker | Other AMR Genes | Contig Size [bp] | Plasmid Marker | Other AMR Genes | |
| Unicycler-NextSeq | 42,601 | IncX3 | - | 5348 | - | - | 13,373 | - | - | 1762 | - | - | 8821 | - | - |
| Flye-PacBio | 46,338 | IncX3 | blaSHV-12 | 107,341 | IncI1_1_α | blaTEM-1 | 103,789 | IncY | tet(A), blaCTX_M-15, blaTEM-1, aph(3″)-ib, sul2, dfrA14, tet(A), aph(6)-Id | 109,877 | IncI1_1α | aadA2, lnu(F) | 47,133 | IncX1, IncX3 | blaTEM-1 |
| Flye-ONT | 46,207 | IncX3 | blaSHV-12 | 107,104 | IncI1_1_α | blaTEM-1 | 103,779 | IncY | tet(A), blaCTX_M-15, blaTEM-1, aph(3″)-ib, sul2, dfrA14, tet(A), aph(6)-Id | 118,872 | IncI1_1α | aadA2, lnu(F) | 46,996 | IncX1, IncX3 | blaTEM-1 |
| Unicycler-PacBio/NextSeq | 46,338 | IncX3 | blaSHV-12 | 107,350 | IncI1_1_α | blaTEM-1 | 103,978 | IncY | tet(A), blaCTX_M-15, blaTEM-1, aph(3″)-ib, sul2, dfrA14, tet(A) | 109,877 | IncI1_1α | aadA2, lnu(F) | 47,132 | IncX1 | blaTEM-1 |
| Unicycler-ONT/NextSeq | 46,338 | IncX3 | blaSHV-12 | 107,350 | IncI1_1_ α | blaTEM-1 | 103,975 | IncY | tet(A), blaCTX_M-15, blaTEM-1, aph(3″)-ib, sul2, dfrA14, tet(A), aph(6)-Id | 109,876 | IncI1_1α | aadA2, lnu(F) | 47,132 | IncX1 | blaTEM-1 |
| PFGE determined size [bp] | 45,000 | 100,000 | 95,000 | 100,000 | 45,000 | ||||||||||
| in vitro conjugational transfer | no | yes | no | yes | yes | ||||||||||
| 17-AB00050 | ||||
| Class of In Silico Type Phenotype | Subclass of In Silico Type Phenotype | Determined Resistance Gene(s) | Determined Phenotype | Class/Subclass of Determined Phenotype |
| Quinolone | Quinolone | qnrS1 | Ciprofloxacin | Fluoroquinolone |
| β-Lactam | Penicillin, Cephalosporin | blaEC-8, blaSHV-12 | Ampicillin | β-Lactam |
| Cefepime | Cephalosporin | |||
| Cefotaxime | Cephalosporin | |||
| Aminoglycoside | Gentamicin | aac(3)-VIa | Gentamicin | Aminoglycoside |
| Sulfonamide | Sulfonamid | sul1 | Sulphamethoxazole | Sulfonamide |
| Aminoglycoside | Streptomycin | aadA1 | Not within the test panel | Not within the test panel |
| 17-AB00090 | ||||
| Class of In Silico Type Phenotype | Subclass of In Silico Type Phenotype | Determined Resistance Gene(s) | Determined Phenotype | Class/Subclass of Determined Phenotype |
| β-Lactam | Penicillin | blaTEM-1, blaTEM-1*1, blaTEM-135*2 | Ampicillin | β-Lactam |
| β-Lactam | Penicillin, Cephalosporine | blaEC-18 | ||
| Quinolone | Quinolone | qnrS1 | Ciprofloxacin | Fluoroquinolone |
| Nalidixic acid | Quinolone | |||
| Tetracycline | Tetracycline | tet(A) | Tetracycline | Tetracycline |
| 17-AB00432 | ||||
| Class of In Silico Type Phenotype | Subclass of In Silico Type Phenotype | Determined Resistance Gene(s) | Determined Phenotype | Class/Subclass of Determined Phenotype |
| β-Lactam | Penicillin | blaTEM-1 | Ampicillin | β-Lactam |
| β-Lactam | Penicillin, Cephalosporin | blaCTX-M-15, blaEC | Cefepime | Cephalosporin |
| Cefotaxime | Cephalosporin | |||
| Ceftazidime | Cephalosporin | |||
| Aminoglycoside | Kanamycin | aph(3′)-Ia | Not within the test panel | Not within the test panel |
| Quinolone | Quinolone | qnrS1 | Ciprofloxacin | Fluoroquinolone |
| Nalidixic acid | Quinolone | |||
| Aminoglycoside | Streptomycin | aph(3″)-Ib, aph(3″)-Ib* | Not within the test panel | Not within the test panel |
| aph(3′)-Ia, | ||||
| aph(6)-Id, aph(6)-Id* | ||||
| Sulfonamide | Sulfonamide | sul2, sul2* | Sulphamethoxazole | Sulfonamide |
| Tetracycline | Tetracycline | tet(A), tet(A)*3, tet(B) | Tetracycline | Tetracycline |
| 17-AB00587 | ||||
| Class of In Silico Type Phenotype | Subclass of In Silico Type Phenotype | Determined Resistance Gene | Determined Phenotype | Class/Subclass of Determined Phenotype |
| β-Lactam | Cephalosporin | blaCTX-M-1, blaEC-15 | Cefepime | Cephalosporin |
| Cefotaxime | Cephalosporin | |||
| Ceftazidime | Cephalosporin | |||
| Lincosamide | Lincosamide | lnu(F) | Not within the test panel | Not within the test panel |
| Macrolide | Macrolide | mph(A) | ||
| Quinolone | Fluoroquinolone | qnrS1 | Ciprofloxacin | Fluoroquinolone |
| Aminoglycoside | Streptomycin | aadA2 | Not within the test panel | Not within the test panel |
| 17-AB00639 | ||||
| Class of In Silico Type Phenotype | Subclass of In Silico Type Phenotype | Determined Resistance Gene(s) | Determined Phenotype | Class/Subclass of Determined Phenotype |
| β-Lactam | Penicillin | blaTEM-1 | Ampicillin | β-Lactam |
| β-Lactam | Penicillin, Cephalosporin*3 | blaCTX-M-1*4, blaEC, | Cefepime | Cephalosporin |
| blaTEM-1*4, blaTEM-1 | Cefotaxime | Cephalosporine | ||
| Ceftazidime | Cephalosporine | |||
| Aminoglycoside | Gentamicin | aac(3)-Iva | Gentamicin | Aminoglycoside |
| Aminoglycoside | Hygromicin | aph(4)-Ia | Not within the test panel | Not within the test panel |
| Macrolide | Macrolide | mph(A) | ||
| Quinolone | Fluoroquinolone | qnrS1 | Ciprofloxacin | Fluoroquinolon |
| Aminoglycoside | Streptomycin | aph(3″)-Ib, aph(3″)-Ib*4, | Not within the test panel | Not within the test panel |
| aph(6)-Id*5 | ||||
| Sulfonamide | Sulfonamide | sul2 | Sulphamethoxazole | Sulfonamide |
| Diaminopyrimidine | Trimethoprim | dfrA5 | Trimethoprim | Diaminopyrimidine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juraschek, K.; Borowiak, M.; Tausch, S.H.; Malorny, B.; Käsbohrer, A.; Otani, S.; Schwarz, S.; Meemken, D.; Deneke, C.; Hammerl, J.A. Outcome of Different Sequencing and Assembly Approaches on the Detection of Plasmids and Localization of Antimicrobial Resistance Genes in Commensal Escherichia coli. Microorganisms 2021, 9, 598. https://doi.org/10.3390/microorganisms9030598
Juraschek K, Borowiak M, Tausch SH, Malorny B, Käsbohrer A, Otani S, Schwarz S, Meemken D, Deneke C, Hammerl JA. Outcome of Different Sequencing and Assembly Approaches on the Detection of Plasmids and Localization of Antimicrobial Resistance Genes in Commensal Escherichia coli. Microorganisms. 2021; 9(3):598. https://doi.org/10.3390/microorganisms9030598
Chicago/Turabian StyleJuraschek, Katharina, Maria Borowiak, Simon H. Tausch, Burkhard Malorny, Annemarie Käsbohrer, Saria Otani, Stefan Schwarz, Diana Meemken, Carlus Deneke, and Jens Andre Hammerl. 2021. "Outcome of Different Sequencing and Assembly Approaches on the Detection of Plasmids and Localization of Antimicrobial Resistance Genes in Commensal Escherichia coli" Microorganisms 9, no. 3: 598. https://doi.org/10.3390/microorganisms9030598
APA StyleJuraschek, K., Borowiak, M., Tausch, S. H., Malorny, B., Käsbohrer, A., Otani, S., Schwarz, S., Meemken, D., Deneke, C., & Hammerl, J. A. (2021). Outcome of Different Sequencing and Assembly Approaches on the Detection of Plasmids and Localization of Antimicrobial Resistance Genes in Commensal Escherichia coli. Microorganisms, 9(3), 598. https://doi.org/10.3390/microorganisms9030598







