How Microbial Food Web Interactions Shape the Arctic Ocean Bacterial Community Revealed by Size Fractionation Experiments
Abstract
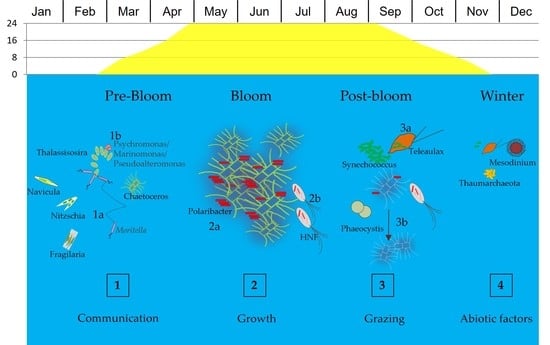
:1. Introduction
2. Materials and Methods
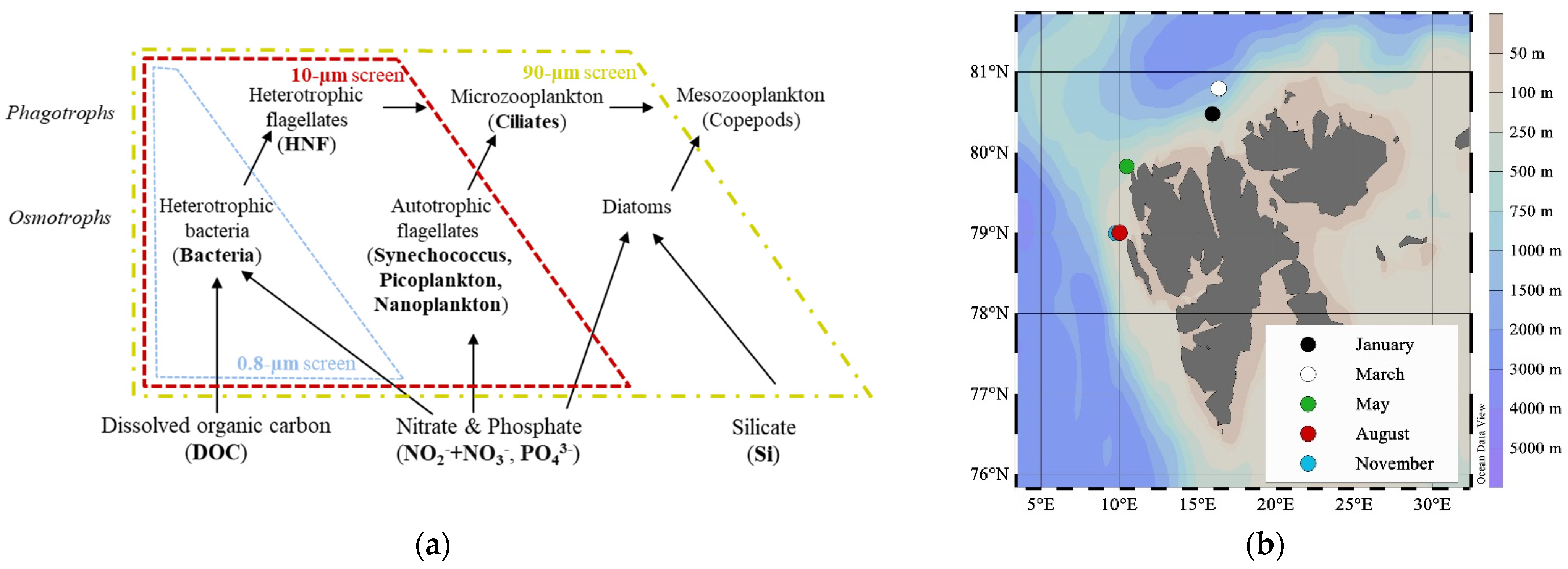
2.1. Study Site
2.2. Experimental Design
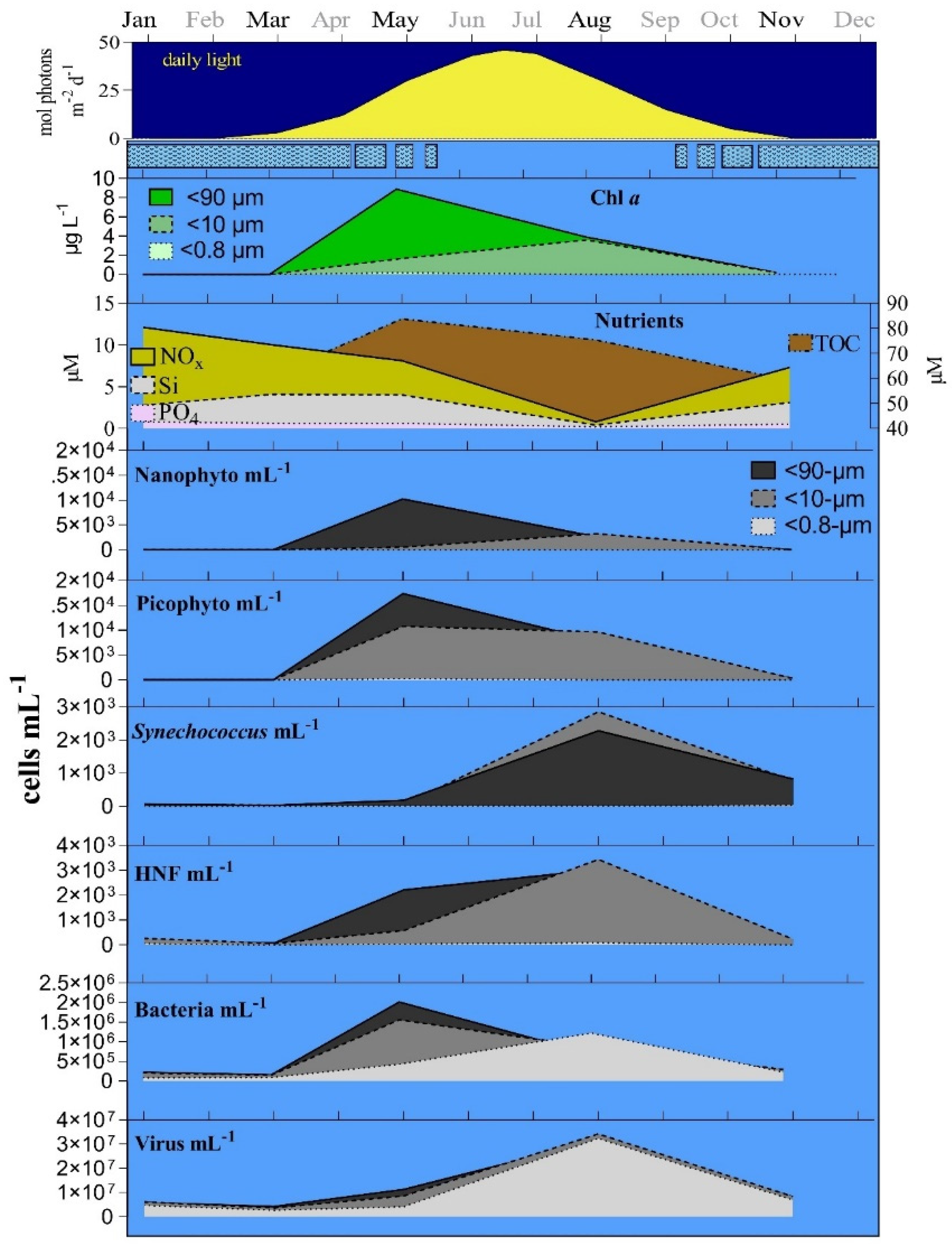
2.3. Environmental Parameters (Nutrients, DOC and Light)
2.4. Chlorophyll a (Chl a)
2.5. Microbial Abundance Measurements Using Flow Cytometry
2.6. Light Microscopy
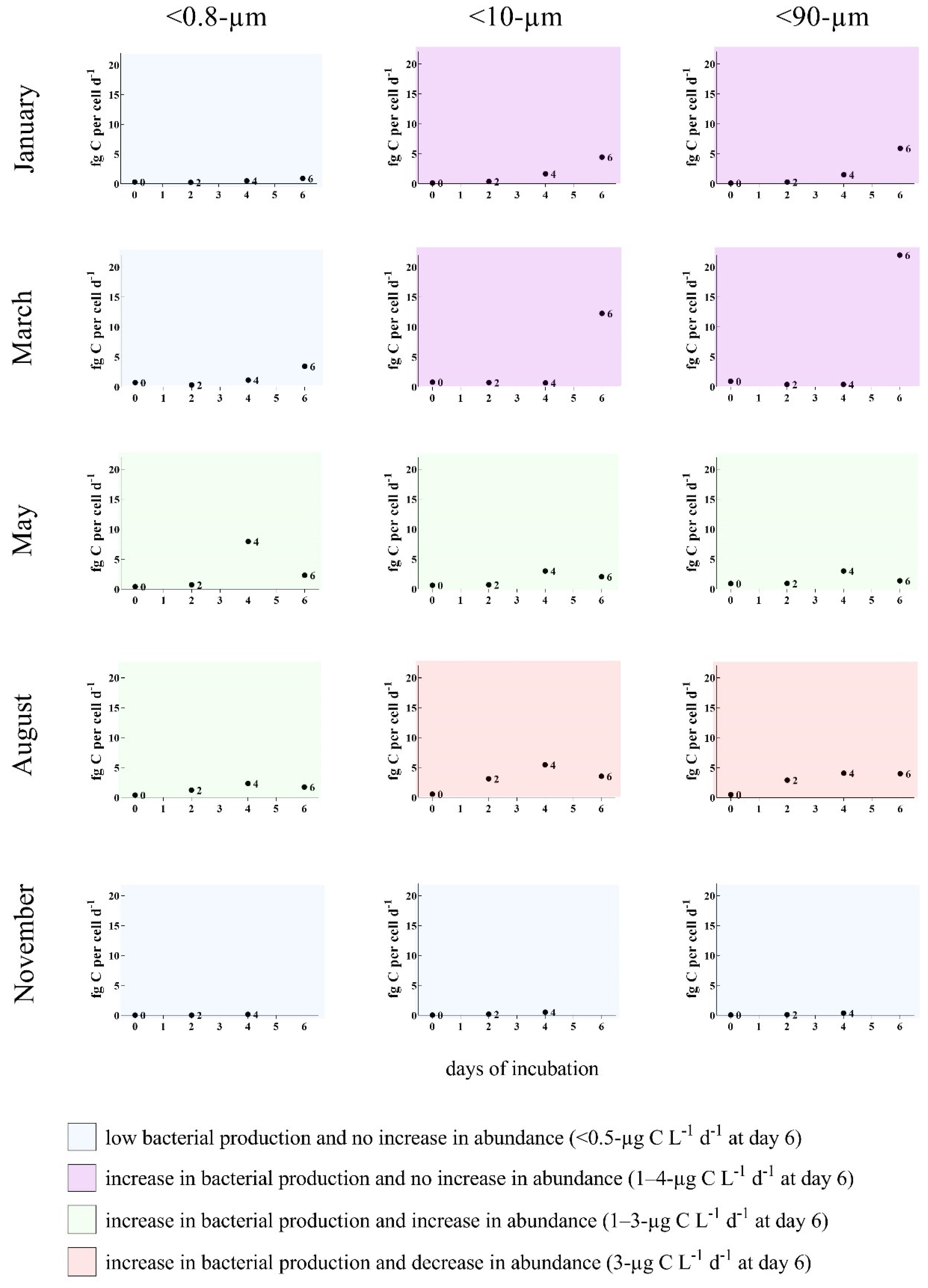
2.7. Bacterial Production (BP)
2.8. Nucleic Acid Extraction and Amplicon Library Preparation
2.9. Sequence Analysis
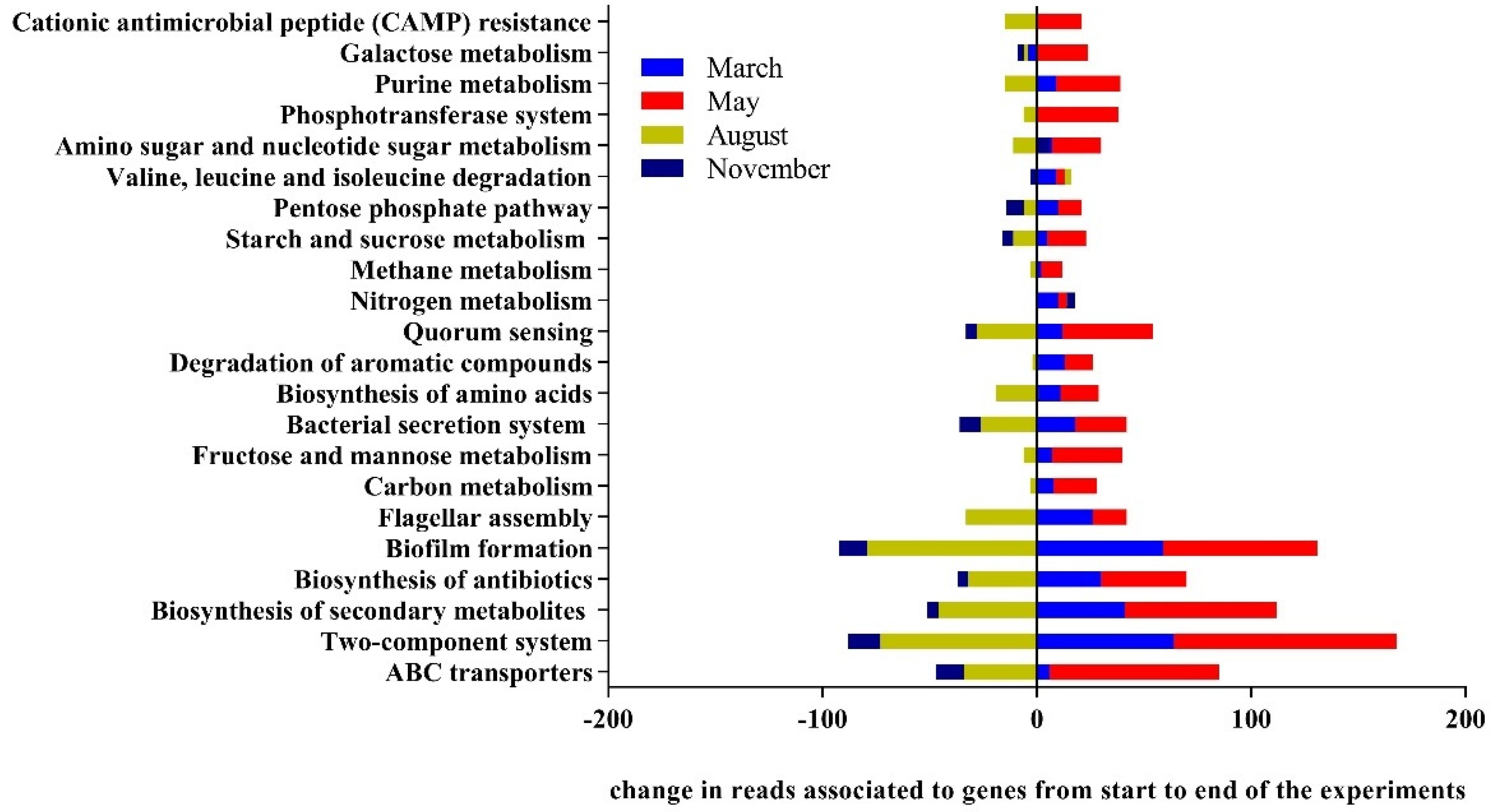
2.10. Functional Prediction with Tax4Fun
2.11. Statistical Analysis
3. Results
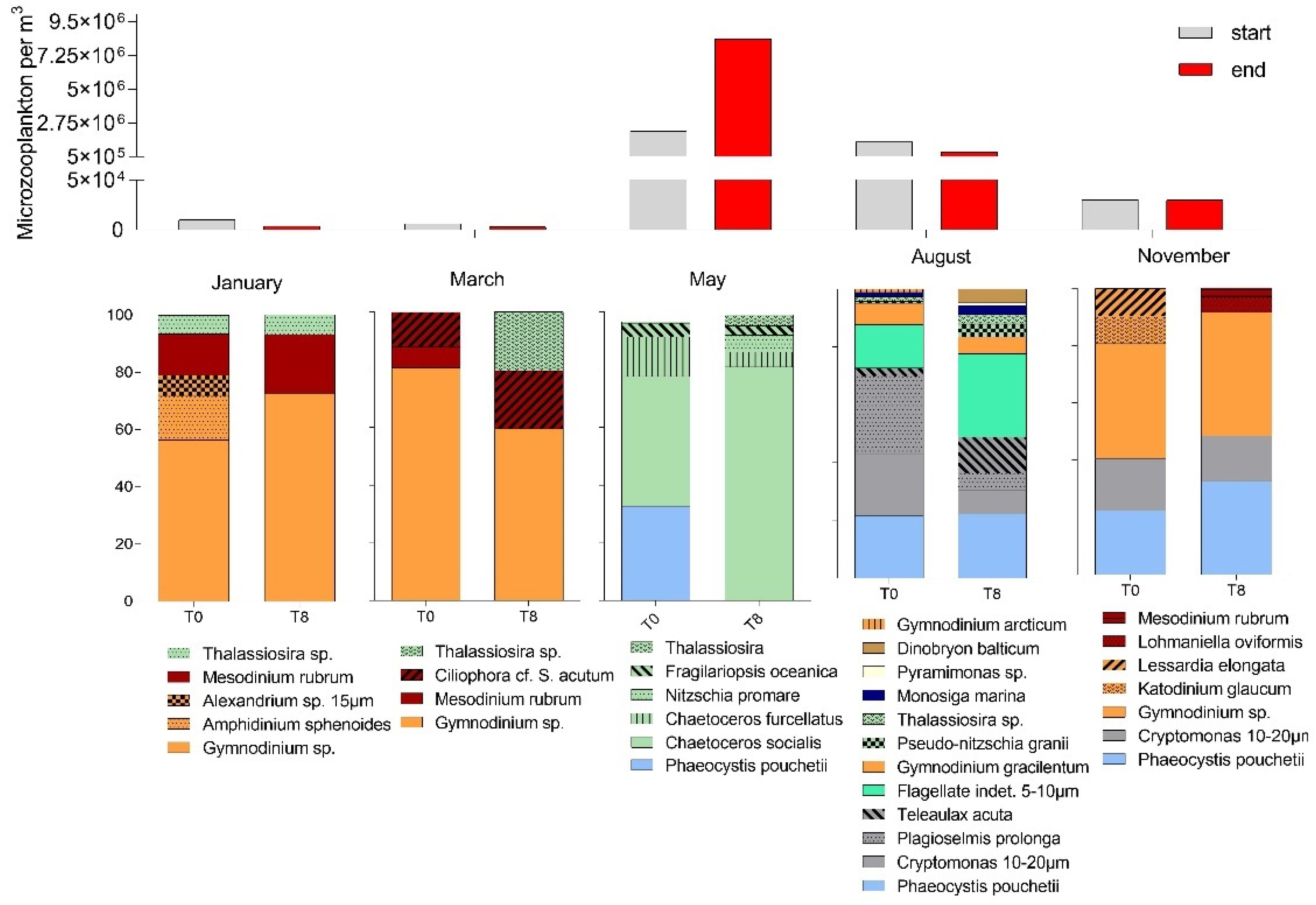
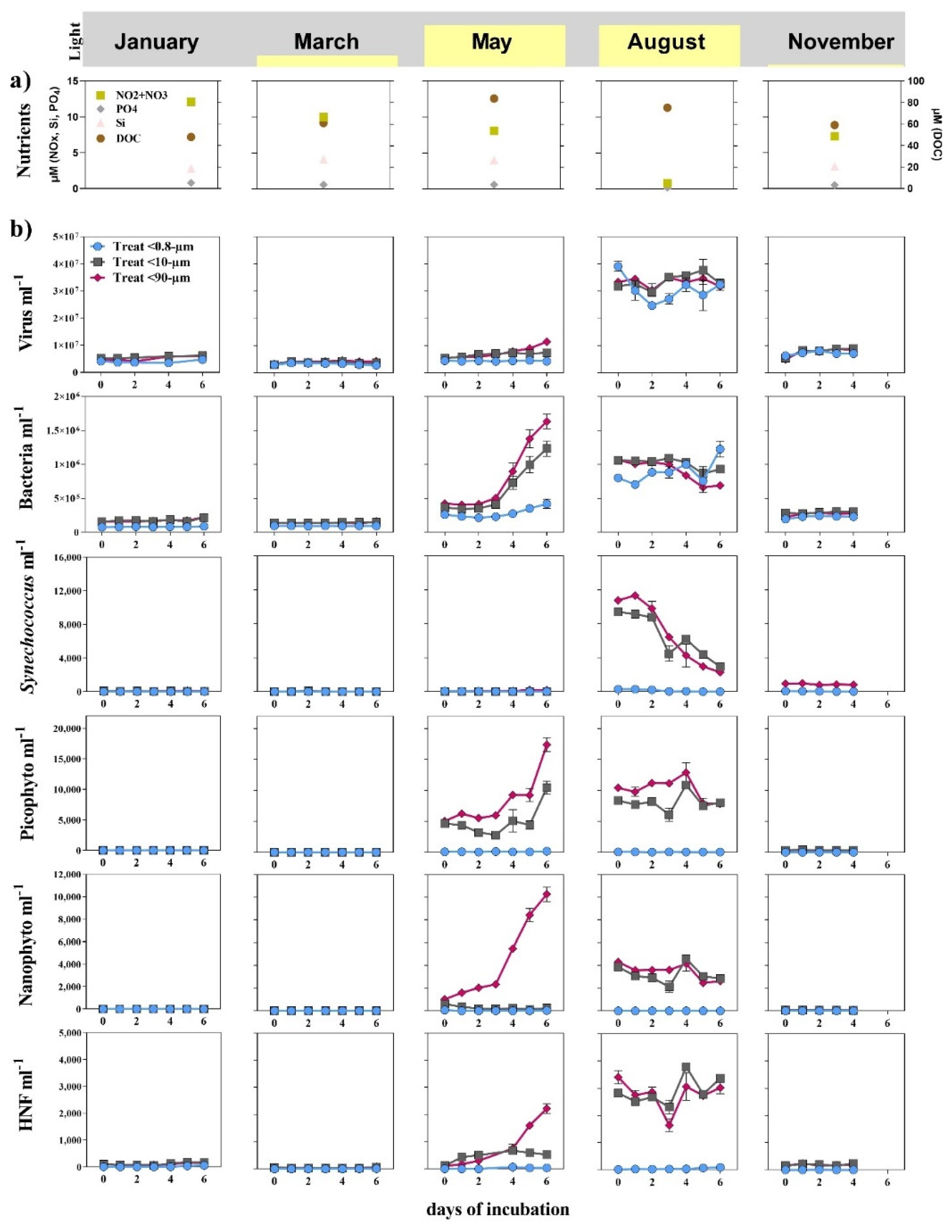
3.1. Seasonal Changes
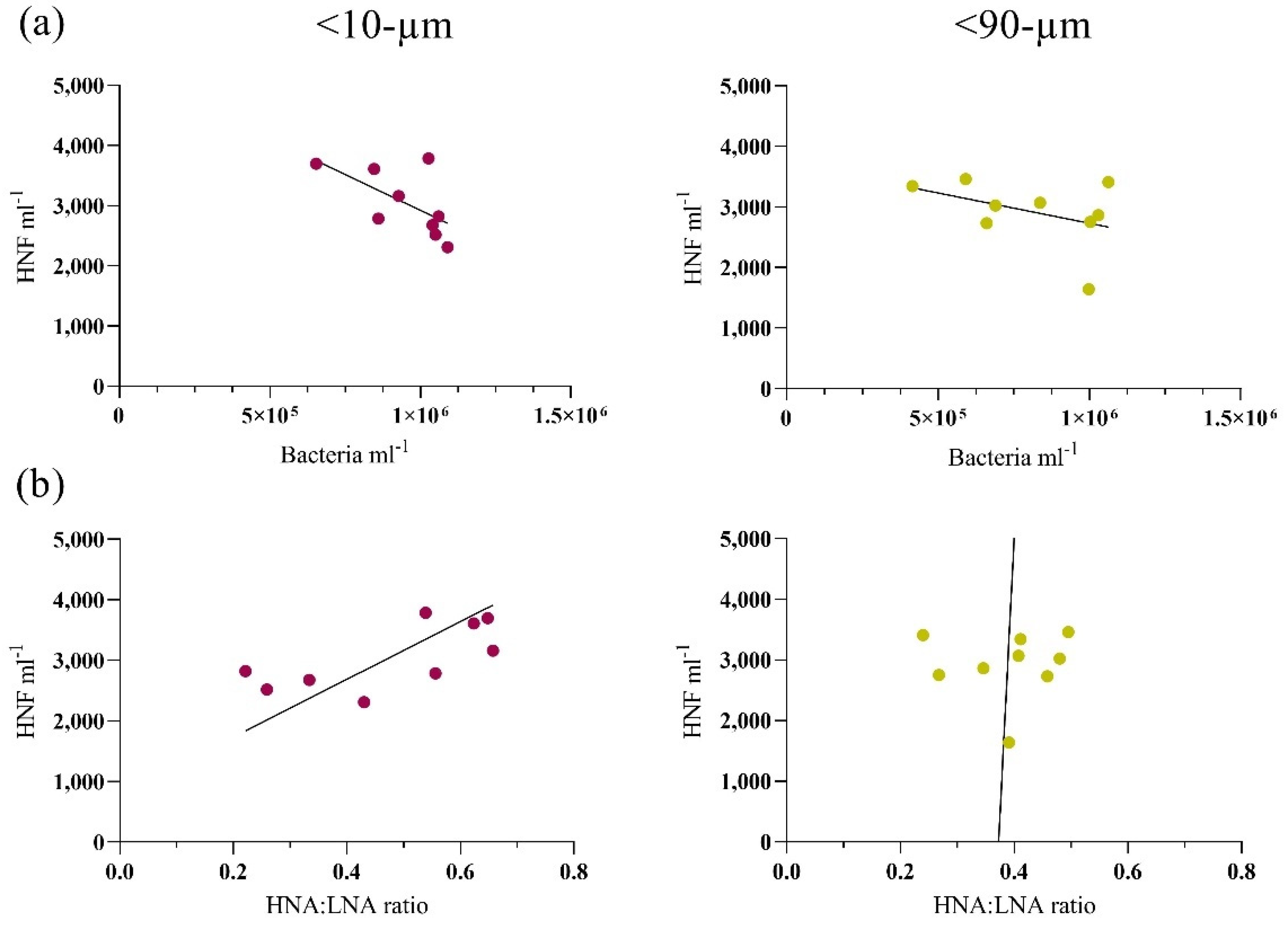
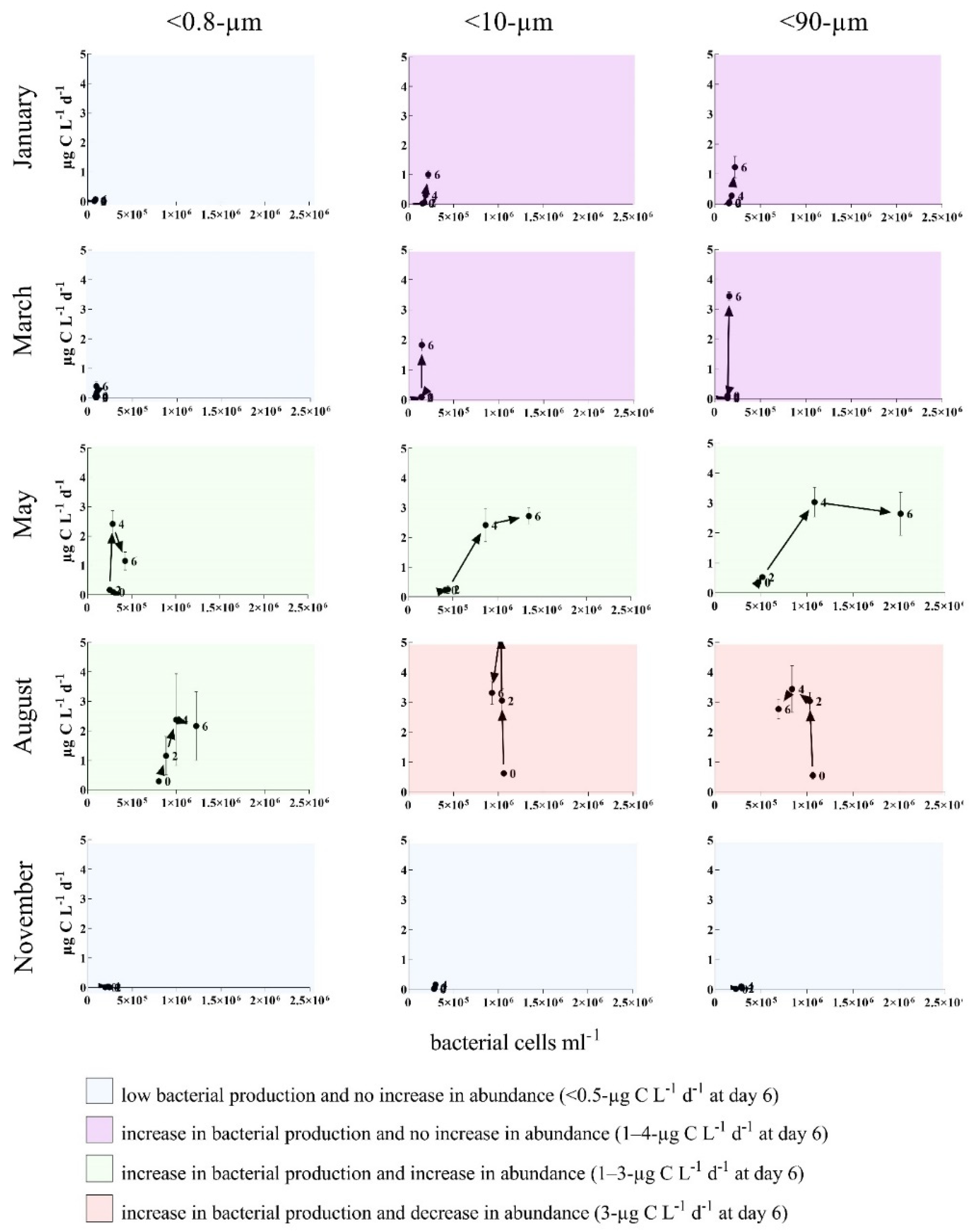
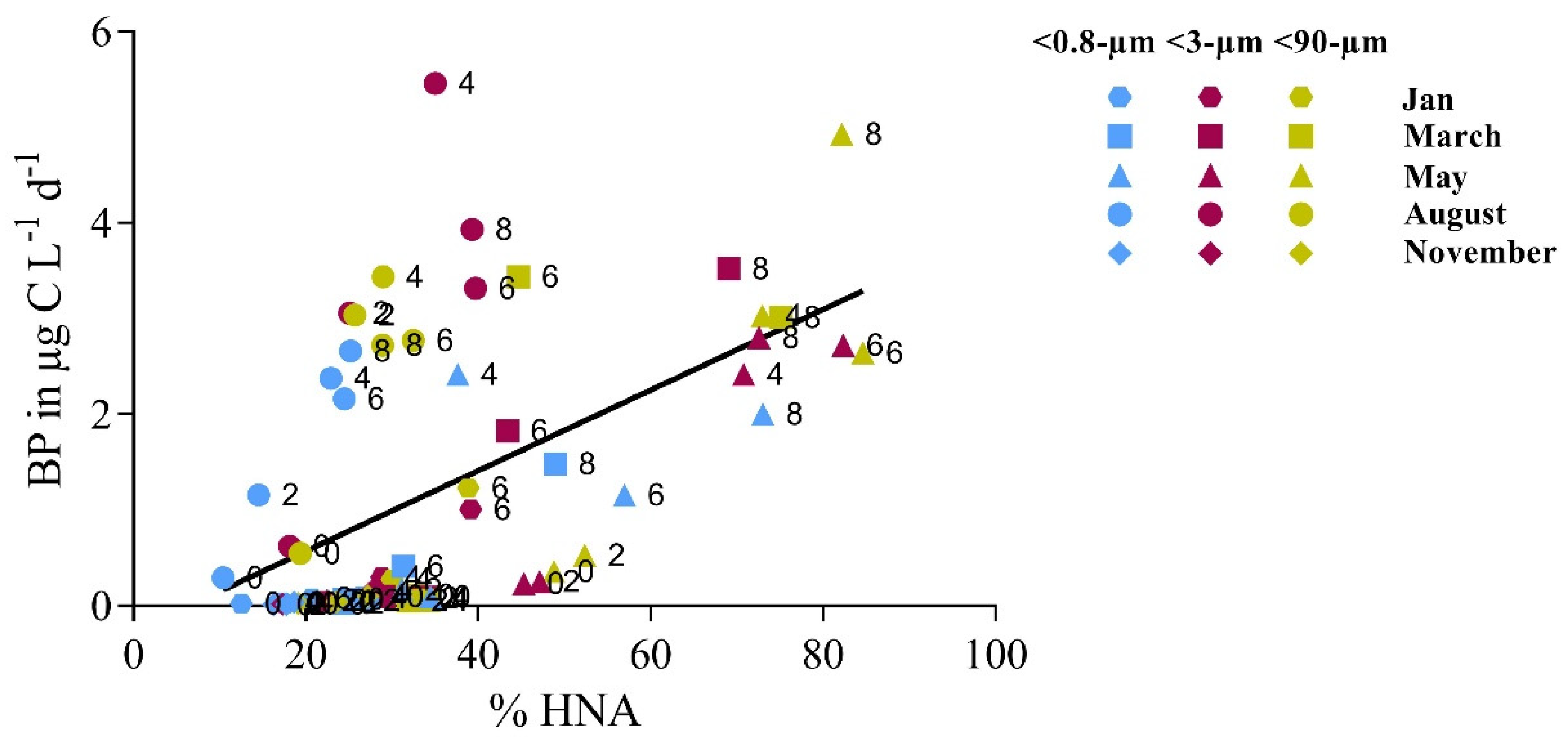
3.2. Bottom-Up and Top-Down Controls of Bacterial Activity
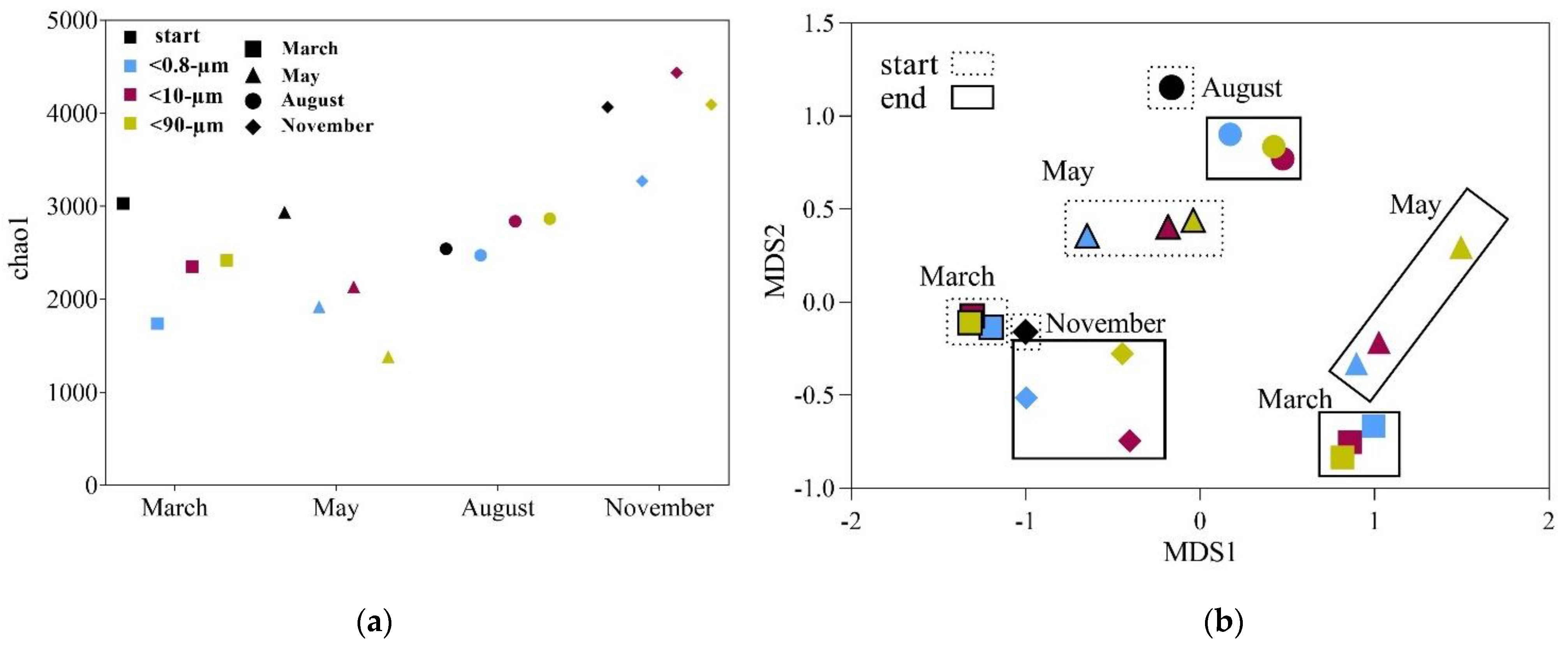
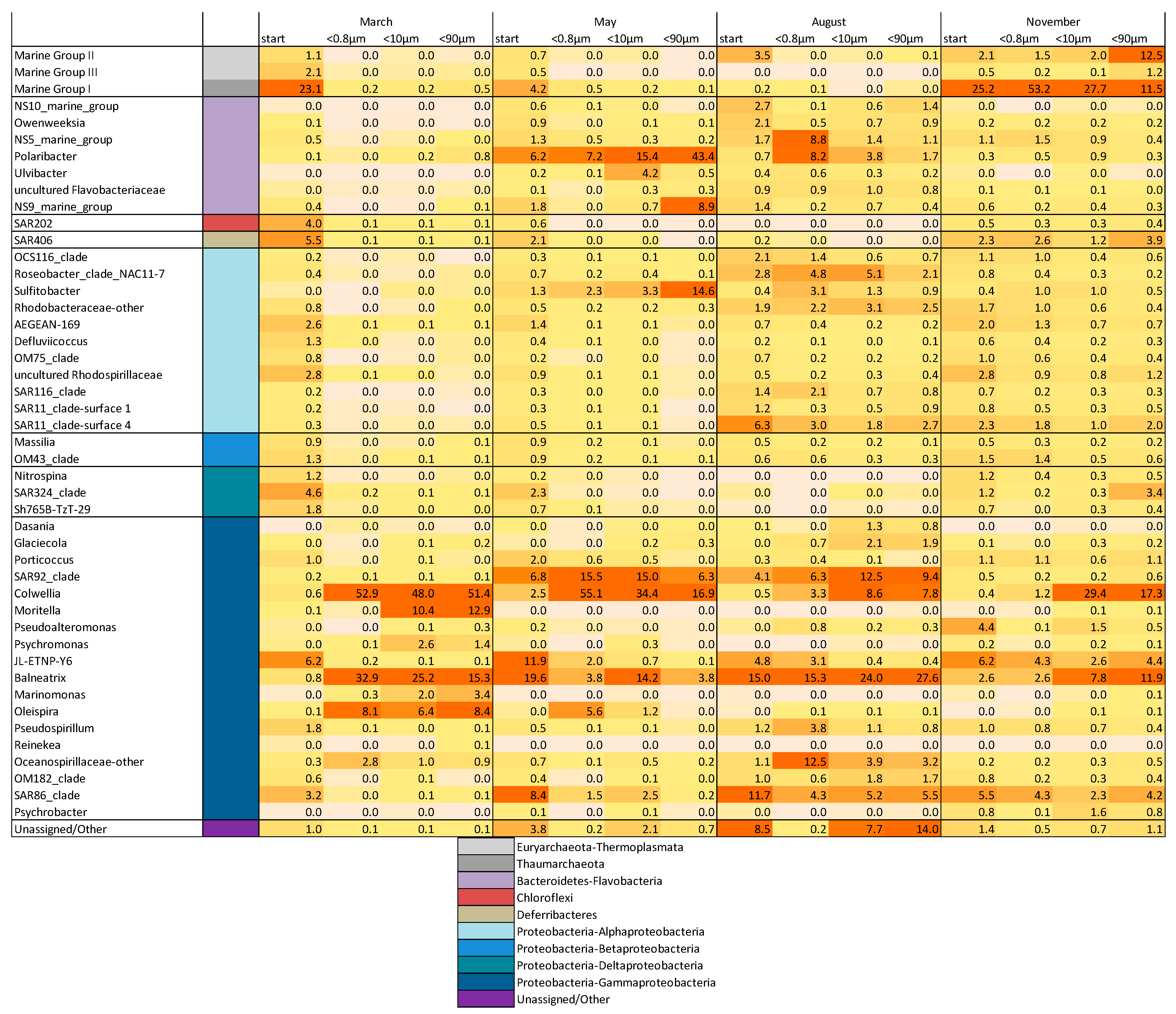
3.3. Drivers of Bacterial Community Composition Changes
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| ΔT8 HNA (Mar+May+Aug) | Polaribacter | Ulvibacter | NS9_marine_group | Sulfitobacter | ||||
| Pearson’s r | p | Pearson’s r | p | Pearson’s r | p | Pearson’s r | p | |
| 0.8176 | 0.0002 | 0.7637 | 0.0009 | 0.5554 | 0.0316 | 0.6985 | 0.0038 | |
References
- Azam, F.; Fenchel, T.; Field, J.G.; Gray, J.S.; Meyer-Reil, L.A.; Thingstad, F. The Ecological Role of Water-Column Microbes in the Sea. Mar. Ecol. Prog. Ser. 1983, 10, 257–263. [Google Scholar] [CrossRef]
- Wilhelm, S.W.; Suttle, C.A. Viruses and Nutrient Cycles in the Sea. Bioscience 1999, 49, 781–788. [Google Scholar] [CrossRef] [Green Version]
- Tsagaraki, T.M.; Pree, B.; Leiknes, Ø.; Larsen, A.; Bratbak, G.; Øvreås, L.; Egge, J.K.; Spanek, R.; Paulsen, M.L.; Olsen, Y.; et al. Bacterial community composition responds to changes in copepod abundance and alters ecosystem function in an Arctic mesocosm study. ISME J. 2018, 12, 2694–2705. [Google Scholar] [CrossRef]
- Thingstad, T.F.; Våge, S.; Bratbak, G.; Egge, J.; Larsen, A.; Nejstgaard, J.C.; Sandaa, R. Reproducing the virus-to-copepod link in Arctic mesocosms using host fitness optimization. Limnol. Oceanogr. 2021, 66, S303–S313. [Google Scholar] [CrossRef]
- Sakshaug, E.; Slagstad, D. Light and productivity of phytoplankton in polar marine ecosystems: A physiological view. Polar Res. 1991, 10, 69–86. [Google Scholar] [CrossRef]
- Ardyna, M.; Arrigo, K.R. Phytoplankton dynamics in a changing Arctic Ocean. Nat. Clim. Chang. 2020, 10, 892–903. [Google Scholar] [CrossRef]
- Wilson, B.; Müller, O.; Nordmann, E.-L.; Seuthe, L.; Bratbak, G.; Øvreås, L. Changes in Marine Prokaryote Composition with Season and Depth Over an Arctic Polar Year. Front. Mar. Sci. 2017, 4, 95. [Google Scholar] [CrossRef] [Green Version]
- Behrenfeld, M.J.; Boss, E.S.; Halsey, K.H. Phytoplankton community structuring and succession in a competition-neutral resource landscape. ISME Commun. 2021, 1, 1–8. [Google Scholar] [CrossRef]
- Tremblay, J.; Michel, C.; Hobson, K.A.; Gosselin, M.; Price, N.M. Bloom dynamics in early opening waters of the Arctic Ocean. Limnol. Oceanogr. 2006, 51, 900–912. [Google Scholar] [CrossRef] [Green Version]
- Leu, E.; Mundy, C.; Assmy, P.; Campbell, K.; Gabrielsen, T.; Gosselin, M.; Juul-Pedersen, T.; Gradinger, R. Arctic spring awakening—Steering principles behind the phenology of vernal ice algal blooms. Prog. Oceanogr. 2015, 139, 151–170. [Google Scholar] [CrossRef]
- Sakshaug, E. Primary and Secondary Production in the Arctic Seas. In The Organic Carbon Cycle in the Arctic Ocean; Stein, R., Macdonald, R.W., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 57–81. [Google Scholar]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; Gonzalez, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Legendre, L.; Rassoulzadegan, F. Food-web mediated export of biogenic carbon in oceans:hydrodynamic control. Mar. Ecol. Prog. Ser. 1996, 145, 179–193. [Google Scholar] [CrossRef] [Green Version]
- Hessen, D.O.; Ågren, G.I.; Anderson, T.R.; Elser, J.J.; De Ruiter, P.C. Carbon sequestration in ecosystems: The role of stoichiometry. Ecology 2004, 85, 1179–1192. [Google Scholar] [CrossRef]
- Thingstad, T.F.; Havskum, H.; Zweifel, U.L.; Berdalet, E.; Sala, M.M.; Peters, F.; Alcaraz, M.; Scharek, R.; Perez, M.; Jacquet, S.; et al. Ability of a “minimum” microbial food web model to reproduce response patterns observed in mesocosms manipulated with N and P, glucose, and Si. J. Mar. Syst. 2007, 64, 15–34. [Google Scholar] [CrossRef] [Green Version]
- Polyakov, I.V.; Pnyushkov, A.V.; Alkire, M.B.; Ashik, I.M.; Baumann, T.M.; Carmack, E.C.; Goszczko, I.; Guthrie, J.; Ivanov, V.V.; Kanzow, T.; et al. Greater role for Atlantic inflows on sea-ice loss in the Eurasian Basin of the Arctic Ocean. Science 2017, 356, 285–291. [Google Scholar] [CrossRef] [Green Version]
- Dinasquet, J.; Dinasquet, J.; Ortega-Retuerta, E.; Ortega-Retuerta, E.; Lovejoy, C.; Lovejoy, C.; Obernosterer, I.; Obernosterer, I.; Dinasquet, J.; Dinasquet, J.; et al. Editorial: Microbiology of the Rapidly Changing Polar Environments. Front. Mar. Sci. 2018, 5, 154. [Google Scholar] [CrossRef]
- Onarheim, I.H.; Smedsrud, L.H.; Ingvaldsen, R.B.; Nilsen, F. Loss of sea ice during winter north of Svalbard. Tellus Dyn. Meteorol. Oceanogr. 2014, 66, 23933. [Google Scholar] [CrossRef] [Green Version]
- Randelhoff, A.; Sundfjord, A.; Reigstad, M. Seasonal variability and fluxes of nitrate in the surface waters over the Arctic shelf slope. Geophys. Res. Lett. 2015, 42, 3442–3449. [Google Scholar] [CrossRef] [Green Version]
- Christaki, U.; Vazquez-Dominguez, E.; Courties, C.; LeBaron, P. Grazing impact of different heterotrophic nanoflagellates on eukaryotic (Ostreococcus tauri) and prokaryotic picoautotrophs (Prochlorococcus and Synechococcus). Environ. Microbiol. 2005, 7, 1200–1210. [Google Scholar] [CrossRef]
- Paulsen, M.L.; Müller, O.; Larsen, A.; Møller, E.F.; Middelboe, M.; Sejr, M.K.; Stedmon, C. Biological transformation of Arctic dissolved organic matter in a NE Greenland fjord. Limnol. Oceanogr. 2019, 64, 1014–1033. [Google Scholar] [CrossRef] [Green Version]
- Sato, M.; Yoshikawa, T.; Takeda, S.; Furuya, K. Application of the size-fractionation method to simultaneous estimation of clearance rates by heterotrophic flagellates and ciliates of pico- and nanophytoplankton. J. Exp. Mar. Biol. Ecol. 2007, 349, 334–343. [Google Scholar] [CrossRef]
- Šimek, K.; Chrzanowski, T.H. Direct and Indirect Evidence of Size-Selective Grazing on Pelagic Bacteria by Freshwater Nanoflagellates. Appl. Environ. Microbiol. 1992, 58, 3715–3720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grasshoff, K.; Kremling, K.; Ehrhardt, M. Methods of Seawater Analysis, 3rd ed.; John Wiley & Sons: Hoboken, NJ, USA, 1978; ISBN 3527295895. [Google Scholar]
- Wood, E.D.; Armstrong, F.A.J.; Richards, F.A. Determination of nitrate in sea water by cadmium-copper reduction to nitrite. J. Mar. Biol. Assoc. U. K. 1967, 47, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Cauwet, G. Determination of dissolved organic carbon and nitrogen by high temperature combustion. In Methods of Seawater Analysis, Methods of Seawater Analysis: Third, Completely Revised and Extended Edition; John Wiley & Sons: Hoboken, NJ, USA, 2007; ISBN 9783527613984. [Google Scholar]
- Marie, D.; Brussaard, C.P.D.; Thyrhaug, R.; Bratbak, G.; Vaulot, D. Enumeration of Marine Viruses in Culture and Natural Samples by Flow Cytometry. Appl. Environ. Microbiol. 1999, 65, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Zubkov, M.V.; Burkill, P.H.; Topping, J.N. Flow cytometric enumeration of DNA-stained oceanic planktonic protists. J. Plankton Res. 2007, 29, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Paulsen, M.L.; Doré, H.; Garczarek, L.; Seuthe, L.; Müller, O.; Sandaa, R.-A.; Bratbak, G.; Larsen, A. Synechococcus in the Atlantic Gateway to the Arctic Ocean. Front. Mar. Sci. 2016, 3, 191. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.C.; Azam, F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine 1. Mar. Microb. Food Webs 1992, 6, 107–114. [Google Scholar]
- Simon, M.; Azam, F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 1989, 51, 201–213. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decelle, J.; Romac, S.; Stern, R.F.; Bendif, E.M.; Zingone, A.; Audic, S.; Guiry, M.D.; Guillou, L.; Tessier, D.; Le Gall, F.; et al. PhytoREF: A reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol. Ecol. Resour. 2015, 15, 1435–1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data: Fig. 1. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egge, E.; Elferink, S.; Vaulot, D.; John, U.; Bratbak, G.; Larsen, A.; Edvardsen, B. An 18S V4 rDNA metabarcoding dataset of protist diversity in the Atlantic inflow to the Arctic Ocean, through the year and down to 1000 m depth. Earth Syst. Sci. Data Discuss. 2021, 13, 1–25. [Google Scholar] [CrossRef]
- Simon, M.; Grossart, H.; Schweitzer, B.; Ploug, H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 2002, 28, 175–211. [Google Scholar] [CrossRef] [Green Version]
- Stocker, R. Marine Microbes See a Sea of Gradients. Science 2012, 338, 628–633. [Google Scholar] [CrossRef] [Green Version]
- Reintjes, G.; Fuchs, B.M.; Scharfe, M.; Wiltshire, K.H.; Amann, R.; Arnosti, C. Short-term changes in polysaccharide utilization mechanisms of marine bacterioplankton during a spring phytoplankton bloom. Environ. Microbiol. 2020, 22, 1884–1900. [Google Scholar] [CrossRef]
- Sapp, M.; Wichels, A.; Wiltshire, K.H.; Gerdts, G. Bacterial community dynamics during the winter–spring transition in the North Sea. FEMS Microbiol. Ecol. 2007, 59, 622–637. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Richlen, M.L.; Sehein, T.R.; Kulis, D.M.; Anderson, D.M.; Cai, Z. Microbial Community Structure and Associations During a Marine Dinoflagellate Bloom. Front. Microbiol. 2018, 9, 1201. [Google Scholar] [CrossRef] [PubMed]
- Fontanez, K.M.; Eppley, J.M.; Samo, T.; Karl, D.; Delong, E.F. Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front. Microbiol. 2015, 6, 469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunse, C.; Koch, H.; Breider, S.; Simon, M.; Wietz, M. Sweet spheres: Succession and CAZyme expression of marine bacterial communities colonizing a mix of alginate and pectin particles. Environ. Microbiol. 2021, 23, 3130–3148. [Google Scholar] [CrossRef]
- Machado, H.; Sonnenschein, E.C.; Melchiorsen, J.; Gram, L. Genome mining reveals unlocked bioactive potential of marine Gram-negative bacteria. BMC Genom. 2015, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kube, M.; Chernikova, T.N.; Al-Ramahi, Y.; Beloqui, A.; Lopez-Cortez, N.; Guazzaroni, M.-E.; Heipieper, H.J.; Klages, S.; Kotsyurbenko, O.R.; Langer, I.; et al. Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica. Nat. Commun. 2013, 4, 2156. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Park, S.; Lee, J.M.; Jung, W.; Kang, J.-S.; Joo, H.M.; Seo, K.-W.; Kang, S.-H. Moritella dasanensis sp. nov., a psychrophilic bacterium isolated from the Arctic ocean. Int. J. Syst. Evol. Microbiol. 2008, 58, 817–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [Green Version]
- Cordero, O.X.; Ventouras, L.-A.; DeLong, E.F.; Polz, M.F. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl. Acad. Sci. USA 2012, 109, 20059–20064. [Google Scholar] [CrossRef] [Green Version]
- Crenn, K.; Duffieux, D.; Jeanthon, C. Bacterial Epibiotic Communities of Ubiquitous and Abundant Marine Diatoms Are Distinct in Short- and Long-Term Associations. Front. Microbiol. 2018, 9, 2879. [Google Scholar] [CrossRef] [Green Version]
- Amin, S.A.; Parker, M.; Armbrust, E.V. Interactions between Diatoms and Bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [Green Version]
- Shibl, A.A.; Isaac, A.; Ochsenkühn, M.A.; Cárdenas, A.; Fei, C.; Behringer, G.; Arnoux, M.; Drou, N.; Santos, M.P.; Gunsalus, K.C.; et al. Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc. Natl. Acad. Sci. USA 2020, 117, 27445–27455. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Hmelo, L.R.; Van Tol, H.M.; Durham, B.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.; Djunaedi, B.; et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef]
- Johansson, O.N.; Pinder, M.I.M.; Ohlsson, F.; Egardt, J.; Töpel, M.; Clarke, A.K. Friends With Benefits: Exploring the Phycosphere of the Marine Diatom Skeletonema marinoi. Front. Microbiol. 2019, 10, 1828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherr, E.B.; Sherr, B.F. Significance of predation by protists in aquatic microbial food webs. Antonie Leeuwenhoek 2002, 81, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Vaqué, D.; Boras, J.; Arrieta, J.; Agustí, S.; Duarte, C.; Sala, M. Enhanced Viral Activity in the Surface Microlayer of the Arctic and Antarctic Oceans. Microorganisms 2021, 9, 317. [Google Scholar] [CrossRef] [PubMed]
- Vaqué, D.; Guadayol, O.; Peters, F.; Felipe, J.; Angel-Ripoll, L.; Terrado, R.; Lovejoy, C.; Pedrós-Alió, C. Seasonal changes in planktonic bacterivory rates under the ice-covered coastal Arctic Ocean. Limnol. Oceanogr. 2008, 53, 2427–2438. [Google Scholar] [CrossRef] [Green Version]
- Calbet, A. The trophic roles of microzooplankton in marine systems. ICES J. Mar. Sci. 2008, 65, 325–331. [Google Scholar] [CrossRef] [Green Version]
- Levinsen, H.; Nielsen, T.G. The trophic role of marine pelagic ciliates and heterotrophic dinoflagellates in arctic and temperate coastal ecosystems: A cross-latitude comparison. Limnol. Oceanogr. 2002, 47, 427–439. [Google Scholar] [CrossRef]
- Smetacek, V. The annual cycle of protozooplankton in the Kiel Bight. Mar. Biol. 1981, 63, 1–11. [Google Scholar] [CrossRef]
- Ladau, J.; Sharpton, T.J.; Finucane, M.M.; Jospin, G.; Kembel, S.; O’Dwyer, J.; Koeppel, A.F.; Green, J.L.; Pollard, K.S. Global marine bacterial diversity peaks at high latitudes in winter. ISME J. 2013, 7, 1669–1677. [Google Scholar] [CrossRef]
- Müller, O.; Wilson, B.; Paulsen, M.L.; Rumińska, A.; Armo, H.R.; Bratbak, G.; Øvreås, L. Spatiotemporal Dynamics of Ammonia-Oxidizing Thaumarchaeota in Distinct Arctic Water Masses. Front. Microbiol. 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, O.; Seuthe, L.; Pree, B.; Bratbak, G.; Larsen, A.; Paulsen, M.L. How Microbial Food Web Interactions Shape the Arctic Ocean Bacterial Community Revealed by Size Fractionation Experiments. Microorganisms 2021, 9, 2378. https://doi.org/10.3390/microorganisms9112378
Müller O, Seuthe L, Pree B, Bratbak G, Larsen A, Paulsen ML. How Microbial Food Web Interactions Shape the Arctic Ocean Bacterial Community Revealed by Size Fractionation Experiments. Microorganisms. 2021; 9(11):2378. https://doi.org/10.3390/microorganisms9112378
Chicago/Turabian StyleMüller, Oliver, Lena Seuthe, Bernadette Pree, Gunnar Bratbak, Aud Larsen, and Maria Lund Paulsen. 2021. "How Microbial Food Web Interactions Shape the Arctic Ocean Bacterial Community Revealed by Size Fractionation Experiments" Microorganisms 9, no. 11: 2378. https://doi.org/10.3390/microorganisms9112378
APA StyleMüller, O., Seuthe, L., Pree, B., Bratbak, G., Larsen, A., & Paulsen, M. L. (2021). How Microbial Food Web Interactions Shape the Arctic Ocean Bacterial Community Revealed by Size Fractionation Experiments. Microorganisms, 9(11), 2378. https://doi.org/10.3390/microorganisms9112378