Microbial Communities Present in Hydrothermal Sediments from Deception Island, Antarctica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Geological Study and Determination of Physicochemical Parameters
2.2.1. Ionic and Atomic Compositional Analysis: Inductively Coupled Plasma Atomic Emission Spectroscopy (ICP-OES) and Ionic Chromatography (IC)
2.2.2. Structural Analysis by X-ray Powder Diffraction (XRD)
2.3. Microbial Diversity Analysis
2.3.1. Metataxonomic Study
2.3.2. Isolation and Identification of Viable Microorganisms
2.3.3. Phenotypical Characterization of the Bacterial Isolates
2.3.4. Study of Scanning Electron Microscopy (SEM)
3. Results
3.1. Geological Study and Determination of Physicochemical Parameters
3.2. Microbial Diversity Analysis
3.2.1. Metataxonomic Study
3.2.2. Isolation and Identification of Viable Microorganisms
3.2.3. Phenotypical Characterization of the Bacterial Isolates
3.2.4. Study of Scanning Electron Microscopy (SEM)
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Marti, J.; Vila, J.; Rey, J. Deception Island (Bransfield Strait, Antarctica): An example of a volcanic caldera developed by extensional tectonics. Geol. Soc. Lond. Spec. Publ. 1996, 110, 253–265. [Google Scholar] [CrossRef]
- Barker, D.H.N.; Austin, J.A. Rift propagation, detachment faulting, and associated magmatism in Bransfield Strait, Antarctic Peninsula. J. Geophys. Res. Solid Earth 1998, 103, 24017–24043. [Google Scholar] [CrossRef]
- Cooper, A.P.R.; Smellie, J.L.; Maylin, J. Evidence for shallowing and uplift from bathymetric records of Deception Island, Antarctica. Antarct. Sci. 2004, 10, 455–461. [Google Scholar] [CrossRef]
- Herbold, C.W.; McDonald, I.R.; Cary, S.C. Microbial Ecology of Geothermal Habitats in Antarctica. In Antarctic Terrestrial Microbiology; Springer: Berlin/Heidelberg, Germany, 2014; pp. 181–215. [Google Scholar] [CrossRef]
- Smith, K.; Baldwin, R.; Kaufmann, R.; Sturz, A. Ecosystem studies at Deception Island, Antarctica: An overview. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2003, 50, 1595–1609. [Google Scholar] [CrossRef]
- Marquez, S.L.; Blamey, J.M. Isolation and partial characterization of a new moderate thermophilic Albidovulum sp. SLM16 with transaminase activity from Deception Island, Antarctica. Biol. Res. 2019, 52, 5. [Google Scholar] [CrossRef] [Green Version]
- Pepi, M.; Agnorelli, C.; Bargagli, R. Iron demand by thermophilic and mesophilic bacteria isolated from an antarctic geothermal soil. Biometals 2005, 18, 529–536. [Google Scholar] [CrossRef]
- Bottos, E.M.; Scarrow, J.W.; Archer, S.D.; McDonald, I.R.; Cary, S.C. Bacterial Community Structures of Antarctic Soils. In Antarctic Terrestrial Microbiology; Springer: Berlin/Heidelberg, Germany, 2014; pp. 9–33. [Google Scholar]
- Darling, C.A.; Siple, P.A. Bacteria of Antarctica. J. Bacteriol. 1941, 42, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, E.A.; Stout, J.D. Microbiology of Some Soils from Antarctica. Nature 1960, 188, 767–768. [Google Scholar] [CrossRef]
- Horowitz, N.H.; Cameron, R.E.; Hubbard, J.S. Microbiology of the dry valleys of antarctica. Science 1972, 176, 242–245. [Google Scholar] [CrossRef]
- Smith, J.J.; Tow, L.A.; Stafford, W.; Cary, C.; Cowan, D.A. Bacterial diversity in three different Antarctic Cold Desert mineral soils. Microb. Ecol. 2006, 51, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Cowan, D.A.; Russell, N.J.; Mamais, A.; Sheppard, D.M. Antarctic Dry Valley mineral soils contain unexpectedly high levels of microbial biomass. Extremophiles 2002, 6, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Bendia, A.G.; Araujo, G.G.; Pulschen, A.A.; Contro, B.; Duarte, R.T.; Rodrigues, F.; Galante, D.; Pellizari, V.H. Surviving in hot and cold: Psychrophiles and thermophiles from Deception Island volcano, Antarctica. Extremophiles 2018, 22, 917–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendia, A.G.; Signori, C.N.; Franco, D.C.; Duarte, R.T.; Bohannan, B.J.; Pellizari, V.H. A mosaic of geothermal and marine features shapes microbial community structure on deception Island Volcano, Antarctica. Front. Microbiol. 2018, 9, 899. [Google Scholar] [CrossRef] [Green Version]
- Figueredo, H.M.; Gonçalves, V.N.; Godinho, V.M.; Lopes, D.V.; Oliveira, F.S.; Rosa, L.H. Diversity and ecology of cultivable fungi isolated from the thermal soil gradients in Deception Island, Antarctica. Extremophiles 2020, 24, 219–225. [Google Scholar] [CrossRef]
- Rosa, L.H.; da Silva, T.H.; Ogaki, M.B.; Pinto, O.H.B.; Stech, M.; Convey, P.; Carvalho-Silva, M.; Rosa, C.A.; Câmara, P.E. DNA metabarcoding uncovers fungal diversity in soils of protected and non-protected areas on Deception Island, Antarctica. Sci. Rep. 2020, 10, 21986. [Google Scholar] [CrossRef]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From Vineyard Soil to Wine Fermentation: Microbiome Approximations to Explain the “terroir”. Concept. Front. Microbiol. 2017, 8, 821. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From raw reads to community analyses. F1000Res 2016, 5, 1492. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glockner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M. Handbook of Media for Environmental Microbiology; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Cenis, J. Rapid extraction of fungal DNA for PCR amplification. J. Nucleic Acids Res. 1992, 20, 2380. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrov, V.; Blagodyr, R.; Ilev, I. Liberation of phosphoric acid from apatite by silicate bacteria. J. Mikrobiol. Z. 1967, 29, 111–114. [Google Scholar]
- Olney, M. MIRACLE. Microfossil Image Recovery and Circulation for Learning and Education. Available online: https://www.ucl.ac.uk/GeolSci/micropal/index.html (accessed on 29 October 2020).
- Scott, F.J.; Marchant, H.J. Antarctic Marine Protists; Australian Biological Resources Study and Australian Antarctic Division: Canberra, Australia, 2005. [Google Scholar]
- Spaulding, S.A.; Bishop, I.W.; Edlund, M.B.; Lee, S.; Furey, P.; Jovanovska, E.; Potapova, M. Diatoms of North America. Available online: https://diatoms.org/ (accessed on 29 October 2020).
- Rey, J.; Somoza, L.; Martínez-Frías, J. Tectonic, volcanic, and hydrothermal event sequence on Deception Island (Antarctica). J. Geo-Mar. Lett. 1995, 15, 1–8. [Google Scholar] [CrossRef]
- Lezcano, M.A.; Moreno-Paz, M.; Carrizo, D.; Prieto-Ballesteros, O.; Fernandez-Martinez, M.A.; Sanchez-Garcia, L.; Blanco, Y.; Puente-Sanchez, F.; de Diego-Castilla, G.; Garcia-Villadangos, M.; et al. Biomarker Profiling of Microbial Mats in the Geothermal Band of Cerro Caliente, Deception Island (Antarctica): Life at the Edge of Heat and cold. Astrobiology 2019, 119, 1490–1504. [Google Scholar] [CrossRef]
- Di Giglio, S.; Agüera, A.; Pernet, P.; M’Zoudi, S.; Angulo-Preckler, C.; Avila, C.; Dubois, P. Effects of ocean acidification on acid-base physiology, skeleton properties, and metal contamination in two echinoderms from vent sites in Deception Island, Antarctica. Sci. Total Environ. 2020, 765, 142669. [Google Scholar] [CrossRef]
- Martinez-Alonso, E.; Pena-Perez, S.; Serrano, S.; Garcia-Lopez, E.; Alcazar, A.; Cid, C. Taxonomic and functional characterization of a microbial community from a volcanic englacial ecosystem in Deception Island, Antarctica. Sci. Rep. 2019, 9, 12158. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, R.; Vila, J.; García, A.; Camacho, A.; Diez, J.; Aparicio, A.; Soto, R.; Viramonte, J.; Risso, C.; Menegatt, N. Geophysical Features of Deception Island. In Recent Progress in Antarctic Earth Science; Terrapub: Tokyo, Japan, 1992; pp. 143–152. [Google Scholar]
- Elderfield, H. Effects of volcanism on water chemistry, Deception Island, Antarctica. J. Mar. Geol. 1972, 13, M1–M6. [Google Scholar] [CrossRef]
- Wilkinson, J.F.G. Classification and Average Chemical-Compositions of Common Basalts and Andesites. J. Petrol. 1986, 27, 31–62. [Google Scholar] [CrossRef]
- Centurion, V.; Lacerda-Júnior, G.; Duarte, A.; Silva, T.; Silva, L.; Rosa, L.; Oliveira, V. Dynamics of microbial stress responses driven by abiotic changes along a temporal gradient in Deception Island, Maritime Antarctica. Sci. Total Environ. 2021, 758, 143671. [Google Scholar] [CrossRef]
- Nicolaus, B.; Marsiglia, F.; Esposito, E.; Trincone, A.; Lama, L.; Sharp, R.; Di Prisco, G.; Gambacorta, A. Isolation of five strains of thermophilic eubacteria in Antarctica. J. Polar Biol. 1991, 11, 425–429. [Google Scholar] [CrossRef]
- Wisotzkey, J.D.; Jurtshuk, P., Jr.; Fox, G.E.; Deinhard, G.; Poralla, K. Comparative sequence analyses on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus, Alicyclobacillus gen. nov. Int. J. Syst. Bacteriol. 1992, 42, 263–269. [Google Scholar] [CrossRef]
- Coorevits, A.; Dinsdale, A.E.; Halket, G.; Lebbe, L.; De Vos, P.; Van Landschoot, A.; Logan, N.A. Taxonomic revision of the genus Geobacillus: Emendation of Geobacillus, G. stearothermophilus, G. jurassicus, G. toebii, G. thermodenitrificans and G. thermoglucosidans (nom. corrig., formerly ‘thermoglucosidasius’); transfer of Bacillus thermantarcticus to the genus as G. thermantarcticus comb. nov.; proposal of Caldibacillus debilis gen. nov., comb. nov.; transfer of G. tepidamans to Anoxybacillus as A. tepidamans comb. nov.; and proposal of Anoxybacillus caldiproteolyticus sp. nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 1470–1485. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, P.A.; Flores, P.A.; Boehmwald, F.A.; Blamey, J.M. Thermophilic bacteria present in a sample from Fumarole Bay, Deception Island. Antarct. Sci. 2011, 23, 549–555. [Google Scholar] [CrossRef]
- Llarch, A.; Logan, N.A.; Castellvi, J.; Prieto, M.J.; Guinea, J. Isolation and Characterization of Thermophilic Bacillus spp. from Geothermal Environments on Deception Island, South Shetland Archipelago. Microb. Ecol. 1997, 34, 58–65. [Google Scholar] [CrossRef]
- Thomas, D.; Dieckmann, G. Antarctic sea ice—A habitat for extremophiles. J. Sci. 2002, 295, 641–644. [Google Scholar] [CrossRef] [Green Version]
- Yukimura, K.; Nakai, R.; Kohshima, S.; Uetake, J.; Kanda, H.; Naganuma, T. Spore-forming halophilic bacteria isolated from Arctic terrains: Implications for long-range transportation of microorganisms. J. Polar Sci. 2009, 3, 163–169. [Google Scholar] [CrossRef] [Green Version]
- Vasileva-Tonkova, E.; Romanovskaya, V.; Gladka, G.; Gouliamova, D.; Tomova, I.; Stoilova-Disheva, M.; Tashyrev, O. Ecophysiological properties of cultivable heterotrophic bacteria and yeasts dominating in phytocenoses of Galindez Island, maritime Antarctica. World J. Microbiol. Biotechnol. 2014, 30, 1387–1398. [Google Scholar] [CrossRef]
- Aislabie, J.M.; Chhour, K.-L.; Saul, D.J.; Miyauchi, S.; Ayton, J.; Paetzold, R.F.; Balks, M.R. Dominant bacteria in soils of Marble Point and Wright Valley, Victoria Land, Antarctica. Soil Biol. Biochem. 2006, 38, 3041–3056. [Google Scholar] [CrossRef]
- Alves, I.M.; Gonçalves, V.N.; Oliveira, F.S.; Schaefer, C.E.; Rosa, C.A.; Rosa, L.H. The diversity, distribution, and pathogenic potential of cultivable fungi present in rocks from the South Shetlands archipelago, Maritime Antarctica. Extremophiles 2019, 23, 327–336. [Google Scholar] [CrossRef]
- Tesei, D.; Marzban, G.; Zakharova, K.; Isola, D.; Selbmann, L.; Sterflinger, K. Alteration of protein patterns in black rock inhabiting fungi as a response to different temperatures. Fungal Biol. 2012, 116, 932–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wentzel, L.C.P.; Inforsato, F.J.; Montoya, Q.V.; Rossin, B.G.; Nascimento, N.R.; Rodrigues, A.; Sette, L.D. Fungi from Admiralty Bay (King George Island, Antarctica) Soils and Marine Sediments. Microb. Ecol. 2019, 77, 12–24. [Google Scholar] [CrossRef] [Green Version]
- Baeza, M.; Barahona, S.; Alcaíno, J.; Cifuentes, V. Amplicon-metagenomic analysis of fungi from Antarctic terrestrial habitats. Front. Microbiol. 2017, 8, 2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fermani, P.; Mataloni, G.; Van de Vijver, B. Soil microalgal communities on an antarctic active volcano (Deception Island, South Shetlands). Polar Biol. 2007, 30, 1381–1393. [Google Scholar] [CrossRef]
- Sabater, S. Diatoms. Encyclopedia of Inland Waters; Elsevier: Oxford, UK, 2009. [Google Scholar]
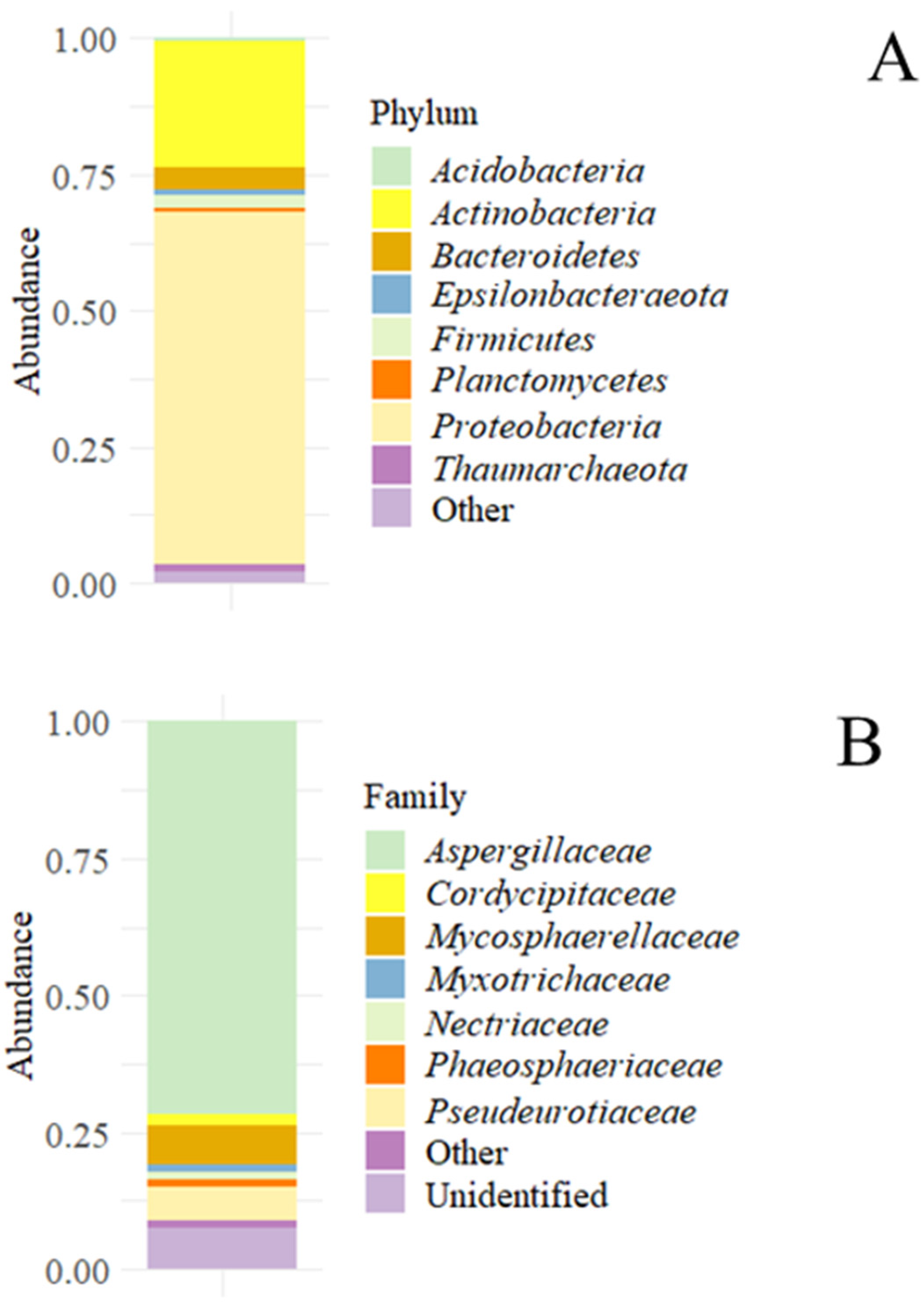
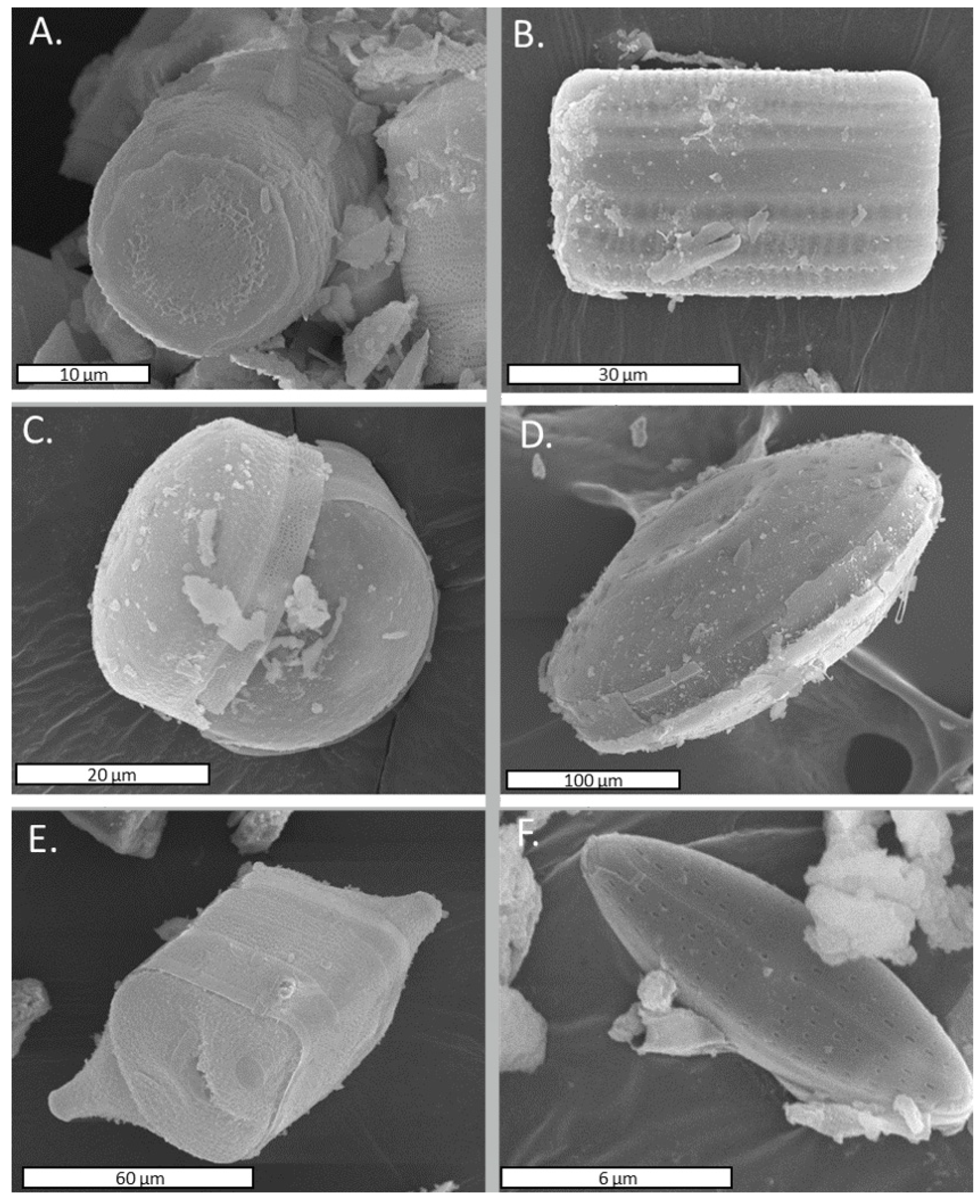
| Ion | Concentration (μg/g) |
|---|---|
| Cl− | 4075 ± 408 |
| SO42− | 1009 ± 202 |
| Br− | 14 ± 1 |
| NO3− | 6 ± 1 |
| F− | 0.42 ± 0.04 |
| Elements | Percentage (%) | Trace Elements | Amount (μg/g) |
|---|---|---|---|
| Si | 17.49 ± 0.71 | Sr | 351.64 ± 43.84 |
| Al | 8.14 ± 0.35 | S | 329.27 ± 40.31 |
| Fe | 6.34 ± 0.07 | Ni | 172.00 ± 0.00 |
| Na | 3.4 ± 0.00 | Zn | 87.98 ± 2.83 |
| Ca | 2.29 ± 0.17 | Cu | 55.32 ± 6.36 |
| K | 0.77 ± 0.01 | Ba | 52.21 ± 7.18 |
| P | 0.17 ± 0.01 | Co | 45.99 ± 1.41 |
| Mn | 0.11 ± 0.00 | Li | 18.89 ± 2.82 |
| Mg | 0.11 ± 0.03 | Mo | 17.66 ± 7.76 |
| Ti | 0.07 ± 0.00 | ||
| Isolates | Identification | Enrichment Media | Temperature (°C) | Enzymatic Activity | Salinity Resistance | NCBI Accesion Number |
|---|---|---|---|---|---|---|
| DIP-1 | Bacillus sp. | SW | 32 | - | 8 | MZ600240 |
| DIP-2 | Bacillus sp. | SW | 32 | - | 8 | MZ600241 |
| DIP-3 | Bacillus sp. | SW | 32 | AMIL, PROT | 10 | MZ600242 |
| DIP-4 | Bacillus sp. | SW | 32 | PHOS | 8 | MZ600243 |
| DIP-5 | Bacillus sp. | SW | 32 | - | 8 | MZ600244 |
| DIP-6 | Bacillus sp. | SW | 32 | PROT | 16 | MZ600245 |
| DIP-7 | Bacillus sp. | SW | 32 | NIT, PHOS, AMIL, PROT | 10 | MZ600246 |
| DIP-8 | Bacillus sp. | SW | 32 | - | 16 | MZ600247 |
| DIP-9 | Bacillus sp. | SW | 32 | PHOS | 10 | MZ600248 |
| DIP-10 | Bacillus cereus | SW | 32 | PHOS, AMIL, PROT | 12 | MZ600249 |
| DIP-11 | Bacillus sp. | SW | 32 | PHOS, AMIL, PROT | 10 | MZ600250 |
| DIP-12 | Bacillus sp. | SW | 32 | PHOS, AMIL, PROT | 16 | MZ600251 |
| DIP-13 | Bacillus megaterium | SW | 32 | PHOS, AMIL, PROT | 10 | MZ600252 |
| DIP-14 | Bacillus sp. | SW | 32 | PHOS, AMIL, PROT | 16 | MZ600253 |
| DIP-15 | Bacillus sp. | SW | 32 | PHOS, AMIL, PROT | 12 | MZ600254 |
| DIP-16 | Bacillus simplex | SW | 32 | PHOS, AMIL, PROT | 10 | MZ600255 |
| DIP-17 | Bacillus sp. | SW | 32 | PHOS, PROT | 10 | MZ600256 |
| DIP-18 | Bacillus sp. | SW | 32 | PHOS, POT, AMIL, PROT | 16 | MZ600257 |
| DIP-19 | Bacillus megaterium | SW | 32 | PHOS, AMIL, PROT | 16 | MZ600258 |
| DIP-20 | Bacillus sp. | SW | 32 | PHOS, AMIL, PROT | 16 | MZ600259 |
| DIP-21 | Bacillus mycoides | SW | 32 | PHOS, AMIL, PROT | 12 | MZ600260 |
| DIP-22 | Bacillus simplex | SW | 32 | AMIL | 16 | MZ600261 |
| DIP-23 | Bacillus circulans | SW | 32 | PHOS, AMIL, PROT | 14 | MZ600262 |
| DIP-24 | Bacillus aryabhattai | SW | 32 | PHOS, PROT | 16 | MZ600263 |
| DIP-25 | Bacillus sp. | SW | 32 | PHOS, AMIL, PROT | 14 | MZ600264 |
| DIP-26 | Bacillus sp. | SW | 32 | PHOS, AMIL, PROT | 10 | MZ600265 |
| DIP-27 | Bacillus sp. | SW | 32 | PHOS, AMIL, PROT | 12 | MZ600266 |
| DIP-28 | Brevibacillus thermoruber | SW | 60 | PHOS | 12 | MZ600267 |
| DIP-29 | Geobacillus sp. | SW | 60 | - | 10 | MZ600268 |
| DIP-30 | Bacillus sp. | SW | 60 | PHOS | 10 | MZ600269 |
| Isolates | Identification | Enrichment Media | Temperature (°C) | NCBI Accesion Number |
|---|---|---|---|---|
| DIF-1 | Aspergillus sp. | PDB | 15 | MZ602115 |
| DIF-2 | Penicillium chrysogenum | YMB | 15 | MZ602116 |
| DIF-3 | Aspergillus sp. | PDB | 15 | MZ602117 |
| DIF-4 | Aspergillus sydowii | PDB | 15 | MZ602118 |
| DIF-5 | Penicillium sp. | PDB | 15 | MZ602119 |
| DIF-6 | Penicillium sp. | YMB | 32 | MZ602120 |
| DIF-7 | Penicillium sp. | PDB | 32 | MZ602121 |
| DIF-8 | Penicillium crustosum | PDB | 32 | MZ602122 |
| DIF-9 | Exophiala sp. | PDB | 32 | MZ602123 |
| DIF-10 | Penicillium chrysogenum | PDB | 32 | MZ602124 |
| DIF-11 | Penicillium sp. | PDB | 32 | MZ602125 |
| DIF-12 | Penicillium crustosum | YMB | 32 | MZ602126 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vicente, J.; de Celis, M.; Alonso, A.; Marquina, D.; Santos, A. Microbial Communities Present in Hydrothermal Sediments from Deception Island, Antarctica. Microorganisms 2021, 9, 1631. https://doi.org/10.3390/microorganisms9081631
Vicente J, de Celis M, Alonso A, Marquina D, Santos A. Microbial Communities Present in Hydrothermal Sediments from Deception Island, Antarctica. Microorganisms. 2021; 9(8):1631. https://doi.org/10.3390/microorganisms9081631
Chicago/Turabian StyleVicente, Javier, Miguel de Celis, Alejandro Alonso, Domingo Marquina, and Antonio Santos. 2021. "Microbial Communities Present in Hydrothermal Sediments from Deception Island, Antarctica" Microorganisms 9, no. 8: 1631. https://doi.org/10.3390/microorganisms9081631
APA StyleVicente, J., de Celis, M., Alonso, A., Marquina, D., & Santos, A. (2021). Microbial Communities Present in Hydrothermal Sediments from Deception Island, Antarctica. Microorganisms, 9(8), 1631. https://doi.org/10.3390/microorganisms9081631






