Epidemiological Studies of Pan-Azole Resistant Aspergillus fumigatus Populations Sampled during Tulip Cultivation Show Clonal Expansion with Acquisition of Multi-Fungicide Resistance as Potential Driver
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Strain Isolation
2.2. Panel of Clinical Isolates
2.3. Fungicide Sensitivity Testing
2.4. DNA Extractions
2.5. PCR Amplification and Sequencing of Fungicide Resistant Alleles
2.6. Cell Surface Protein Typing
2.7. Microsatellite Typing Based on Short Tandem Repeats in A. fumigatus (STRAf)
3. Results
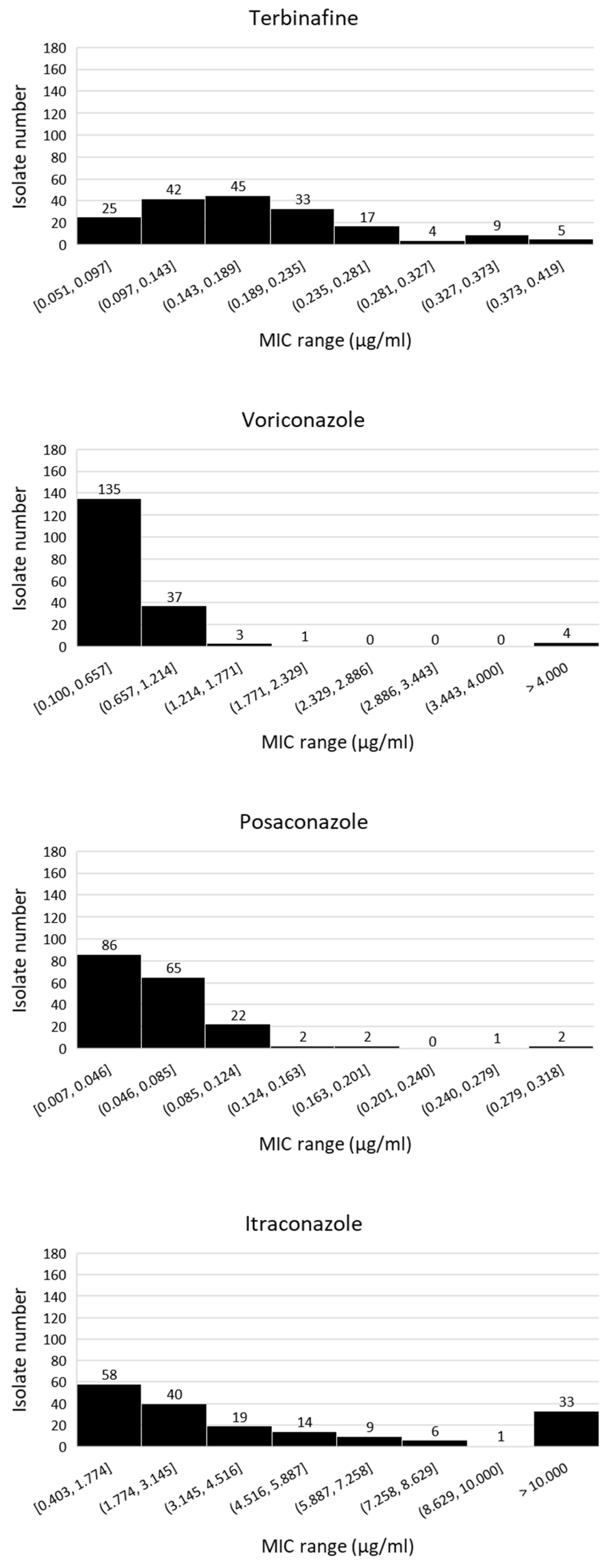
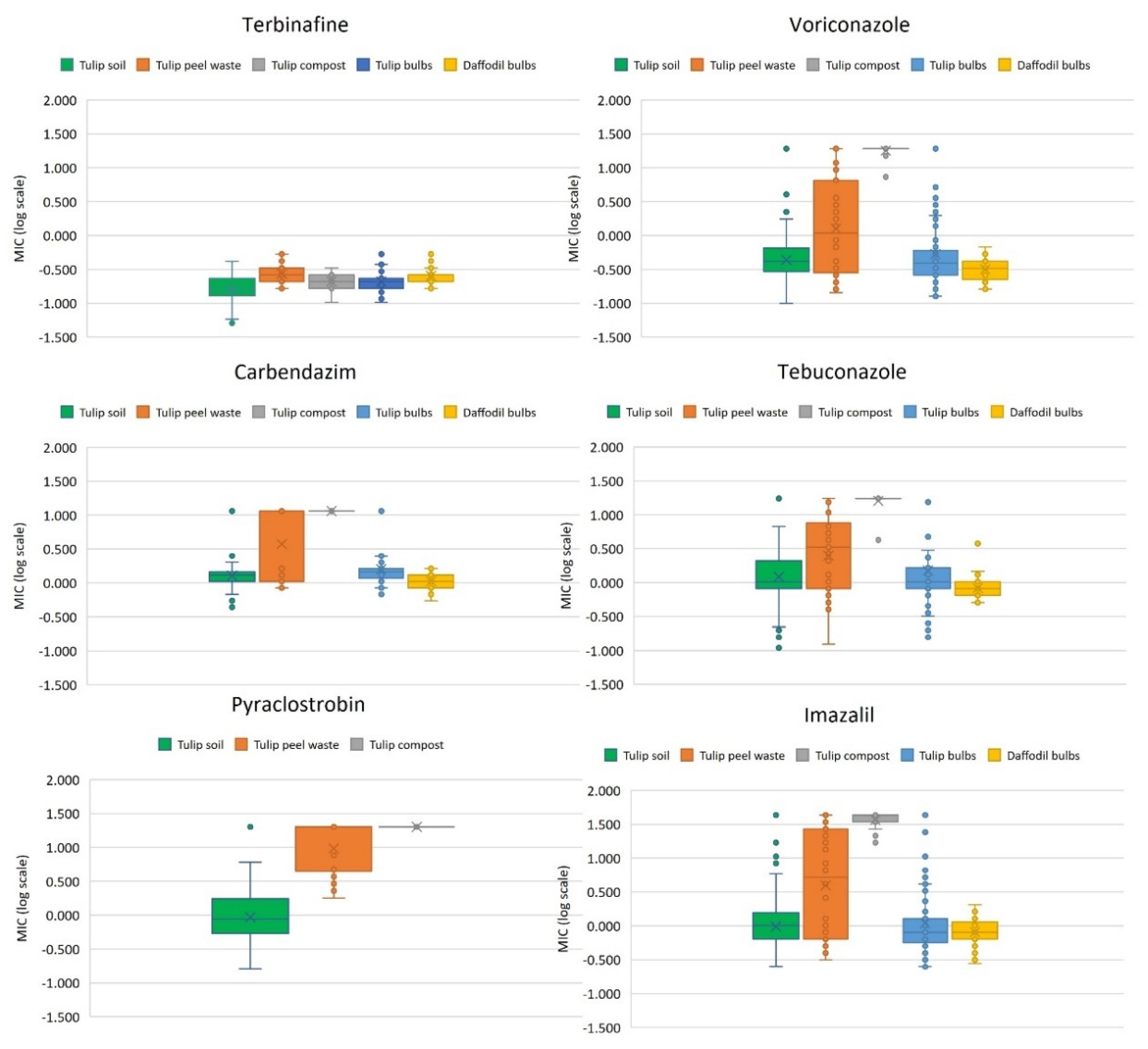
3.1. Isolation and Fungicide Sensitivity Testing of A. fumigatus Isolates from Tulip Field Soils
3.2. Isolation and Fungicide Sensitivity Testing of A. fumigatus Isolates from Flower Bulbs, Tulip Peel Waste Heaps and Compost
3.3. Azole Resistance Phenotype-to-Genotype Relationship, Cell Surface Protein and Mating Typing of A. fumigatus Isolates from the Environment with a Focus on Tulip Cultivation
3.4. Azole Resistance Phenotype-to-Genotype Relationship, Cell Surface Protein and Mating Typing of A. fumigatus Isolates from the Clinical Setting
3.5. Genetic Diversity of Azole-Resistant Aspergillus fumigatus Isolated from the Clinical Setting and the Wider Environment with a Focus on Tulip Cultivation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Denning, D.W.; Venkateswarlu, K.; Oakley, K.L.; Anderson, M.J.; Manning, N.J.; Stevens, D.A.; Warnock, D.W.; Kelly, S.L. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 1997, 41, 1364–1368. [Google Scholar] [CrossRef] [Green Version]
- Snelders, E.; van der Lee, H.A.L.; Kuijpers, J.; Rijs, A.J.M.M.; Varga, J.; Samson, R.A.; Mellado, E.; Donders, A.R.T.; Melchers, W.J.G.; Verweij, P.E. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 2008, 5, e219. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.J.; Cerar, D.; Anderson, M.J.; Albarrag, A.; Fisher, M.C.; Pasqualotto, A.C.; Laverdiere, M.; Arendrup, M.C.; Perlin, D.S.; Denning, D.W. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 2009, 15, 1068–1076. [Google Scholar] [CrossRef]
- Lestrade, P.P.A.; Bentvelsen, R.; Schauwvlieghe, A.F.A.D.; Schalekamp, S.; van der Velden, W.J.F.M.; Kuiper, E.J.; van Paassen, J.; van der Hoven, B.; van der Lee, H.A.; Melchers, W.J.G.; et al. Voriconazole resistance and mortality in invasive aspergillosis: A multicentre retrospective cohort study. Clin. Infect. Dis. 2019, 68, 1463–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraczek, M.G.; Bromley, M.; Buied, A.; Moore, C.B.; Rajendran, R.; Rautemaa, R.; Ramage, G.; Denning, D.W.; Bowyer, P. The crd1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 2013, 68, 1486–1496. [Google Scholar] [CrossRef]
- Hagiwara, D.; Arai, T.; Takahashi, H.; Kusuya, Y.; Watanabe, A.; Kamei, K. Non-cyp51A azole-resistant Aspergillus fumigatus isolates with mutation in HMG-CoA reductase. Emerg. Infect. Dis. 2018, 24, 1889–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Chen, P.; Gao, R.; Li, Y.; Zhang, A.; Liu, F.; Lu, L. Screening and characterization of a non-cyp51A mutation in an Aspergillus fumigatus cox10 strain conferring azole resistance. Antimicrob. Agents Chemother. 2017, 61, e02101-16. [Google Scholar] [CrossRef] [Green Version]
- Rivero-Menendez, O.; Alastruey-Izquierdo, A.; Mellado, E.; Cuenca-Estrella, M. Triazole Resistance in Aspergillus spp.: A Worldwide Problem? J. Fungi 2016, 2, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, S.; Chazli, Y.E.; Babu, A.F.; Coste, A.T. Azole resistance in Aspergillus fumigatus: A consequence of antifungal use in agriculture? Front. Microbiol. 2017, 8, 1024. [Google Scholar] [CrossRef] [Green Version]
- Lazzarini, C.; Esposto, M.C.; Prigitano, A.; Cogliati, M.; De Lorenzis, G.; Tortorano, A.M. Azole resistance in Aspergillus fumigatus clinical isolates from an Italian culture collection. Antimicrob. Agents Chemother. 2016, 60, 682–685. [Google Scholar] [CrossRef] [Green Version]
- Lockhart, S.R.; Frade, J.P.; Etienne, K.A.; Pfaller, M.A.; Diekema, D.J.; Balajee, S.A. Azole resistance in Aspergillus fumigatus isolates from the ARTEMIS global surveillance study is primarily due to the TR/L98H mutation in the cyp51A gene. Antimicrob. Agents Chemother. 2011, 55, 4465–4468. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, Z.; Han, X.; Tian, S.; Zhao, J.; Chen, F.; Su, X.; Zhao, J.; Zou, Z.; Gong, Y.; et al. Elevated MIC Values of Imidazole Drugs against Aspergillus fumigatus Isolates with TR34/L98H/S297T/F495I Mutation. Antimicrob. Agents Chemother. 2018, 62, e01549-17. [Google Scholar] [CrossRef] [Green Version]
- Wiederhold, N.P.; Gil, V.G.; Gutierrez, F.; Lindner, J.R.; Albataineh, M.T.; McCarthy, D.I.; Sanders, C.; Fan, H.; Fothergill, A.W.; Sutton, D.A. First detection of TR34 L98H and TR46 Y121F T289A cyp51 mutations in Aspergillus fumigatus isolates in the United States. J. Clin. Microbiol. 2016, 54, 168–171. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Snelders, E.; Zwaan, B.J.; Schoustra, S.E.; Meis, J.F.; van Dijk, K.; Hagen, F.; van der Beek, M.T.; Kampinga, G.A.; Zoll, J.; et al. A novel environmental azole resistance mutation in Aspergillus fumigatus and a possible role of sexual reproduction in its emergence. MBio 2017, 8, e00791-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Lopez Jimenez, L.; Snelders, E.; Debets, A.J.M.; Rietveld, A.G.; Zwaan, B.J.; Verweij, P.E.; Schoustra, S.E. 2021. Dynamics of Aspergillus fumigatus in azole fungicide-containing plant waste in the Netherlands (2016–2017). Appl. Environ. Microbiol. 2021, 87, e02295-20. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Tashiro, M.; Urano, R.; Kikuchi, M.; Ito, N.; Moriya, E.; Shirahige, T.; Mishima, M.; Takazono, T.; Miyazaki, T.; et al. Characteristics of azole-resistant Aspergillus fumigatus attached to agricultural products imported to Japan. J. Infect. Chemother. 2020, 26, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Fraaije, B.; Atkins, S.; Hanley, S.; Macdonald, A.; Lucas, J. The Multi-Fungicide Resistance Status of Aspergillus fumigatus Populations in Arable Soils and the Wider European Environment. Front. Microbiol. 2020, 11, 599233. [Google Scholar] [CrossRef]
- Kang, S.E.; Sumabat, L.G.; Melie, T.; Mangum, B.; Momany, M.; Brewer, M.T. Evidence for the agricultural origin of antimicrobial resistance in a fungal pathogen of humans. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gonzalez-Jimenez, I.; Garcia-Rubio, R.; Monzon, S.; Lucio, J.; Cuesta, I.; Mellado, E. 2021. Multiresistance to nonazole fungicides in Aspergillus fumigatus TR34/L98H azole-resistant isolates. Antimicrob. Agents Chemother. 2021, 65, e00642-21. [Google Scholar] [CrossRef]
- Lemaire, B.; Normand, A.C.; Forel, J.M.; Cassir, N.; Piarroux, R.; Ranque, S. Hospitalized Patient as Source of Aspergillus fumigatus, 2015. Emerg. Infect. Dis. 2018, 24, 1524–1527. [Google Scholar] [CrossRef]
- Engel, T.G.P.; Erren, E.; Van den Driessche, K.S.J.; Melchers, W.J.G.; Reijers, M.H.; Merkus, P.; Verweij, P.E. Aerosol Transmission of Aspergillus fumigatus in Cystic Fibrosis Patients in the Netherlands. Emerg. Infect. Dis. 2019, 25, 797–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verweij, P.E.; Lucas, J.A.; Arendrup, M.C.; Bowyer, P.; Brinkmann, A.J.F.; Denning, D.W.; Dyer, P.S.; Fisher, M.C.; Geenen, P.L.; Gisi, U.; et al. The one health problem of azole resistance in Aspergillus fumigatus: Current insights and future research agenda. Fungal Biol. Rev. 2020, 34, 202–214. [Google Scholar] [CrossRef]
- Dunne, K.; Hagen, F.; Pomeroy, N.; Meis, J.F.; Rogers, T.R. Intercountry transfer of triazole-resistant Aspergillus fumigatus on plant bulbs. Clin. Infect. Dis. 2017, 65, 147–149. [Google Scholar] [CrossRef] [Green Version]
- Hagiwara, D. Isolation of azole-resistant Aspergillus fumigatus from imported plant bulbs in Japan and the effect of fungicide treatment. J. Pestic. Sci. 2020, 45, 147–150. [Google Scholar] [CrossRef]
- Schoustra, S.E.; Debets, A.J.M.; Rijs, A.J.M.M.; Zhang, J.; Snelders, E.; Leendertse, P.C.; Melchers, W.J.G.; Rietveld, A.G.; Zwaan, B.J.; Verweij, P.E. Environmental hotspots for azole resistance selection of Aspergillus fumigatus, the Netherlands. Emerg. Infect. Dis. 2019, 25, 1347–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klaassen, C.H.W.; de Valk, H.A.; Balajee, S.A.; Meis, J.F.G.M. Utility of CSP typing to sub-type clinical Aspergillus fumigatus isolates and proposal for a new CSP type nomenclature. J. Microbiol. Methods 2009, 77, 292–296. [Google Scholar] [CrossRef]
- De Valk, H.A.; Meis, J.F.G.M.; Curfs, I.M.; Muehlethaler, K.; Mouton, J.W.; Klaassen, C.H.W. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J. Clin. Microbiol. 2005, 43, 4112–4120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sewell, T.R.; Zhu, J.; Rhodes, J.; Hagen, F.; Meis, J.F.; Fisher, M.C.; Jombart, T. Nonrandom distribution of azole resistance across the global population of Aspergillus fumigatus. MBio 2019, 10, e00392-19. [Google Scholar] [CrossRef] [Green Version]
- Balajee, S.A.; Tay, S.T.; Lasker, B.A.; Hurst, S.F.; Rooney, A.P. Characterization of a novel gene for strain typing reveals substructuring of Aspergillus fumigatus across North America. Eukaryot. Cell 2007, 6, 1392–1399. [Google Scholar] [CrossRef] [Green Version]
- De Groot, T.; Meis, J.F. Microsatellite stability in STR analysis Aspergillus fumigatus depends on number of repeat units. Front. Cell Infect. Microbiol. 2019, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
- Bruvo, R.; Michiels, N.K.; D’Souza, T.G.; Schulenburg, H. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol. Ecol. 2004, 13, 2101–2106. [Google Scholar] [CrossRef] [PubMed]
- Toyotome, T.; Hagiwara, D.; Kida, H.; Ogi, T.; Watanabe, A.; Wada, T.; Komatsu, R.; Kamei, K. First clinical isolation report of azole-resistant Aspergillus fumigatus with TR34/L98H-type mutation in Japan. J. Infect. Chemother. 2017, 23, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, R.A.; Morio, F.; Favennec, L.; Dominique, S.; Meis, J.F.; Gargala, G.; Verweij, P.E.; Le Pape, P. First description of azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation in France. Antimicrob. Agents Chemother. 2015, 59, 4331–4335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhary, A.; Sharma, C.; van den Boom, M.; Yntema, J.B.; Hagen, F.; Verweij, P.E.; Meis, J.F. Multi-Azole-Resistant Aspergillus fumigatus in the Environment in Tanzania. J. Antimicrob. Chem. 2014, 69, 2979–2983. [Google Scholar] [CrossRef]
- Fernández-Ortuño, D.; Torés, J.A.; de Vicente, A.; Pérez-García, A. Mechanisms of resistance to QoI fungicides in phytopathogenic fungi. Int. Microbiol. 2008, 11, 1–9. [Google Scholar] [PubMed]
- Chowdhary, A.; Kathuria, S.; Xu, J.; Sharma, C.; Sundar, G.; Singh, P.K.; Gaur, S.N.; Hagen, F.; Klaassen, C.H.; Meis, J.F. Clonal expansion and emergence of environmental multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR₃₄/L98H mutations in the cyp51A gene in India. PLoS ONE 2012, 7, e52871. [Google Scholar] [CrossRef]
- Ahangarkani, F.; Badali, H.; Abbasi, K.; Nabili, M.; Khodavaisy, S.; de Groot, T.; Meis, J.F. Clonal Expansion of Environmental Triazole Resistant Aspergillus fumigatus in Iran. J. Fungi 2020, 6, 199. [Google Scholar] [CrossRef]
- Cho, S.-Y.; Lee, D.-G.; Kim, W.-B.; Chun, H.-S.; Park, C.; Myong, J.-P.; Park, Y.-J.; Choi, J.-K.; Lee, H.-J.; Kim, S.-H.; et al. Epidemiology and antifungal susceptibility profile of Aspergillus species: Comparison between environmental and clinical isolates from patients with hematologic malignancies. J. Clin. Microbiol. 2019, 57, e02023-18. [Google Scholar] [CrossRef] [Green Version]
- Rhodes, J.; Abdolrasouli, A.; Dunne, K.; Sewell, T.R.; Zhang, Y.; Ballard, E.; Brackin, A.P.; van Rhijn, N.; Tsitsopoulou, A.; Posso, R.B.; et al. Tracing patterns of evolution and acquisition of drug resistant Aspergillus fumigatus infection from the environment using population genomics. bioRxiv 2021. [Google Scholar] [CrossRef]
- Shelton, J.M.G.; Collins, R.; Uzzell, C.B.; Alghamdi, A.; Dyer, P.S.; Singer, A.C.; Fisher, M.C. Citizen-science surveillance of triazole-resistant Aspergillus fumigatus in UK residential garden soils. bioRxiv 2021. [Google Scholar] [CrossRef]
- Cools, H.J.; Fraaije, B.A. Update on mechanisms of azole resistance in Mycosphaerella graminicola and implications for future control. Pest Manag. Sci. 2013, 69, 150–155. [Google Scholar] [CrossRef]
- Koenraadt, H.; Somerville, S.C.; Jones, A.L. Characterisation of mutations in the β-tubulin gene of benomyl-resistant field strains of Venturia inaequalis and other plant pathogenic fungi. Phytopathology 1992, 82, 1348–1354. [Google Scholar] [CrossRef]
- Chen, Y.; Dong, F.; Zhao, J.; Fan, H.; Qin, C.; Li, R.; Verweij, P.E.; Zheng, Y.; Han, L. High Azole Resistance in Aspergillus fumigatus Isolates from Strawberry Fields, China, 2018. Emerg. Infect. Dis. 2020, 26, 81–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Rubio, R.; Escribano, P.; Gomez, A.; Guinea, J.; Mellado, E. Comparison of Two Highly Discriminatory Typing Methods to Analyze Aspergillus fumigatus Azole Resistance. Front. Microbiol. 2018, 9, 1626. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, S.; Sewell, T.R.; Valot, B.; Godeau, C.; Laboissiere, A.; Millon, L.; Fisher, M.C. Molecular Epidemiology of Azole-Resistant Aspergillus fumigatus in France Shows Patient and Healthcare Links to Environmentally Occurring Genotypes. Front. Cell. Infect. Microbiol. 2021, 11, 729476. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, Y.; Korfanty, G.A.; Archer, M.; Xu, J. Genome-Wide Association Analysis for Triazole Resistance in Aspergillus fumigatus. Pathogens 2021, 10, 701. [Google Scholar] [CrossRef]
- Slavin, M.A.; Chen, Y.C.; Cordonnier, C.; Cornely, O.A.; Cuenca-Estrella, M.; Donnelly, J.P.; Groll, A.H.; Lortholary, O.; Marty, F.M.; Nucci, M.; et al. When to change treatment of acute invasive aspergillosis: An expert viewpoint. J. Antimicrob. Chemother. 2021, dkab317. [Google Scholar] [CrossRef]


| Sample 1 | Origin and Year | Frequency of Azole Resistant Isolates 2 |
|---|---|---|
| Soil tulip field 1 | Biddinghuizen, NL (2016) | 0/30 |
| Soil tulip field 2 | Dronten, NL (2016) | 1/30 |
| Soil tulip field 3 | Venhuizen, NL (2016) | 1/30 |
| Soil tulip field 4 | Westwoud, NL (2016) | 0/30 |
| Soil tulip field 5 | Westwoud, NL (2016) | 5/30 |
| Soil tulip field 6 | Zyperdijk, NL (2016) | 3/30 |
| Bulbs 1 Tulipa ‘Stresa’ | Noordwijkerhout, NL (2015) | 4/10 |
| Bulbs 2 Tulipa ‘Apeldoorn Yellow’ | Vaassen, NL (2015) | 4/10 |
| Bulbs 3 Tulipa greigii ‘Roodkapje’ | Vaassen, NL (2015) | 4/10 |
| Bulbs 4 Tulipa ‘Praestans Shogun’ | Vaassen, NL (2015) | 3/10 |
| Bulbs 5 Tulipa ‘Claudia’ | Noordwijkerhout, NL (2015) | 6/10 |
| Bulbs 6 Tulipa ‘Triumph Hotpants’ | Millbrook, UK (2015) | 1/8 |
| Bulbs 7 Tulipa ‘Mickey Mouse’ | Canterbury, UK (2015) | 1/10 |
| Bulbs 8 Tulipa ‘Gavota’ | Wickford, UK (2015) | 0/10 |
| Bulbs 9 Tulipa ‘Guiseppi Verdi’ | Horsham, UK (2015) | 0/10 |
| Bulbs 10 Tulipa ‘White Marvel’ | Preston, UK (2015) | 1/10 |
| Bulbs 11 Tulipa ‘Red Impression’ | Hillegom, NL (2015) | 1/10 |
| Bulbs 12 Tulipa ‘Negrita’ | Vaassen, NL (2017) | 0/10 |
| Bulbs 13 Tulipa ‘Rembrand’ | Vaassen, NL (2017) | 0/10 |
| Bulbs 14 Narcissus ‘Pink Pride’ | Vaassen, NL (2017) | 0/10 |
| Bulbs 15 Narcissus ‘Jetfire’ | Vaassen, NL (2017) | 0/10 |
| Bulbs 16 Narcissus mix | Hillegom, NL (2017) | 0/10 |
| Bulb peel waste heap tulip grower 1 | NL (2018) | 6/11 |
| Bulb peel waste heap tulip grower 2 | NL (2018) | 1/1 |
| Bulb peel waste heap tulip grower 3 | NL (2018) | 1/2 |
| Bulb peel waste heap tulip grower 4 | NL (2018) | 3/4 |
| Bulb peel waste heap tulip grower 5 | NL (2018) | 11/19 |
| Bulb peel waste heap tulip grower 6 | NL (2018) | 1/5 |
| Compost heap tulip grower A | NL (2018) | 8/8 |
| Compost heap tulip grower B | NL (2018) | 1/1 |
| Compost heap tulip grower C | NL (2018) | 10/10 |
| Isolate | CYP51A | VRC | IMA | TEB | CAR | PYR | BOS | CSP | Mating Type |
|---|---|---|---|---|---|---|---|---|---|
| STNL1-A8 | WT | 0.292 | 0.714 | 0.814 | >11.464 | 1.110 | - | t04A | MAT1-1 |
| T8-2 | WT | 0.471 | 0.803 | 0.643 | 1.169 | 0.872 | 0.253 | t11 | MAT1-1 |
| STNL5-C5 | TR34/L98H/S297T/F495I | 0.531 | 8.367 | 4.227 | >11.464 | 0.685 | 0.199 | t02 | MAT1-2 |
| T6-3 | TR34/L98H | 0.675 | 5.236 | >17.349 | 2.013 | 0.477 | - | t11 | MAT1-1 |
| STNL2-C9 | F46Y/M172V/E427K | 0.760 | 1.443 | 2.970 | 0.843 | 6.017 | - | t02B | MAT1-1 |
| STNL3-C8 | TR34/L98H | 0.857 | 1.283 | 4.227 | 1.303 | >20.120 | - | t04B | MAT1-1 |
| TP UT1A-2 | TR34/L98H | 0.857 | 8.367 | 5.348 | >11.464 | >20.120 | 0.253 | t02 | MAT1-2 |
| STNL6-B2 | TR34/L98H | 1.381 | 2.592 | 4.227 | 1.169 | 0.538 | 0.157 | - | MAT1-1 |
| STNL6-A3 | TR34/L98H | 1.381 | 3.277 | 6.768 | 1.303 | 0.538 | 0.199 | t01 | MAT1-2 |
| T3-6 | TR34/L98H | 1.381 | 6.619 | >17.349 | 2.013 | 1.110 | 0.253 | t11 | MAT1-1 |
| TP UT1B-1 | TR34/L98H | 1.754 | 6.619 | >17.349 | >11.464 | >20.120 | 0.517 | t11 | MAT1-1 |
| TP UT5C-5 | TR34/L98H | 1.754 | 6.619 | 4.227 | 0.679 | >20.12 | 0.224 | t02 | MAT1-2 |
| STNL2-B8 | TR34/L98H | 1.754 | 10.577 | 4.227 | 2.013 | 0.294 | 0.321 | t04B | MAT1-1 |
| STNL5-B6 | TR34/L98H | 2.227 | 1.622 | 3.341 | 1.619 | 0.872 | 0.321 | t11 | MAT1-2 |
| T2-1 | TR34/L98H | 2.227 | 4.142 | >17.349 | 1.619 | 1.110 | 0.321 | t11 | MAT1-1 |
| STNL6-B1 | TR34/L98H | 4.047 | 5.887 | >17.349 | >11.464 | >20.120 | >18.469 | t01 | MAT1-2 |
| T11-8 | TR34/L98H | 5.139 | 10.577 | >17.349 | >11.464 | >20.120 | 0.517 | t02 | MAT1-1 |
| STNL5-B7 | TR46/Y121F/T289A | >19.120 | 16.901 | >17.349 | >11.464 | >20.120 | 0.157 | t02 | MAT1-1 |
| T7-9 | TR46/Y121F/T289A | >19.120 | 24.020 | >17.349 | >11.464 | >20.120 | - | t01 | MAT1-1 |
| TP UT4A-1 | TR46/Y121F/T289A | >19.120 | 34.138 | >17.349 | >11.464 | >20.120 | 0.407 | t01 | - |
| STNL5-C1 | TR46/Y121F/T289A | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | 0.177 | t06A | MAT1-1 |
| STNL5-C8 | TR46/Y121F/T289A | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | 0.199 | - | MAT1-2 |
| T3-5 | TR46/Y121F/T289A | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | - | t01 | MAT1-1 |
| T4-9 | TR46/Y121F/T289A | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | - | t01 | MAT1-1 |
| T4-10 | TR46/Y121F/T289A | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | - | t01 | MAT1-1 |
| T5-1 | TR46/Y121F/T289A | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | 0.285 | t01 | MAT1-1 |
| T5-2 | TR46/Y121F/T289A | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | - | t01 | MAT1-1 |
| T5-5 | TR46/Y121F/T289A | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | - | t01 | MAT1-1 |
| TP TEB5C-2 | TR34/L98H/T289A/I364V/G448S | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | 0.321 | t02 | MAT1-2 |
| TC TEB6B-1 | TR34/L98H/T289A/I364V/G448S | >19.120 | >43.153 | >17.349 | >11.464 | >20.120 | >18.469 | t02 | MAT1-2 |
| Isolate | CYP51A | VOR | IMA | TEB | CAR | PYR | BOS | CSP | Mating Type |
|---|---|---|---|---|---|---|---|---|---|
| AF65 | WT | 0.329 | 1.141 | 0.814 | 1.619 | 1.413 | 0.157 | t02 | MAT1-2 |
| AF293 | F46Y/M172V/N284T/D255E/E427K | 0.531 | 2.915 | 1.649 | 1.303 | 0.538 | - | t06A | MAT1-2 |
| ARAF013 | TR34/L98H/S297T/F495I | 0.760 | 13.370 | 4.227 | >11.464 | 0.332 | 0.253 | t11 | MAT1-2 |
| Asp 251 | TR34/L98H/S297T/F495I | 1.088 | 19.002 | 5.348 | >11.464 | 1.413 | 0.157 | t02 | MAT1-1 |
| CYP_15_63 | TR34/L98H/S297T/F495I | 1.088 | 13.370 | 6.768 | >11.464 | 1.110 | 1.702 | t01 | MAT1-1 |
| D007 | TR34/L98H/S297T/F495I | 1.088 | 15.032 | 3.341 | 0.843 | 0.100 | 0.285 | t04A | MAT1-1 |
| CXH_07 | TR34/L98H | 1.381 | 5.887 | 6.768 | 1.303 | 0.332 | 0.199 | t04A | MAT1-1 |
| Asp 267 | TR34/L98H | 1.381 | 4.142 | 4.227 | 1.619 | 0.161 | 0.157 | t11 | MAT1-2 |
| CXH_06 | TR34/L98H | 1.557 | 5.236 | 8.563 | 1.303 | 0.423 | 0.321 | t04B | MAT1-2 |
| Asp 164 | TR34/L98H | 1.976 | 6.619 | 6.768 | 1.619 | 1.110 | 0.224 | t11 | MAT1-2 |
| Asp 168 | TR34/L98H | 1.976 | 5.236 | 8.563 | 2.013 | 0.607 | 0.199 | t04B | MAT1-2 |
| OKH50 | TR34/L98H | 2.227 | 4.142 | 5.348 | 0.757 | 0.332 | 0.407 | t02 | MAT1-2 |
| ARAF017 | TR34/L98H | 4.560 | 13.370 | >17.349 | >11.464 | >20.12 | >18.469 | t04A | MAT1-2 |
| CYP_15_46 | TR34/L98H | 5.791 | 8.367 | 13.711 | >11.464 | >20.12 | >18.469 | t02 | MAT1-1 |
| CYP_15_80 | TR46/Y121F/T289A | 9.338 | >43.153 | 8.563 | >11.464 | >20.12 | 0.199 | t02 | MAT1-1 |
| CYP_15_2 | TR46/Y121F/T289A | >19.12 | >43.153 | 13.711 | >11.464 | >20.12 | 0.253 | t01 | MAT1-2 |
| CYP_15_7 | TR46/Y121F/T289A | >19.12 | >43.153 | >17.349 | >11.464 | >20.12 | 0.253 | t01 | MAT1-2 |
| CYP_15_38 | TR46/Y121F/T289A | >19.12 | >43.153 | >17.349 | >11.464 | >20.12 | >18.469 | t09 | MAT1-2 |
| V093-26 | TR46/Y121F/T289A | >19.12 | 27.006 | 8.563 | >11.464 | >20.12 | 0.517 | t01 | MAT1-2 |
| V094-54 | TR46/Y121F/T289A | >19.12 | >43.153 | >17.349 | >11.464 | >20.12 | 0.285 | t01 | MAT1-2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fraaije, B.A.; Atkins, S.L.; Santos, R.F.; Hanley, S.J.; West, J.S.; Lucas, J.A. Epidemiological Studies of Pan-Azole Resistant Aspergillus fumigatus Populations Sampled during Tulip Cultivation Show Clonal Expansion with Acquisition of Multi-Fungicide Resistance as Potential Driver. Microorganisms 2021, 9, 2379. https://doi.org/10.3390/microorganisms9112379
Fraaije BA, Atkins SL, Santos RF, Hanley SJ, West JS, Lucas JA. Epidemiological Studies of Pan-Azole Resistant Aspergillus fumigatus Populations Sampled during Tulip Cultivation Show Clonal Expansion with Acquisition of Multi-Fungicide Resistance as Potential Driver. Microorganisms. 2021; 9(11):2379. https://doi.org/10.3390/microorganisms9112379
Chicago/Turabian StyleFraaije, Bart A., Sarah L. Atkins, Ricardo F. Santos, Steven J. Hanley, Jonathan S. West, and John A. Lucas. 2021. "Epidemiological Studies of Pan-Azole Resistant Aspergillus fumigatus Populations Sampled during Tulip Cultivation Show Clonal Expansion with Acquisition of Multi-Fungicide Resistance as Potential Driver" Microorganisms 9, no. 11: 2379. https://doi.org/10.3390/microorganisms9112379
APA StyleFraaije, B. A., Atkins, S. L., Santos, R. F., Hanley, S. J., West, J. S., & Lucas, J. A. (2021). Epidemiological Studies of Pan-Azole Resistant Aspergillus fumigatus Populations Sampled during Tulip Cultivation Show Clonal Expansion with Acquisition of Multi-Fungicide Resistance as Potential Driver. Microorganisms, 9(11), 2379. https://doi.org/10.3390/microorganisms9112379







