Reliable Identification of Environmental Pseudomonas Isolates Using the rpoD Gene
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
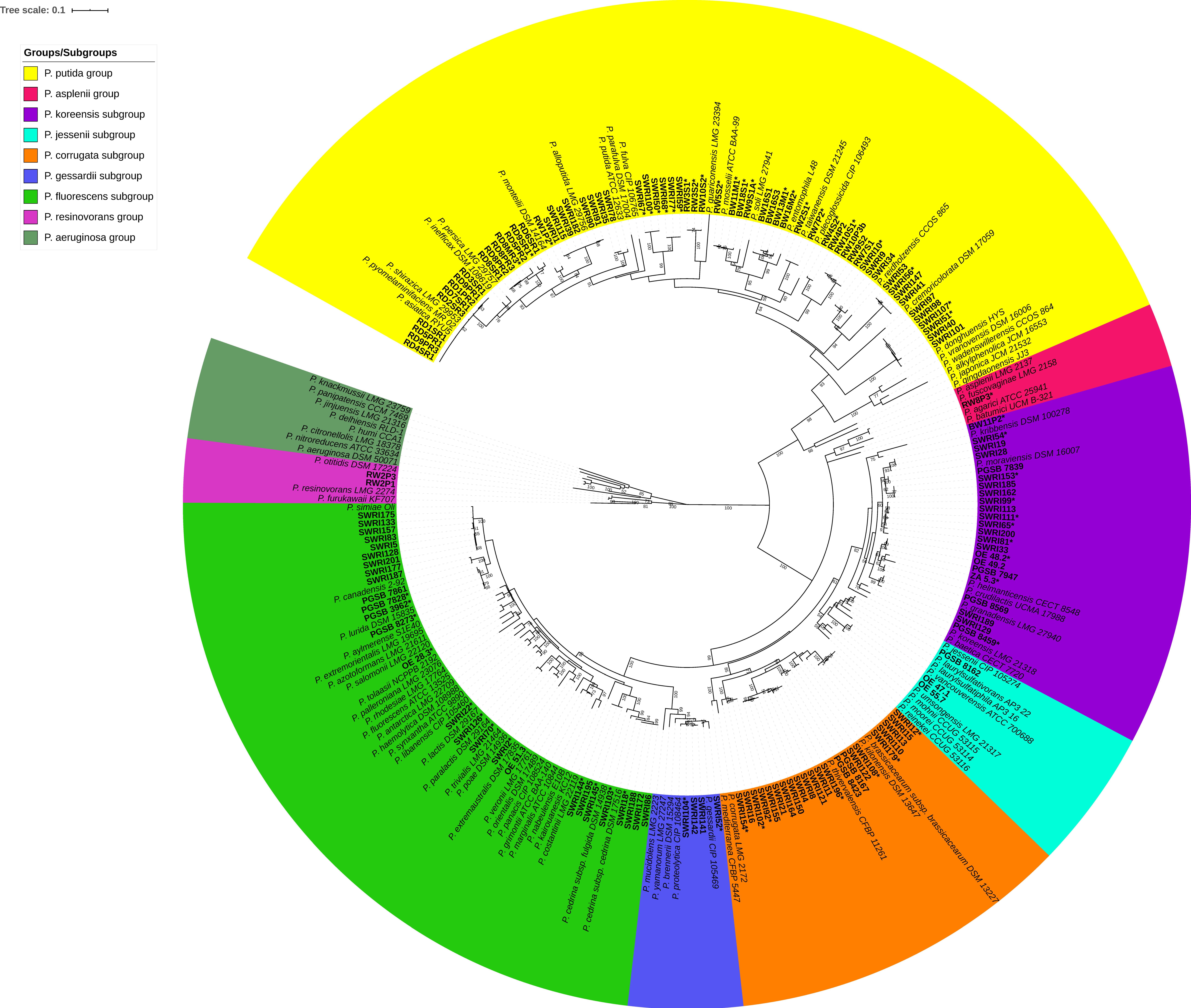
3.1. rpoD Phylogeny
3.2. Species Delineation
3.3. Identification of Environmental Isolates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bollinger, A.; Thies, S.; Katzke, N.; Jaeger, K. The biotechnological potential of marine bacteria in the novel lineage of Pseudomonas pertucinogena. Microb. Biotechnol. 2020, 13, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höfte, M.; De Vos, P. Plant pathogenic Pseudomonas species. In Plant-Associated Bacteria; Gnanamanickam, S.S., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 507–533. [Google Scholar]
- Wiklund, T. Pseudomonas anguilliseptica infection as a threat to wild and farmed fish in the Baltic Sea. Microbiol. Aust. 2016, 37, 135. [Google Scholar] [CrossRef] [Green Version]
- Beaton, A.; Lood, C.; Cunningham-Oakes, E.; MacFadyen, A.; Mullins, A.J.; Bestawy, W.E.; Botelho, J.; Chevalier, S.; Coleman, S.; Dalzell, C.; et al. Community-led comparative genomic and phenotypic analysis of the aquaculture pathogen Pseudomonas baetica a390T sequenced by Ion semiconductor and Nanopore technologies. FEMS Microbiol. Lett. 2018, 365, fny069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasi, S.; Tabrez, S.; Ahmad, M. Use of Pseudomonas spp. for the bioremediation of environmental pollutants: A review. Environ. Monit. Assess. 2013, 185, 8147–8155. [Google Scholar] [CrossRef] [PubMed]
- Weller, D.M. Pseudomonas biocontrol agents of soilborne pathogens: Looking back over 30 years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef] [Green Version]
- Gross, H.; Loper, J.E. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef]
- Mende, D.R.; Sunagawa, S.; Zeller, G.; Bork, P. Accurate and universal delineation of prokaryotic species. Nat. Methods 2013, 10, 881–884. [Google Scholar] [CrossRef]
- Moore, E.R.B.; Mau, M.; Arnscheidt, A.; Böttger, E.C.; Hutson, R.A.; Collins, M.D.; Van De Peer, Y.; De Wachter, R.; Timmis, K.N. The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto and estimation of the natural intrageneric relationships. Syst. Appl. Microbiol. 1996, 19, 478–492. [Google Scholar] [CrossRef]
- Brosch, R.; Lefèvre, M.; Grimont, F.; Grimont, P.A.D. Taxonomic diversity of Pseudomonads revealed by computer-interpretation of ribotyping data. Syst. Appl. Microbiol. 1996, 19, 541–555. [Google Scholar] [CrossRef]
- Lalucat, J.; Mulet, M.; Gomila, M.; García-Valdés, E. Genomics in bacterial taxonomy: Impact on the Genus Pseudomonas. Genes 2020, 11, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Bodilis, J.; Nsigue-Meilo, S.; Besaury, L.; Quillet, L. Variable copy number, intra-genomic heterogeneities and lateral transfers of the 16S rRNA gene in Pseudomonas. PLoS ONE 2012, 7, e35647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [PubMed] [Green Version]
- Gomila, M.; Peña, A.; Mulet, M.; Lalucat, J.; García-Valdés, E. Phylogenomics and systematics in Pseudomonas. Front. Microbiol. 2015, 6, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, L.D.; Davis, E.W.; Pereira e Silva, M.d.C.; Weisberg, A.J.; Bresciani, L.; Chang, J.H.; Loper, J.E.; Andreote, F.D. Tropical soils are a reservoir for fluorescent Pseudomonas spp. biodiversity. Environ. Microbiol. 2018, 20, 62–74. [Google Scholar] [CrossRef]
- Biessy, A.; Novinscak, A.; Blom, J.; Léger, G.; Thomashow, L.S.; Cazorla, F.M.; Josic, D.; Filion, M. Diversity of phytobeneficial traits revealed by whole-genome analysis of worldwide-isolated phenazine-producing Pseudomonas spp. Environ. Microbiol. 2019, 21, 437–455. [Google Scholar] [CrossRef] [Green Version]
- Oni, F.E.; Geudens, N.; Omoboye, O.O.; Bertier, L.; Hua, H.G.K.; Adiobo, A.; Sinnaeve, D.; Martins, J.C.; Höfte, M. Fluorescent Pseudomonas and cyclic lipopeptide diversity in the rhizosphere of cocoyam (Xanthosoma sagittifolium). Environ. Microbiol. 2019, 21, 1019–1034. [Google Scholar] [CrossRef] [Green Version]
- Keshavarz-Tohid, V.; Vacheron, J.; Dubost, A.; Prigent-Combaret, C.; Taheri, P.; Tarighi, S.; Taghavi, S.M.; Moënne-Loccoz, Y.; Muller, D. Genomic, phylogenetic and catabolic re-assessment of the Pseudomonas putida clade supports the delineation of Pseudomonas alloputida sp. nov., Pseudomonas inefficax sp. nov., Pseudomonas persica sp. nov., and Pseudomonas shirazica sp. nov. Syst. Appl. Microbiol. 2019, 42, 468–480. [Google Scholar] [CrossRef]
- Camiade, M.; Bodilis, J.; Chaftar, N.; Riah-Anglet, W.; Gardères, J.; Buquet, S.; Ribeiro, A.F.; Pawlak, B. Antibiotic resistance patterns of Pseudomonas spp. isolated from faecal wastes in the environment and contaminated surface water. FEMS Microbiol. Ecol. 2020, 96, fiaa008. [Google Scholar] [CrossRef]
- Tran, P.N.; Savka, M.A.; Gan, H.M. In-silico taxonomic classification of 373 genomes reveals species misidentification and new genospecies within the genus Pseudomonas. Front. Microbiol. 2017, 8, 1296. [Google Scholar] [CrossRef] [PubMed]
- Mulet, M.; Bennasar, A.; Lalucat, J.; García-Valdés, E. An rpoD-based PCR procedure for the identification of Pseudomonas species and for their detection in environmental samples. Mol. Cell. Probe. 2009, 23, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Mulet, M.; David, Z.; Nogales, B.; Bosch, R.; Lalucat, J.; García-Valdés, E. Pseudomonas diversity in crude-oil-contaminated intertidal sand samples obtained after the prestige oil spill. Appl. Environ. Microbiol. 2011, 77, 1076–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkinson, N.; Bryant, R.; Bew, J.; Elphinstone, J. Rapid phylogenetic identification of members of the Pseudomonas syringae species complex using the rpoD locus. Plant Pathol. 2011, 60, 338–344. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Ghequire, M.G.K.; Swings, T.; Michiels, J.; Gross, H.; De Mot, R. Draft genome sequence of Pseudomonas putida BW11M1, a banana rhizosphere isolate with a diversified antimicrobial armamentarium. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.D.; Melnik, A.V.; Koyama, N.; Lu, X.; Schorn, M.; Fang, J.; Aguinaldo, K.; Lincecum, T.L.; Ghequire, M.G.K.; Carrion, V.J.; et al. Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat. Microbiol. 2016, 2, 16197. [Google Scholar] [CrossRef] [Green Version]
- Zarvandi, S.; Bahrami, T.; Pauwels, B.; Asgharzadeh, A.; Hosseini-Mazinani, M.; Salari, F.; Girard, L.; De Mot, R.; Rokni-Zadeh, H. Draft genome sequence of cyclic lipopeptide producer Pseudomonas sp. SWRI103 isolated from wheat rhizosphere. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tohya, M.; Watanabe, S.; Tada, T.; Tin, H.H.; Kirikae, T. Genome analysis-based reclassification of Pseudomonas fuscovaginae and Pseudomonas shirazica as later heterotypic synonyms of Pseudomonas asplenii and Pseudomonas asiatica, respectively. Int. J. Syst. Evol. Microbiol. 2020, 70, 3547–3552. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Kolthoff, J.P.; Klenk, H.-P.; Göker, M. Taxonomic use of DNA G+C content and DNA–DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [Green Version]
- Hesse, C.; Schulz, F.; Bull, C.T.; Shaffer, B.T.; Yan, Q.; Shapiro, N.; Hassan, K.A.; Varghese, N.; Elbourne, L.D.H.; Paulsen, I.T.; et al. Genome-based evolutionary history of Pseudomonas spp. Environ. Microbiol. 2018, 20, 2142–2159. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Vlassak, K.; Van Holm, L.; Duchateau, L.; Vanderleyden, J.; De Mot, R. Isolation and characterization of fluorescent Pseudomonas associated with the roots of rice and banana grown in Sri Lanka. Plant Soil 1992, 145, 51–63. [Google Scholar] [CrossRef]
- Rasouli Sadaghiani, M.H.; Khavazi, K.; Rahimian, H.; Malakouti, M.J.; Asadi Rahmani, H. An evaluation of the potentials of indigenous fluorescent Pseudomonads of wheat rhizosphere for producing siderophore. Iran. J. Soil Water Sci. 2006, 20, 133–143. [Google Scholar]

| Pseudomonas Species | rpoD | ANIb | Reclassification | |
|---|---|---|---|---|
| P. gallaeciensis | P. abyssi | 99.24 | 97.56 | P. gallaeciensis |
| P. citronellolis | P. humi | 99.38 | 96.7 | P. citronellolis |
| P. flexibilis | P. tuomuerensis | 98.01 | 98.69 | P. flexibilis |
| P. fluvialis | P. pharmacofabricae | 98.44 | 98.61 | P. fluvialis |
| P. chengduensis | P. sihuiensis | 98.79 | 96.24 | P. chengduensis |
| P. oleovoransa | P. pseudoalcaligenesb | 99.85 | 97.16 | P. oleovorans |
| P. luteola | P. zeshuii | 98.92 | 97.87 | P. luteola |
| P. asiatica | P. pyomelaninifaciens | 100 | 99.03 | P. asiatica |
| P. shirazica | 99.85 | 99.17 | ||
| P. amygdali | P. ficuserectae | 99.08 | 97.42 | P. amygdali |
| P. meliae | 99.23 | 98.27 | ||
| P. savastanoi | 99.54 | 98.75 | ||
| P. asplenii | P. fuscovaginae | 99.38 | 98.23 | P. asplenii |
| Pseudomonas Species | rpoD | 4-Genes MLSA | ANIb | Reclassification | |
|---|---|---|---|---|---|
| P. chengduensis | P. toyotomiensis | 98.76 | 97.74 | 94.48 | - |
| P. indoloxidans | P. oleovoransa | 98.61 | 97.51 | 95.78 c | - |
| P. pseudoalcaligenesb | 98.45 | 97.48 | 95.42 c | - | |
| P. chloritidismutans | P. kunmingensis | 94.79 | 98.24 | 96.47 d | P. chloritidismutans |
| P. oryzihabitans | P. psychrotolerans | 97.4 | 98.60 | 98.22 | P. oryzihabitans |
| P. grimontii | P. marginalis | 99.23 | 98.41 | 93.46 | - |
| P. panacis | 98.31 | 97.78 | 88.20 | - | |
| P. veronii | P. panacis | 94.3 | 95.96 | 99.95 | P. veronii |
| P. tremae | P. coronafaciens | 92.45 | 93.86 | 98.73 | P. tremae |
| P. tremae | P. amygdali | 99.54 | 99.20 | 85.85 | - |
| P. ficuserectae | 98.92 | 97.72 | 85.74 | - | |
| P. meliae | 99.08 | 98.40 | 85.99 | - | |
| P. savastanoi | 99.38 | 97.58 | 85.91 | - | |
| P. libanensis | P. synxantha | 98.15 | 98.49 | 95.25 c | - |
| P. guguanensis | P. mendocina | 98.14 | 94.99 | 89.21 | - |
| Group/Subgroup | Strain | Closest-Related Strain | rpoD | ANIb | Taxonomic Affiliation |
|---|---|---|---|---|---|
| P. putida group | RD8MR3 | RD9SR1 | 97.84 | 94.70 | Pseudomonas sp. #1 |
| RD9SR1 | RD8MR3 | 97.84 | 94.70 | Pseudomonas sp. #2 | |
| RW1P2 | P. monteilii | 98.00 | 95.77 * | Pseudomonas sp. #3 | |
| SWRI67 | SWRI68 | 93.03 | <95 | Pseudomonas sp. #4 | |
| SWRI100 | SWRI77 | ||||
| SWRI50 | SWRI59 | ||||
| SWRI68 | SWRI67 | 93.03 | <95 | Pseudomonas sp. #5 | |
| SWRI77 | SWRI100 | ||||
| SWRI59 | SWRI50 | ||||
| RW3S1 | BW13M1 | 84.07 | 85.36 | Pseudomonas sp. #6 | |
| RW3S2 | 84.07 | 85.32 | |||
| RW10S2 | 83.92 | 85.44 | |||
| RW5S2 | P. mosselii | 99.85 | 99.01 | P. mosselii | |
| BW11M1 | 99.69 | 99.17 | |||
| BW18S1 | 99.23 | 97.36 | |||
| RW9S1A | BW18S1 | 92.94 | 89.14 | Pseudomonas sp. #7 | |
| BW13M1 | BW18S1 | 91.06 | 94.81 | Pseudomonas sp. #8 | |
| BW16M2 | 90.91 | 94.62 | |||
| RW2S1 | P. taiwanensis | 100 | 99.63 | P. taiwanensis | |
| RW7P2 | 100 | 99.67 | |||
| RW4S2 | P. plecoglossicida | 85.69 | 87.10 | Pseudomonas sp. #9 | |
| RW10S1 | RW4S2 | 89.52 | 86.57 | Pseudomonas sp. #10 | |
| SWRI10 | P. reidholzensis | 92.91 | 86.65 | Pseudomonas sp. #11 | |
| SWRI56 | P. reidholzensis | 85.56 | 85.12 | Pseudomonas sp. #12 | |
| SWRI107 | BW13M1 | 83.82 | 85.62 | Pseudomonas sp. #13 | |
| SWRI51 | 83.82 | 85.66 | |||
| P. asplenii group | RW8P3 | P. asplenii | 89.55 | 88.26 | Pseudomonas sp. #14 |
| P. koreensis subgroup | BW11P2 | P. kribbensis | 91.06 | 91.61 | Pseudomonas sp. #15 |
| SWRI54 | P. moraviensis | 96.28 | 91.95 | Pseudomonas sp. #16 | |
| SWRI153 | SWRI54 | 95.05 | 89 | Pseudomonas sp. #17 | |
| SWRI99 | SWRI54 | 93.99 | 88.44 | Pseudomonas sp. #18 | |
| SWRI111 | 94.58 | 88.56 | |||
| SWRI65 | 94.43 | 88.57 | |||
| SWRI81 | 93.99 | 88.55 | |||
| OE 48.2 | P. crudilactis | 96.74 | 91.60 | Pseudomonas sp. #19 | |
| ZA 5.3 | P. crudilactis | 96.89 | 93.47 | Pseudomonas sp. #20 | |
| PGSB 8459 | P. koreensis | 94.61 | 89.11 | Pseudomonas sp. #21 | |
| P. corrugata subgroup | SWRI12 | P. thivervalensis | 95.20 | 92.84 | Pseudomonas sp. #22 |
| SWRI179 | 95.20 | 92.85 | |||
| SWRI108 | P. kilonensis | 95.41 | 95.73* | Pseudomonas sp. #23 | |
| SWRI196 | P. kilonensis | 92.15 | 88.02 | Pseudomonas sp. #24 | |
| SWRI92 | P. kilonensis | 91.69 | 87.92 | Pseudomonas sp. #25 | |
| SWRI102 | 91.69 | 87.89 | |||
| SWRI154 | 90.92 | 87.66 | |||
| P. gessardii subgroup | SWRI52 | P. gessardii | 96.92 | 92.50 | Pseudomonas sp. #26 |
| SWRI104 | P. proteolytica | 99.38 | 98.35 | P. proteolytica | |
| P. fluorescens subgroup | SWRI18 | P. cedrina subsp. cedrina | 98.92 | 96.94 | P. cedrina subsp. cedrina |
| SWRI103 | P. cedrina subsp. cedrina | 98.61 | 94.19 | Pseudomonas sp. #27 | |
| SWRI145 | P. cedrina subsp. cedrina | 93.07 | 88.51 | Pseudomonas sp. #28 | |
| SWRI144 | 93.07 | 88.46 | |||
| SWRI2 | P. poae | 99.54 | 98.85 | P. poae | |
| SWRI70 | P. paralactis | 99.23 | 98.54 | P. paralactis | |
| SWRI126 | P. lactis | 98.92 | 95.09* | Pseudomonas sp. #29 | |
| SWRI22 | P. lactis | 97.99 | 94.45 | Pseudomonas sp. #30 | |
| OE 28.3 | P. salomonii | 98.31 | 94.10 | Pseudomonas sp. #31 | |
| PGSB 8273 | P. lurida | 99.85 | 99.19 | P. lurida | |
| PGSB 3962 | 99.69 | 99.26 | |||
| PGSB 7828 | 99.54 | 99.27 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girard, L.; Lood, C.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. Reliable Identification of Environmental Pseudomonas Isolates Using the rpoD Gene. Microorganisms 2020, 8, 1166. https://doi.org/10.3390/microorganisms8081166
Girard L, Lood C, Rokni-Zadeh H, van Noort V, Lavigne R, De Mot R. Reliable Identification of Environmental Pseudomonas Isolates Using the rpoD Gene. Microorganisms. 2020; 8(8):1166. https://doi.org/10.3390/microorganisms8081166
Chicago/Turabian StyleGirard, Léa, Cédric Lood, Hassan Rokni-Zadeh, Vera van Noort, Rob Lavigne, and René De Mot. 2020. "Reliable Identification of Environmental Pseudomonas Isolates Using the rpoD Gene" Microorganisms 8, no. 8: 1166. https://doi.org/10.3390/microorganisms8081166
APA StyleGirard, L., Lood, C., Rokni-Zadeh, H., van Noort, V., Lavigne, R., & De Mot, R. (2020). Reliable Identification of Environmental Pseudomonas Isolates Using the rpoD Gene. Microorganisms, 8(8), 1166. https://doi.org/10.3390/microorganisms8081166






