Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Cultivation of Strains
2.2. 16. S-23S rRNA Gene ITS and 16S rRNA Gene Sequencing
2.3. Genome Sequencing
2.4. Phenotypic Analysis
2.5. Fatty Acid Analysis
2.6. Antimicrobial Resistance
2.7. Bioinformatics
3. Results and Discussion
3.1. Isolation and Basic Characterization of Strains AV382 and AV436
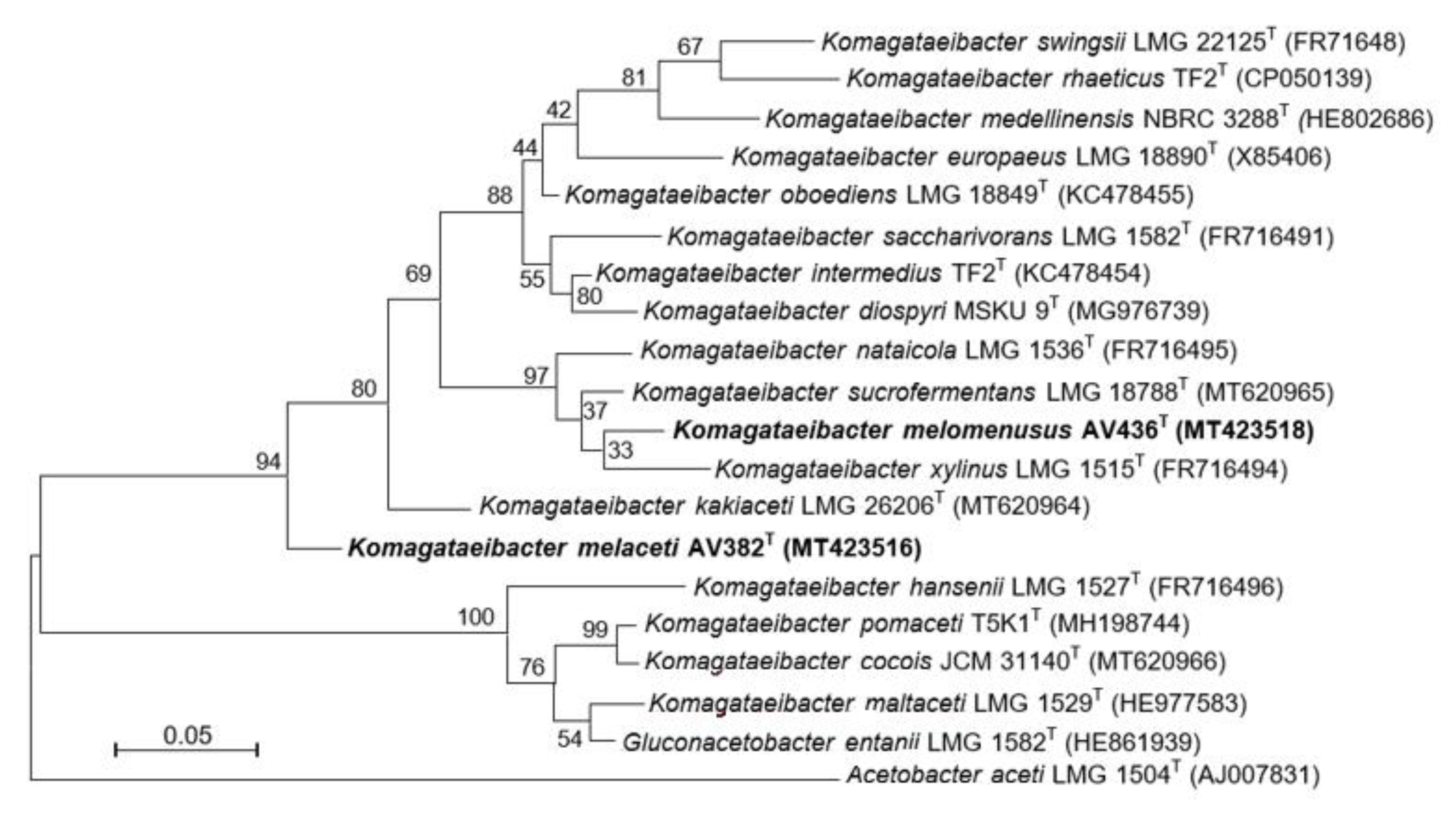
3.2. Phylogenetic Affiliation of Strains AV382 and AV436
3.3. Phenotypic Characterization of Strains AV382 and AV436
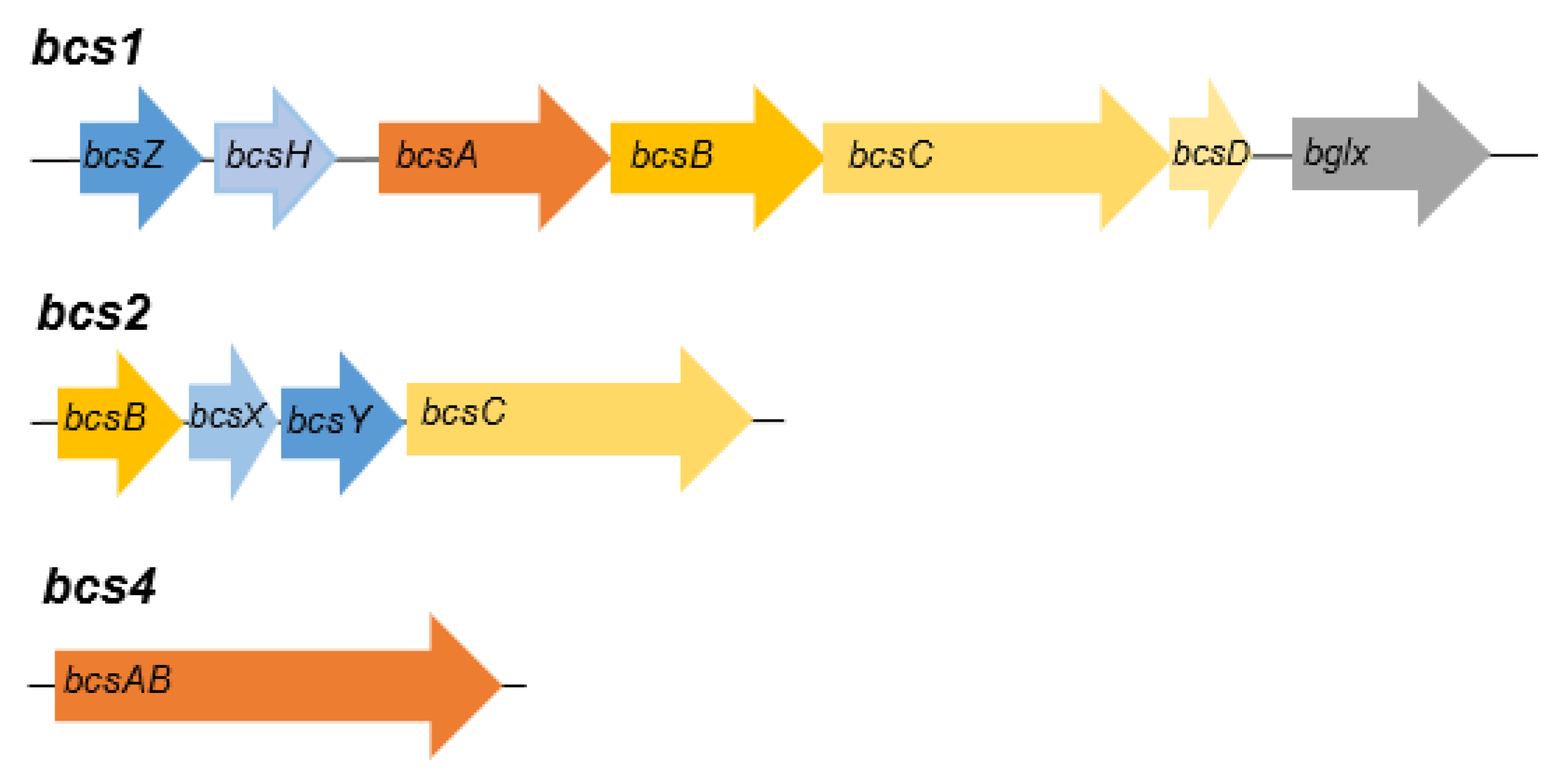
3.4. Genome analysis of strains AV382 and AV436
4. Conclusions
4.1. Description of Komagataeibacter melaceti sp. nov.
4.2. Description of Komagataeibacter melomenusus sp. nov.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Trček, J.; Barja, F. Updates on quick identification of acetic acid bacteria with a focus on the 16S-23S rRNA gene internal transcribed spacer and the analysis of cell proteins by MALDI-TOF mass spectrometry. Int. J. Food Microbiol. 2015, 196, 137–144. [Google Scholar] [CrossRef]
- Cleenwerck, I.; De Vos, P. Polyphasic taxonomy of acetic acid bacteria: An overview of the currently applied methodology. Int. J. Food Microbiol. 2008, 125, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Yukphan, P.; Vu, H.T.L.; Muramatsu, Y.; Ochaikul, D.; Tanasupawat, S.; Nakagawa, Y. Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 2012, 58, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Euzéby, J. List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 2013, 63, 1–5. [Google Scholar] [CrossRef]
- Yamada, Y.; Yukphan, P.; Vu, H.T.L.; Muramatsu, Y.; Ochaikul, D.; Nakagawa, Y. Subdivision of the genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: The proposal of Komagatabacter gen. nov., for strains accommodated to the Gluconacetobacter xylinus group in the α-Proteobacteria. Ann. Microbiol. 2012, 62, 849–859. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of prokaryotic names with standing in nomenclature (Bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 69, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Boesch, C.; Trček, J.; Sievers, M.; Teuber, M. Acetobacter intermedius, sp. nov. Syst. Appl. Microbiol. 1998, 21, 220–229. [Google Scholar] [CrossRef]
- Castro, C.; Cleenwerck, I.; Trček, J.; Zuluaga, R.; de Vos, P.; Caro, G.; Aguirre, R.; Putaux, J.-L.; Gañán, P. Gluconacetobacter medellinensis sp. nov., cellulose- and non-cellulose-producing acetic acid bacteria isolated from vinegar. Int. J. Syst. Evol. Microbiol. 2013, 63, 1119–1125. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.X.; Liu, S.X.; Wang, Y.M.; Bi, J.C.; Chen, H.M.; Deng, J.; Zhang, C.; Hu, Q.S.; Li, C.F. Komagataeibacter cocois sp. nov., a novel cellulose-producing strain isolated from coconut milk. Int. J. Syst. Evol. Microbiol. 2018, 68, 3125–3131. [Google Scholar] [CrossRef]
- Naloka, K.; Yukphan, P.; Matsutani, M.; Matsushita, K.; Theeragool, G. Komagataeibacter diospyri sp. nov., a novel species of thermotolerant bacterial nanocellulose-producing bacterium. Int. J. Syst. Evol. Microbiol. 2020, 70, 251–258. [Google Scholar] [CrossRef]
- Lisdiyanti, P.; Navarro, R.R.; Uchimura, T.; Komagata, K. Reclassification of Gluconacetobacter hansenii strains and proposals of Gluconacetobacter saccharivorans sp. nov. and Gluconacetobacter nataicola sp. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 2101–2111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slapšak, N.; Cleenwerck, I.; De Vos, P.; Trček, J. Gluconacetobacter maltaceti sp. nov., a novel vinegar producing acetic acid bacterium. Syst. Appl. Microbiol. 2013, 36, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Škraban, J.; Cleenwerck, I.; Vandamme, P.; Fanedl, L.; Trček, J. Genome sequences and description of novel exopolysaccharides producing species Komagataeibacter pomaceti sp. nov. and reclassification of Komagataeibacter kombuchae (Dutta and Gachhui 2007) Yamada et al., 2013 as a later heterotypic synonym of Komagataeibacter hansenii (Gosselé et al. 1983) Yamada et al., 2013. Syst. Appl. Microbiol. 2018, 41, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Gorgieva, S.; Trček, J. Bacterial cellulose: Production, modification and perspectives in biomedical applications. Nanomaterials 2019, 9, 1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorgieva, S. Bacterial cellulose as a versatile platform for research and development of biomedical materials. Processes 2020, 8, 624. [Google Scholar] [CrossRef]
- Cleenwerck, I.; De Vos, P.; De Vuyst, L. Phylogeny and differentiation of species of the genus Gluconacetobacter and related taxa based on multilocus sequence analyses of housekeeping genes and reclassification of Acetobacter xylinus subsp. sucrofermentans as Gluconacetobacter sucrofermentans (Toyosaki et al. 1996) sp. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 2277–2283. [Google Scholar] [CrossRef] [Green Version]
- De Vuyst, L.; Camu, N.; De Winter, T.; Vandemeulebroecke, K.; Van de Perre, V.; Vancanneyt, M.; De Vos, P.; Cleenwerck, I. Validation of the (GTG)5-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. Int. J. Food Microbiol. 2008, 125, 79–90. [Google Scholar] [CrossRef]
- Andrés-Barrao, C.; Benagli, C.; Chappuis, M.; Ortega Pérez, R.; Tonolla, M.; Barja, F. Rapid identification of acetic acid bacteria using MALDI-TOF mass spectrometry fingerprinting. Syst. Appl. Microbiol. 2013, 36, 75–81. [Google Scholar] [CrossRef]
- Trček, J.; Teuber, M. Genetic and restriction analysis of the 16S-23S rDNA internal transcribed spacer regions of the acetic acid bacteria. FEMS Microbiol. Lett. 2002, 208, 69–75. [Google Scholar] [CrossRef]
- Trcek, J. Quick identification of acetic acid bacteria based on nucleotide sequences of the 16S-23S rDNA internal transcribed spacer region and of the PQQ-dependent alcohol dehydrogenase gene. Syst. Appl. Microbiol. 2005, 28, 735–745. [Google Scholar] [CrossRef]
- Sokollek, S.J.; Hammes, W.P. Description of a starter culture preparation for vinegar fermentation. Syst. Appl. Microbiol. 1997, 20, 481–491. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gosselé, F.; Swings, J.; De Ley, J. A rapid, simple and simultaneous detection of 2-keto-, 5-keto-and 2,5-diketogluconic acids by thin-layer chromatography in culture media of acetic acid bacteria. Zentralblatt für Bakteriol. I. Abt. Orig. C Allg. angew. und ökologische Mikrobiol. 1980, 1, 178–181. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol Author Information; American Society for Microbiology: Washington, DC, USA, 2009; pp. 1–13. [Google Scholar]
- European Society of Clinical Microbiology and Infectious Diseases. Antimicrobial Susceptibility Testing EUCAST Disk Diffusion Method, Version 8.0, January 2020; European Society of Clinical Microbiology and Infectious Diseases: Basel, Switzerland, 2020; pp. 1–21. [Google Scholar]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. ClustalW2 and ClustalX version 2.0. Bioinfomatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arahal, D.R. Whole-genome analyses: Average nucleotide identity. In Methods in Microbiology: New Approaches to Prokaryotic Systematics; Goodfellow, M., Sutcliffe, I.C., Chun, J., Eds.; Elsevier: London, UK, 2014; Volume 41, pp. 103–122. [Google Scholar]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Sievers, M.; Sellmer, S.; Teuber, M. Acetobacter europaeus sp. nov., a main component of industrial vinegar fermenters in central Europe. Syst. Appl. Microbiol. 1992, 15, 386–392. [Google Scholar] [CrossRef]
- Gosselé, F.; Swings, J.; Kersters, K.; Pauwels, P.; De Ley, J. Numerical analysis of phenotypic features and protein gel electrophoregrams of a wide variety of Acetobacter strains. Proposal for the improvement of the taxonomy of the genus Acetobacter Beijerinck 1898, 215. Syst. Appl. Microbiol. 1983, 4, 338–368. [Google Scholar] [CrossRef]
- Iino, T.; Suzuki, R.; Tanaka, N.; Kosako, Y.; Ohkuma, M.; Komagata, K.; Uchimura, T. Gluconacetobacter kakiaceti sp. nov., an acetic acid bacterium isolated from a traditional Japanese fruit vinegar. Int. J. Syst. Evol. Microbiol. 2012, 62, 1465–1469. [Google Scholar] [CrossRef]
- Sokollek, S.J.; Hertel, C.; Hammes, W.P. Description of Acetobacter oboediens sp. nov. and Acetobacter pomorum sp. nov., two new species isolated from industrial vinegar fermentations. Int. J. Syst. Bacteriol. 1998, 48, 935–940. [Google Scholar] [CrossRef] [Green Version]
- Schüller, G.; Hertel, C.; Hammes, W.P. Gluconacetobacter entanii sp. nov., isolated from submerged high-acid industrial vinegar fermentations. Int. J. Syst. Evol. Microbiol. 2000, 50, 2013–2020. [Google Scholar] [CrossRef] [Green Version]
- Komagata, K.; Iino, T.; Yamada, Y. The family Acetobacteraceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., De Long, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–78. [Google Scholar]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [Green Version]
- Thompson, C.C.; Chimetto, L.; Edwards, R.A.; Swings, J.; Stackebrandt, E.; Thompson, F.L. Microbial genomic taxonomy. BMC Genomics 2013, 14, 913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, Y.; Nunoda, M.; Ishikawa, T.; Tahara, Y. The cellular fatty acid composition in acetic acid bacteria. J. Gen. Appl. Microbiol. 1981, 27, 405–417. [Google Scholar] [CrossRef]
- Loganathan, P.; Nair, S. Swaminathania salitolerans gen. nov., sp. nov., a salt-tolerant, nitrogen-fixing and phosphate-solubilizing bacterium from wild rice (Porteresia coarctata Tateoka). Int. J. Syst. Evol. Microbiol. 2004, 54, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Jojima, Y.; Mihara, Y.; Suzuki, S.; Yokozeki, K.; Yamanaka, S.; Fudou, R. Saccharibacter floricola gen. nov., sp. nov., a novel osmophilic acetic acid bacterium isolated from pollen. Int. J. Syst. Evol. Microbiol. 2004, 54, 2263–2267. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Praet, J.; Borremans, W.; Nunes, O.C.; Manaia, C.M.; Cleenwerck, I.; Meeus, I.; Smagghe, G.; De Vuyst, L.; Vandamme, P. Bombella intestini gen. nov., sp. nov., an acetic acid bacterium isolated from bumble bee crop. Int. J. Syst. Evol. Microbiol. 2015, 65, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, P.; Lei, Q.; Li, B.; Sun, Y.; Li, S.; Lei, H.; Xie, N. Metabolic adaptability shifts of cell membrane fatty acids of Komagataeibacter hansenii HDM1-3 improve acid stress resistance and survival in acidic environments. J. Ind. Microbiol. Biotechnol. 2019, 46, 1491–1503. [Google Scholar] [CrossRef]
- Brown, J.L.; Ross, T.; McMeekin, T.A.; Nichols, P.D. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. Int. J. Food Microbiol. 1997, 37, 163–173. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef] [Green Version]
- Shaw, K.J.; Rather, P.N.; Hare, R.S.; Miller, G.H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993, 57, 138–163. [Google Scholar] [CrossRef]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.; Martinez, J. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saichana, N.; Matsushita, K.; Adachi, O.; Frébort, I.; Frebortova, J. Acetic acid bacteria: A group of bacteria with versatile biotechnological applications. Biotechnol. Adv. 2015, 33, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Škraban, J.; Trček, J. Comparative genomics of Acetobacter and other acetic acid bacteria. In Acetic Acid Bacteria: Fundamentals and Food Applications; Sengun, I.Y., Ed.; CRC Press, Taylor and Francis Group: Boca Raton, FL, USA, 2017; pp. 44–70. [Google Scholar]
- Kubiak, K.; Kurzawa, M.; Jedrzejczak-Krzepkowska, M.; Ludwicka, K.; Krawczyk, M.; Migdalski, A.; Kacprzak, M.M.; Loska, D.; Krystynowicz, A.; Bielecki, S. Complete genome sequence of Gluconacetobacter xylinus E25 strain-Valuable and effective producer of bacterial nanocellulose. J. Biotechnol. 2014, 176, 18–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, X.; Chen, X.; Yuan, F.; Sun, B.; Xu, Y.; Yang, J.; Sun, D. Complete genome sequence of the cellulose-producing strain Komagataeibacter nataicola RZS01. Sci. Rep. 2017, 7, 4431. [Google Scholar] [CrossRef]
- Liu, M.; Liu, L.; Jia, S.; Li, S.; Zou, Y.; Zhong, C. Complete genome analysis of Gluconacetobacter xylinus CGMCC 2955 for elucidating bacterial cellulose biosynthesis and metabolic regulation. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Pfeffer, S.; Santos, R.; Ebels, M.; Bordbar, D.; Brown, R.M. Complete genome sequence of Komagataeibacter hansenii LMG 23726T. Genome Announc. 2017, 5, e00168-17. [Google Scholar] [CrossRef] [Green Version]
- Römling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: Diversity of operons, subunits, products, and functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef] [Green Version]
- Griffin, A.M.; Morris, V.J.; Gasson, M.J. Genetic analysis of the acetan biosynthetic pathway in Acetobacter xylinum: Nucleotide sequence analysis of the aceB aceC aceD and aceE genes. Mitochondrial DNA 1996, 6, 275–284. [Google Scholar] [CrossRef]
- Brandt, J.U.; Jakob, F.; Wefers, D.; Bunzel, M.; Vogel, R.F. Characterization of an acetan-like heteropolysaccharide produced by Kozakia baliensis NBRC 16680. Int. J. Biol. Macromol. 2018, 106, 248–257. [Google Scholar] [CrossRef]
- Deeraksa, A.; Moonmangmee, S.; Toyama, H.; Adachi, O.; Matsushita, K. Conversion of capsular polysaccharide, involved in pellicle formation, to extracellular polysaccharide by galE deletion in Acetobacter tropicalis. Biosci. Biotechnol. Biochem. 2006, 70, 2536–2539. [Google Scholar] [CrossRef] [Green Version]
- Deeraksa, A.; Moonmangmee, S.; Toyama, H.; Yamada, M.; Adachi, O.; Matsushita, K. Characterization and spontaneous mutation of a novel gene, polE, involved in pellicle formation in Acetobacter tropicalis SKU1100. Microbiology 2005, 151, 4111–4120. [Google Scholar] [CrossRef] [Green Version]
- Mullins, E.A.; Francois, J.A.; Kappock, T.J. A specialized citric acid cycle requiring succinyl-coenzyme A (CoA): Acetate CoA-transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti. J. Bacteriol. 2008, 190, 4933–4940. [Google Scholar] [CrossRef] [Green Version]
- Tenreiro, S.; Rosa, P.C.; Viegas, C.A.; Sá-Correia, I. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast 2000, 6, 1469–1481. [Google Scholar] [CrossRef]
- Matsushita, K.; Fujii, Y.; Ano, Y.; Toyama, H.; Shinjoh, M.; Tomiyama, N.; Miyazaki, T.; Sugisawa, T.; Hoshino, T.; Adachi, O. 5-keto-D-gluconate production is catalyzed by a quinoprotein glycerol dehydrogenase, major polyol dehydrogenase, in Gluconobacter species. Appl. Environ. Microbiol. 2003, 69, 1959–1966. [Google Scholar] [CrossRef] [Green Version]
- Saichana, I.; Moonmangmee, D.; Adachi, O.; Matsushita, K.; Toyama, H. Screening of thermotolerant Gluconobacter strains for production of 5-keto-D-gluconic acid and disruption of flavin adenine dinucleotide-containing D-gluconate dehydrogenase. Appl. Environ. Microbiol. 2009, 75, 4240–4247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariette, I.; Schwarz, E.; Vogel, R.F.; Hammes, W.P. Characterization by plasmid profile analysis of acetic acid bacteria from wine, spirit and cider acetators for industrial vinegar production. J. Appl. Bacteriol. 1991, 71, 134–138. [Google Scholar] [CrossRef]
- Sievers, M.; Andresen, A.; Teuber, M. Plasmid profiles as tools to characterize the microflora of industrial vinegar fermenters. Food Biotechnol. 1990, 4, 555. [Google Scholar]
- Štornik, A.; Skok, B.; Trček, J. Comparison of cultivable acetic acid bacterial microbiota in organic and conventional apple cider vinegar. Food Technol. Biotechnol. 2016, 54, 113–119. [Google Scholar] [CrossRef]
- Trček, J.; Mahnič, A.; Rupnik, M. Diversity of the microbiota involved in wine and organic apple cider submerged vinegar production as revealed by DHPLC analysis and next-generation sequencing. Int. J. Food Microbiol. 2016, 223, 57–62. [Google Scholar] [CrossRef]


| Fatty Acid | AV382 (mol%) | AV436 (mol%) | K. cocois JCM 31140T (mol%) |
|---|---|---|---|
| C14:0 2-OH | 5.9/6.5 | 13.2/1.7 | 9.5/5.5 |
| C16:0 | 9.2/9.0 | 16.4/2.0 | 13.8/1.7 |
| C16:0 2-OH | 7.1/7.4 | 15.2/2.6 | 8.0/6.3 |
| C16:0 3-OH | 1.1/1.7 | 5.4/2.0 | 1.7/1.7 |
| C17:0 | 8.2/6.7 | 0.8/1.0 | nd/1.9 |
| C17:1 ω6c | 6.6/2.6 | nd/0.4 | nd/0.9 |
| C18:1 ω7c | 36.9/34.4 | 31.4/43.4 | 58.9/61.9 |
| C19:1 cyclo ω8c | 13.0/16.7 | 5.0/4.5 | nd/nd |
| Phenotypic Features | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Formation from D-Glucose | |||||||
| 2-keto-D-Gluconic acid | + | + | + a | + a | + b | + a | - c |
| 5-keto-D-Gluconic acid | + | + | + a | - a | + b | + a | - c |
| Growth on carbon sources: | |||||||
| D-Ribose | + | + | + | + | w | + | w |
| Sorbitol | + | + | + | + | w | + | w |
| D-Mannitol | + | + | + | + | + | + | + |
| Glycerol | + | + | + | + | + | + | w |
| 1-Propanol | + | w | + | + | - | w | + |
| Growth in the presence of 30% D-glucose | - | - | - | - | - | + | + |
| Utilization of ammoniacal nitrogen in: | |||||||
| Hoyer-Frateur medium with: | |||||||
| Ethanol | + | - | + | - | - | - | - |
| Asai medium with: | |||||||
| D-Mannitol | + | + | + | + | + | + | - |
| Growth on RAE medium in the presence of 1% ethanol and acetic acid at: | |||||||
| 4% | + | + | + | + | + | + | + |
| 5% | + | + | - | + | - | - | + |
| Growth on RAE medium in the presence of 3% ethanol and acetic acid at: | |||||||
| 4% | + | + | - | + | - | + | + |
| 5% | + | + | - | + | - | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marič, L.; Cleenwerck, I.; Accetto, T.; Vandamme, P.; Trček, J. Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar. Microorganisms 2020, 8, 1178. https://doi.org/10.3390/microorganisms8081178
Marič L, Cleenwerck I, Accetto T, Vandamme P, Trček J. Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar. Microorganisms. 2020; 8(8):1178. https://doi.org/10.3390/microorganisms8081178
Chicago/Turabian StyleMarič, Leon, Ilse Cleenwerck, Tomaž Accetto, Peter Vandamme, and Janja Trček. 2020. "Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar" Microorganisms 8, no. 8: 1178. https://doi.org/10.3390/microorganisms8081178
APA StyleMarič, L., Cleenwerck, I., Accetto, T., Vandamme, P., & Trček, J. (2020). Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar. Microorganisms, 8(8), 1178. https://doi.org/10.3390/microorganisms8081178





