Staphylococcus aureus Lung Infection Results in Down-Regulation of Surfactant Protein-A Mainly Caused by Pro-Inflammatory Macrophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Human Alveolus-on-a-Chip Model
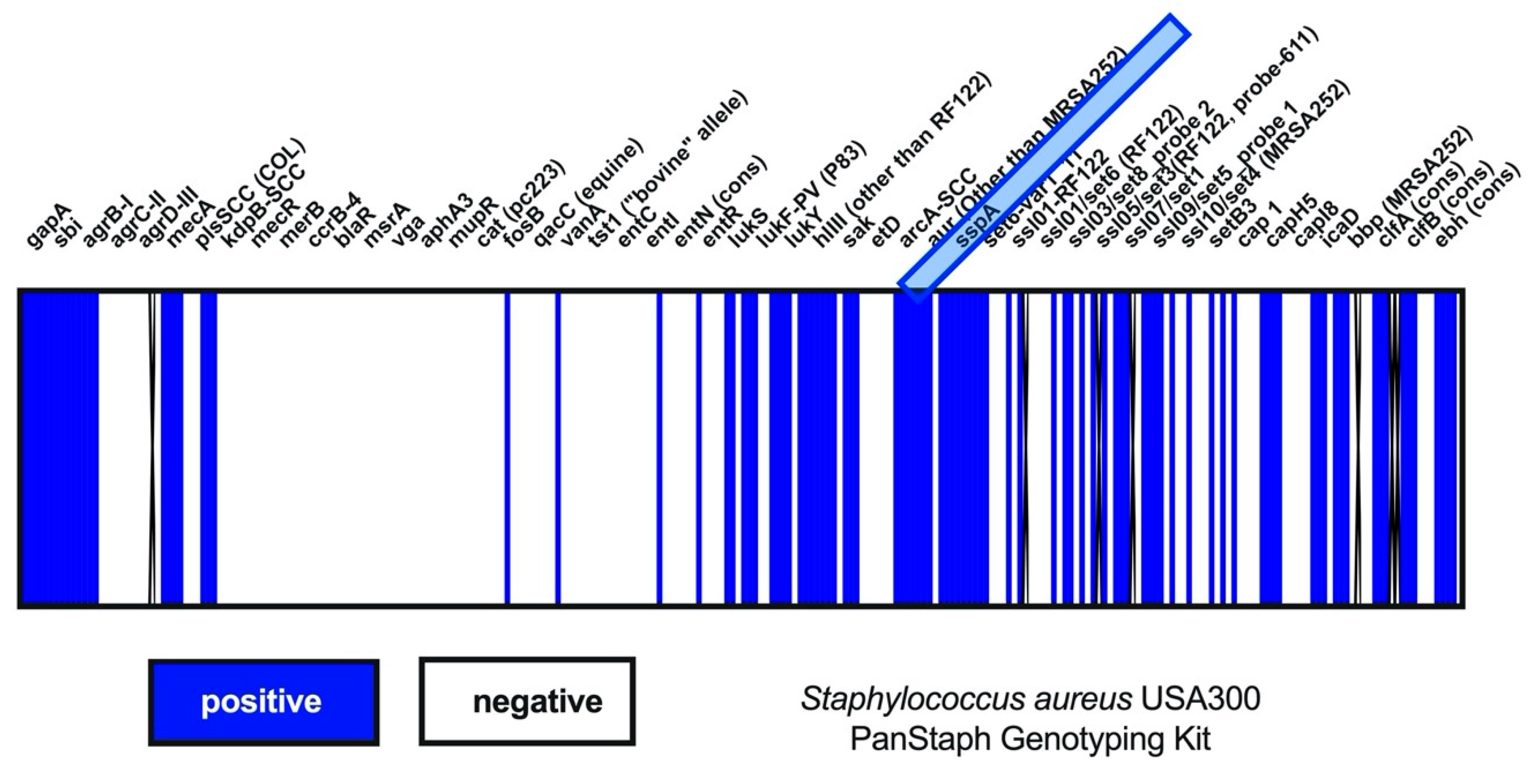
2.3. Pathogens
2.4. In Vitro Infection
2.5. In Vivo Infection
2.6. Quantitative Real-Time PCR
2.7. Protein Analysis
2.8. Immunofluorescence Microscopy
2.9. Scanning Electron Microscopy
2.10. Image Analysis and Quantification
2.11. Statistical Analysis
3. Results
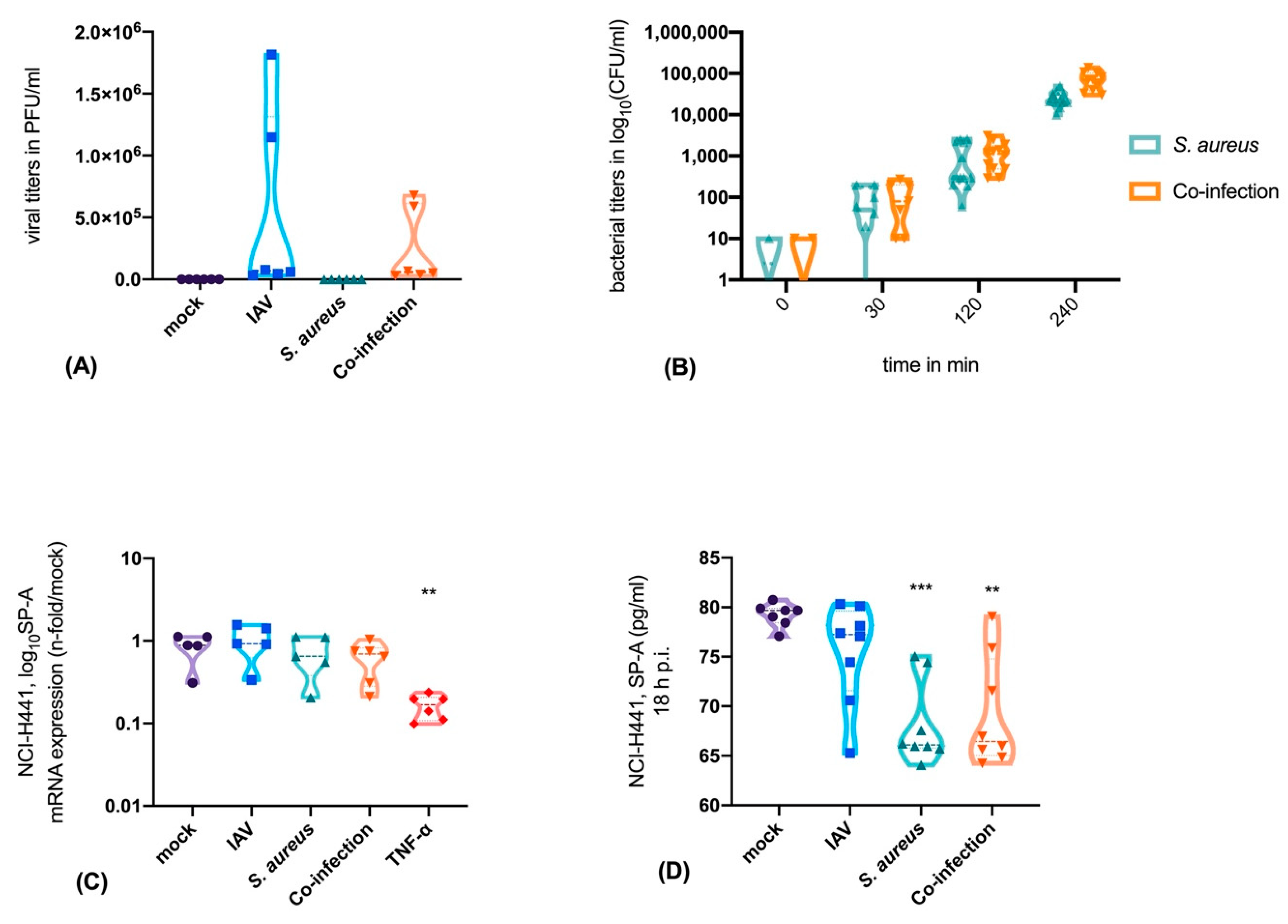
3.1. Neither IAV Nor S. aureus Directly Affects SP-A Expression in Epithelial Cells, But TNF-α is a Strong Inhibitor of SP-A Expression
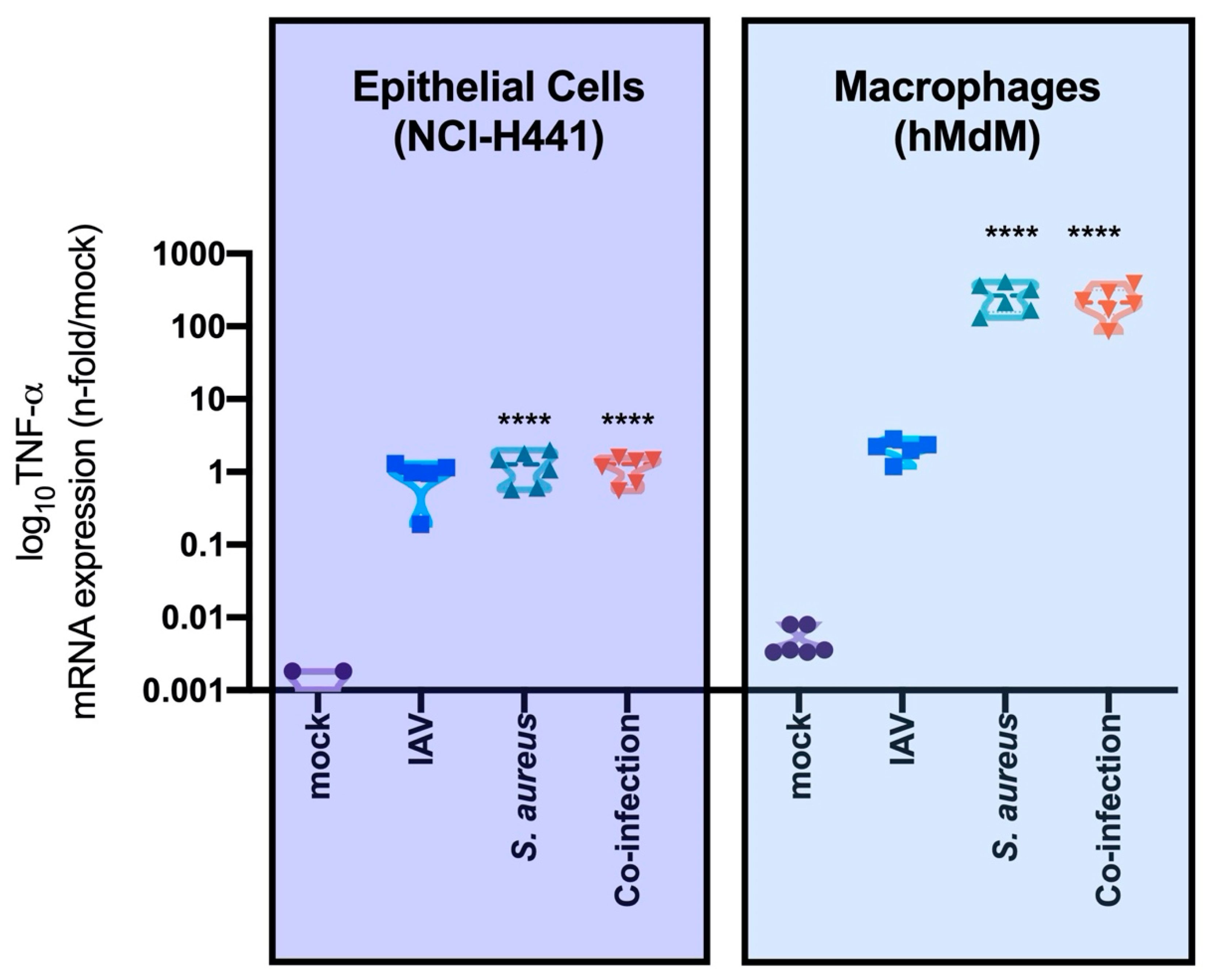
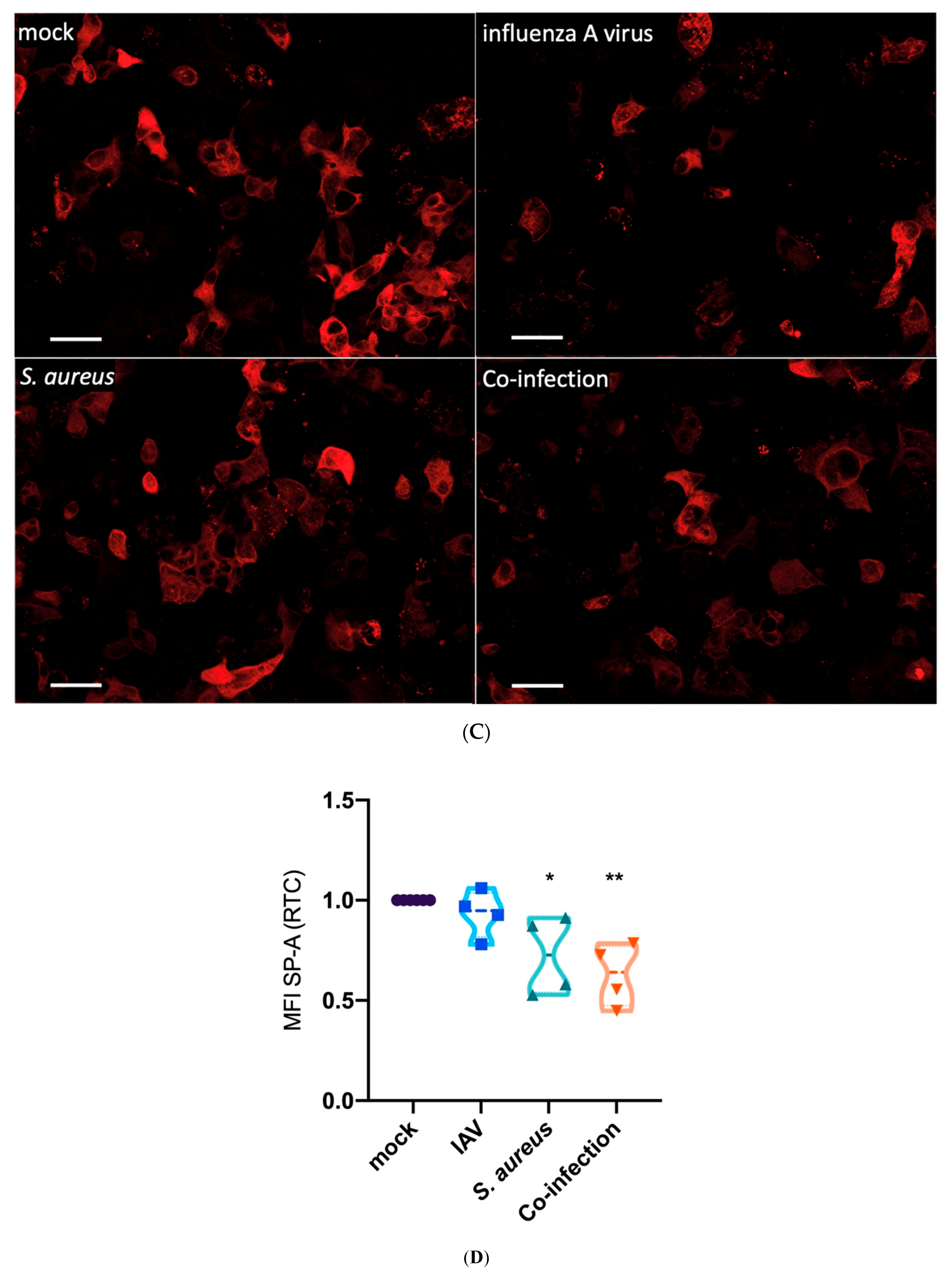
3.2. Interaction of Epithelial, Endothelial and Macrophages Causes SP-A Downregulation Mainly Triggered by Bacteria
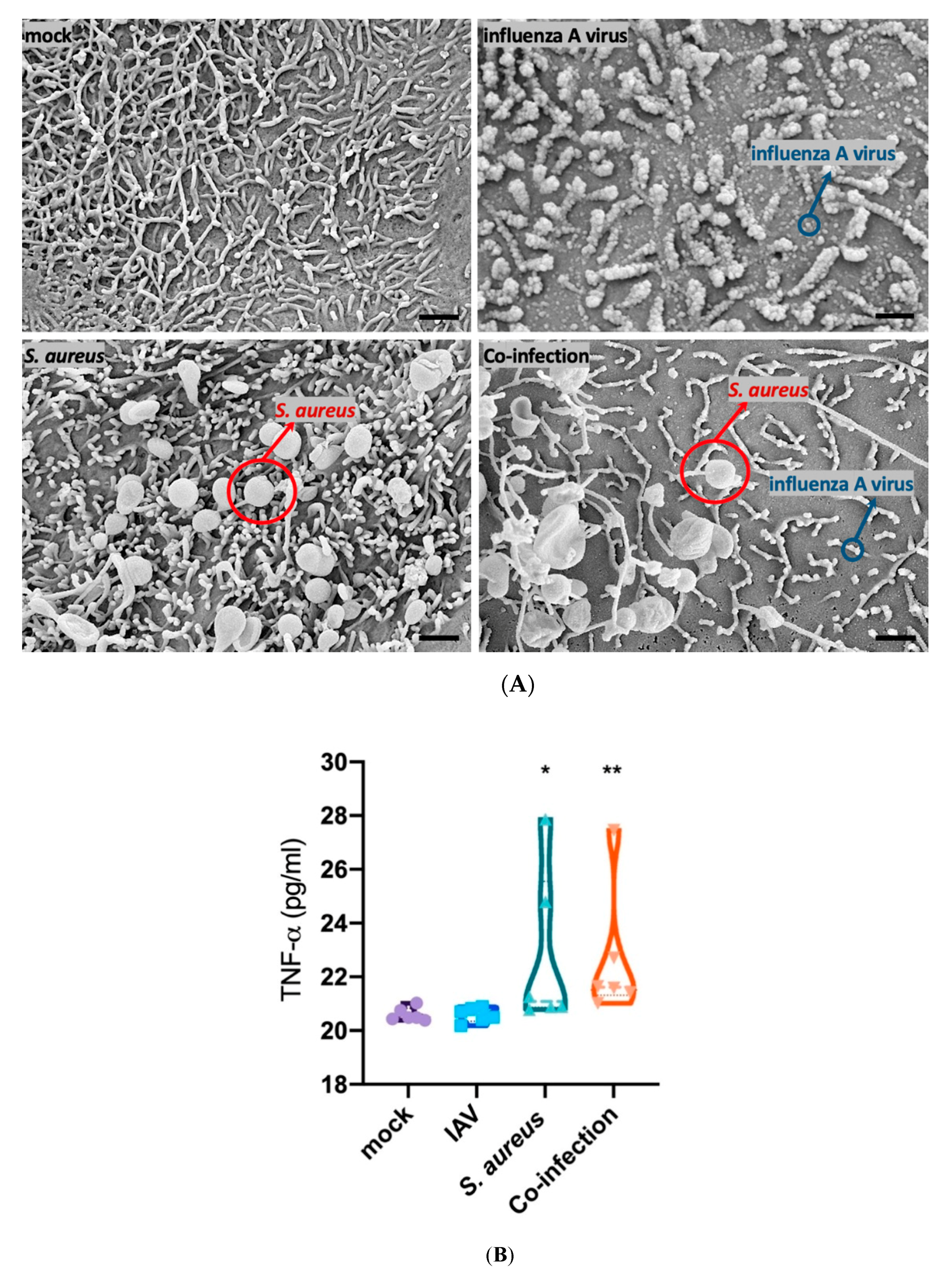
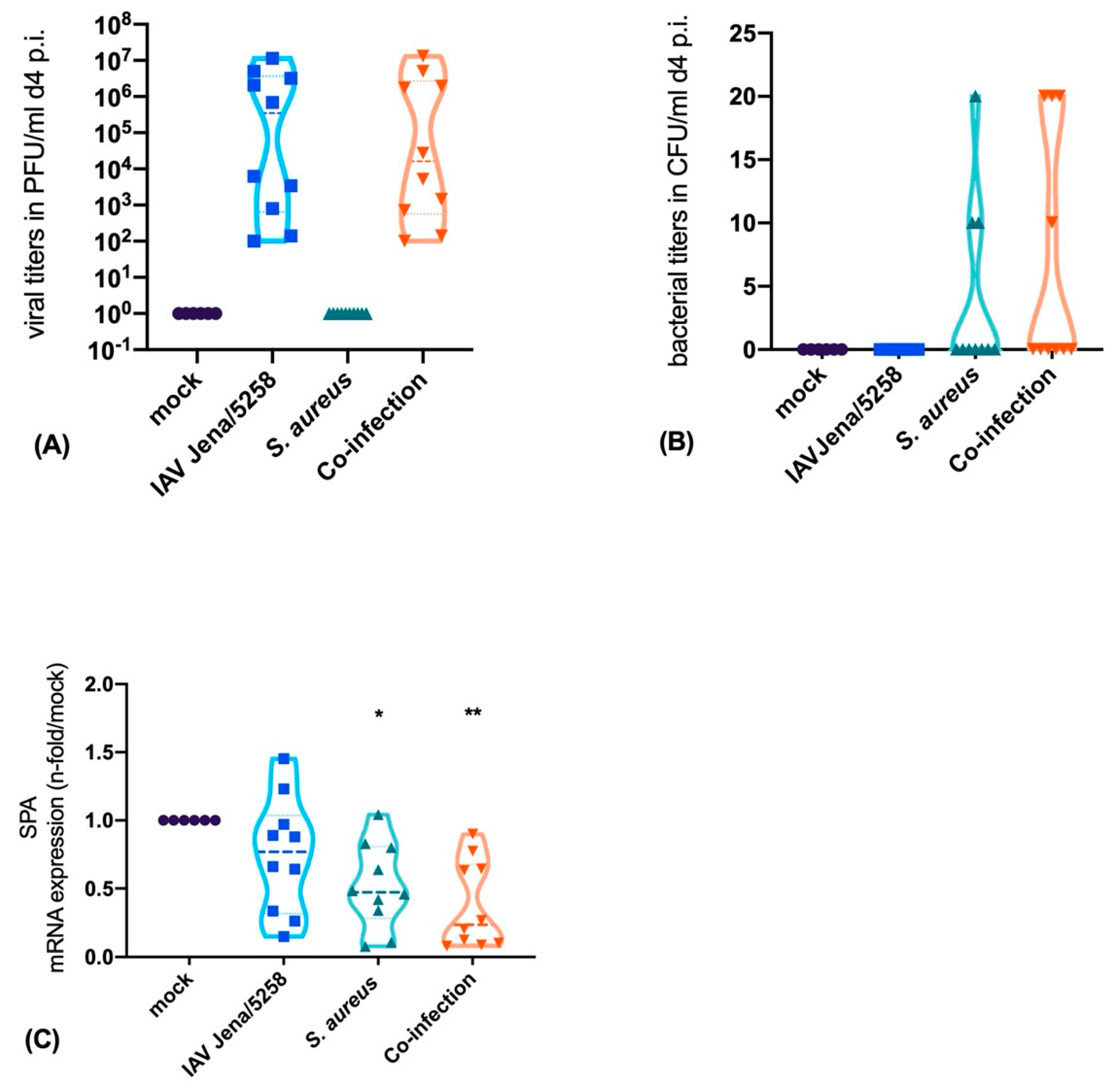
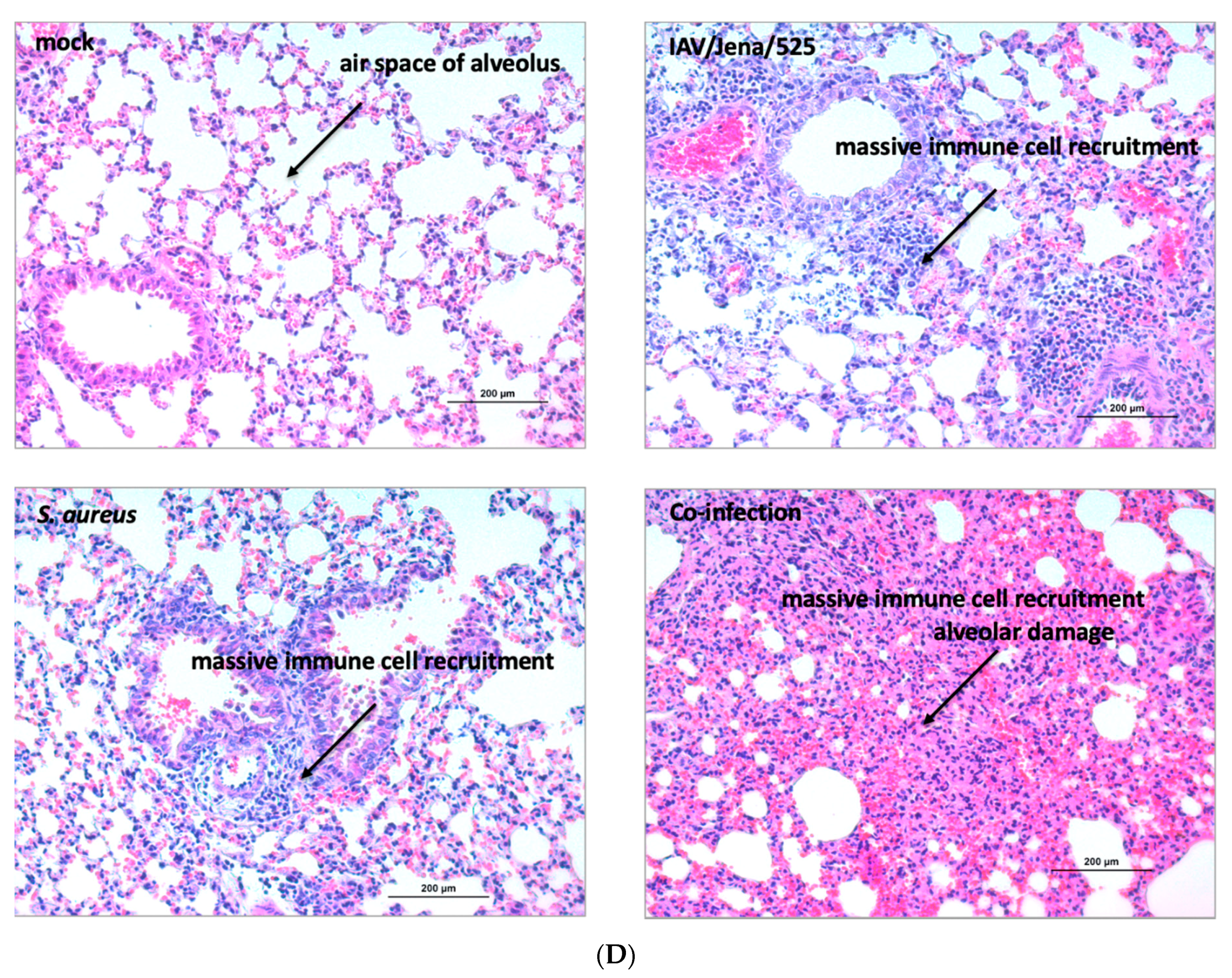
3.3. S. aureus Downregulates SP-A Expression in a Murine Model of Pneumonia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Taubenberger, J.K.; Morens, D.M. The pathology of influenza virus infections. Annu. Rev. Pathol. 2008, 3, 499–522. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Strieter, R.M.; Belperio, J.A.; Keane, M.P. Cytokines in innate host defense in the lung. J. Clin. Investig. 2002, 109, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Strieter, R.M.; Belperio, J.A.; Keane, M.P. Host innate defenses in the lung: The role of cytokines. Curr. Opin. Infect. Dis. 2003, 16, 193–198. [Google Scholar] [CrossRef]
- Wright, J.R. Immunoregulatory functions of surfactant proteins. Nat. Rev. Immunol. 2005, 5, 58–68. [Google Scholar] [CrossRef]
- Wright, J.R. Immunomodulatory functions of surfactant. Physiol. Rev. 1997, 77, 931–962. [Google Scholar] [CrossRef]
- Kishore, U.; Greenhough, T.J.; Waters, P.; Shrive, A.K.; Ghai, R.; Kamran, M.F.; Bernal, A.L.; Reid, K.B.; Madan, T.; Chakraborty, T. Surfactant proteins SP-A and SP-D: Structure, function and receptors. Mol. Immunol. 2006, 43, 1293–1315. [Google Scholar] [CrossRef]
- Han, S.; Mallampalli, R.K. The role of surfactant in lung disease and host defense against pulmonary infections. Ann. Am. Thorac. Soc. 2015, 12, 765–774. [Google Scholar] [CrossRef] [Green Version]
- Benne, C.A.; Kraaijeveld, C.A.; van Strijp, J.A.; Brouwer, E.; Harmsen, M.; Verhoef, J.; van Golde, L.M.; van Iwaarden, J.F. Interactions of surfactant protein A with influenza A viruses: Binding and neutralization. J. Infect. Dis. 1995, 171, 335–341. [Google Scholar] [CrossRef]
- Geertsma, M.F.; Nibbering, P.H.; Haagsman, H.P.; Daha, M.R.; van Furth, R. Binding of surfactant protein A to C1q receptors mediates phagocytosis of Staphylococcus aureus by monocytes. Am. J. Physiol. 1994, 267, L578–L584. [Google Scholar] [CrossRef]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. S4), D49–D53. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, I.N.; De Luna, X.; White, M.R.; Hartshorn, K.L. The role and molecular mechanism of action of surfactant protein D in innate host defense against influenza A virus. Front. Immunol. 2018, 9, 1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sever-Chroneos, Z.; Krupa, A.; Davis, J.; Hasan, M.; Yang, C.H.; Szeliga, J.; Herrmann, M.; Hussain, M.; Geisbrecht, B.V.; Kobzik, L.; et al. Surfactant protein A (SP-A)-mediated clearance of Staphylococcus aureus involves binding of SP-A to the staphylococcal adhesin eap and the macrophage receptors SP-A receptor 210 and scavenger receptor class A. J. Biol. Chem. 2011, 286, 4854–4870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wispe, J.R.; Clark, J.C.; Warner, B.B.; Fajardo, D.; Hull, W.E.; Holtzman, R.B.; Whitsett, J.A. Tumor necrosis factor-alpha inhibits expression of pulmonary surfactant protein. J. Clin. Investig. 1990, 86, 1954–1960. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.N.; Mendelson, C.R. Potential role of nuclear factor κb and reactive oxygen species in camp and cytokine regulation of surfactant protein-a gene expression in lung type II cells. Mol. Endocrinol. 2002, 16, 1428–1440. [Google Scholar] [PubMed] [Green Version]
- Ito, Y.; Mason, R.J. The effect of interleukin-13 (IL-13) and interferon-γ (IFN-γ) on expression of surfactant proteins in adult human alveolar type II cells in vitro. Respir. Res. 2010, 11, 157. [Google Scholar] [CrossRef] [Green Version]
- Väyrynen, O.; Glumoff, V.; Hallman, M. Regulation of surfactant proteins by LPS and proinflammatory cytokines in fetal and newborn lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002, 282, L803–L810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Wang, C.C. Inflammatory response of macrophages in infection. Hepatobiliary Pancreat Dis. Int. 2014, 13, 138–152. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Gong, J.H.; Sprenger, H.; Hinder, F.; Bender, A.; Schmidt, A.; Horch, S.; Nain, M.; Gemsa, D. Influenza A virus infection of macrophages. Enhanced tumor necrosis factor-alpha (TNF-alpha) gene expression and lipopolysaccharide-triggered TNF-alpha release. J. Immunol. 1991, 147, 3507. [Google Scholar]
- Cui, W.; Morrison, D.C.; Silverstein, R. Differential tumor necrosis factor alpha expression and release from peritoneal mouse macrophages in vitro in response to proliferating gram-positive versus gram-negative bacteria. Infect. Immun. 2000, 68, 4422–4429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kantyka, T.; Pyrc, K.; Gruca, M.; Smagur, J.; Plaza, K.; Guzik, K.; Zeglen, S.; Ochman, M.; Potempa, J. Staphylococcus aureus proteases degrade lung surfactant protein A potentially impairing innate immunity of the lung. J. Innate Immun. 2013, 5, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Raasch, M.; Rennert, K.; Jahn, T.; Peters, S.; Henkel, T.; Huber, O.; Schulz, I.; Becker, H.; Lorkowski, S.; Funke, H.; et al. Microfluidically supported biochip design for culture of endothelial cell layers with improved perfusion conditions. Biofabrication 2015, 7, 015013. [Google Scholar] [CrossRef] [PubMed]
- Deinhardt-Emmer, S.; Rennert, K.; Schicke, E.; Cseresnyes, Z.; Windolph, M.; Nietzsche, S.; Heller, R.; Siwczak, F.; Haupt, K.F.; Carlstedt, S.; et al. Co-infection withStaphylococcus aureusafter primary influenza virus infection leads to damage of the endothelium in a human alveolus-on-a-chip model. Biofabrication 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rennert, K.; Heisig, K.; Groeger, M.; Wallert, M.; Funke, H.; Lorkowski, S.; Huber, O.; Mosig, A.S. Recruitment of CD16(+) monocytes to endothelial cells in response to LPS-treatment and concomitant TNF release is regulated by CX3CR1 and interfered by soluble fractalkine. Cytokine 2016, 83, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Seidel, N.; Sauerbrei, A.; Wutzler, P.; Schmidtke, M. Hemagglutinin 222D/G polymorphism facilitates fast intra-host evolution of pandemic (H1N1) 2009 influenza A viruses. PLoS ONE 2014, 9, e104233. [Google Scholar] [CrossRef] [Green Version]
- Manchanda, H.; Seidel, N.; Krumbholz, A.; Sauerbrei, A.; Schmidtke, M.; Guthke, R. Within-host influenza dynamics: A small-scale mathematical modeling approach. Bio Syst. 2014, 118, 51–59. [Google Scholar] [CrossRef]
- Tuchscherr, L.; Medina, E.; Hussain, M.; Volker, W.; Heitmann, V.; Niemann, S.; Holzinger, D.; Roth, J.; Proctor, R.A.; Becker, K.; et al. Staphylococcus aureus phenotype switching: An effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 2011, 3, 129–141. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Bloes, D.A.; Haasbach, E.; Hartmayer, C.; Hertlein, T.; Klingel, K.; Kretschmer, D.; Planz, O.; Peschel, A. Phenol-soluble modulin peptides contribute to influenza a virus-associated staphylococcus aureus pneumonia. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, S.; Kotecha, S. Lung growth and development. Early Hum. Dev. 2007, 83, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Katyal, S.; Amenta, J.; Singh, G.; Silverman, J. Deficient lung surfactant apoproteins in amniotic fluid with mature phospholipid profile from diabetic pregnancies. Am. J. Obstet. Gynecol. 1984, 148, 48–53. [Google Scholar] [CrossRef]
- Chida, S.; Phelps, D.S.; Cordle, C.; Soll, R.; Floros, J.; Taeusch, H.W. Surfactant-associated proteins in tracheal aspirates of infants with respiratory distress syndrome after surfactant therapy. Am. Rev. Respir. Dis. 1988, 137, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Liggins, G.C.; Howie, R.N. A controlled trial of antepartum glucocorticoid treatment for prevention of the respiratory distress syndrome in premature infants. Pediatrics 1972, 50, 515–525. [Google Scholar] [PubMed]
- deLemos, R.A.; Shermeta, D.W.; Knelson, J.H.; Kotas, R.; Avery, M.E. Acceleration of appearance of pulmonary surfactant in the fetal lamb by administration of corticosteroids. Am. Rev. Respir. Dis. 1970, 102, 459–461. [Google Scholar] [PubMed]
- Kotas, R.V.; Avery, M.E. Accelerated appearance of pulmonary surfactant in the fetal rabbit. J. Appl. Physiol. 1971, 30, 358–361. [Google Scholar] [CrossRef]
- Guarino, N.; Oue, T.; Shima, H.; Puri, P. Antenatal dexamethasone enhances surfactant protein synthesis in the hypoplastic lung of nitrofen-induced diaphragmatic hernia in rats. J. Pediatr. Surg. 2000, 35, 1468–1473. [Google Scholar] [CrossRef]
- Konishi, M.; Fujiwara, T.; Naito, T.; Takeuchi, Y.; Ogawa, Y.; Inukai, K.; Fujimura, M.; Nakamura, H.; Hashimoto, T. Surfactant replacement therapy in neonatal respiratory distress syndrome. A multi-centre, randomized clinical trial: Comparison of high- versus low-dose of surfactant TA. Eur. J. Pediatr. 1988, 147, 20–25. [Google Scholar] [CrossRef]
- Rojas-Reyes, M.X.; Morley, C.J.; Soll, R. Prophylactic versus selective use of surfactant in preventing morbidity and mortality in preterm infants. Cochrane Database Syst. Rev. 2012. [Google Scholar] [CrossRef]
- Jobe, A.H.; Mitchell, B.R.; Gunkel, J.H. Beneficial effects of the combined use of prenatal corticosteroids and postnatal surfactant on preterm infants. Am. J. Obstet. Gynecol. 1993, 168, 508–513. [Google Scholar] [CrossRef]
- Gregory, T.; Longmore, W.; Moxley, M.; Whitsett, J.; Reed, C.; Fowler, A.R.; Hudson, L.; Maunder, R.; Crim, C.; Hyers, T. Surfactant chemical composition and biophysical activity in acute respiratory distress syndrome. J. Clin. Investig. 1991, 88, 1976–1981. [Google Scholar] [CrossRef] [PubMed]
- Pison, U.; Obertacke, U.; Brand, M.; Seeger, W.; Joka, T.; Bruch, J.; Schmit-Neuerburg, K. Altered pulmonary surfactant in uncomplicated and septicemia-complicated courses of acute respiratory failure. J. Trauma 1990, 30, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Baughman, R.P.; Sternberg, R.I.; Hull, W.; Buchsbaum, J.A.; Whitsett, J. Decreased surfactant protein A in patients with bacterial pneumonia. Am. Rev. Respir. Dis. 1993, 147, 653–657. [Google Scholar] [CrossRef] [PubMed]
- LeVine, A.M.; Lotze, A.; Stanley, S.; Stroud, C.; O’Donnell, R.; Whitsett, J.; Pollack, M.M. Surfactant content in children with inflammatory lung disease. Crit. Care Med. 1996, 24, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Miakotina, O.L.; Snyder, J.M. TNF-α inhibits SP-A gene expression in lung epithelial cells via p38 MAPK. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002, 283, L418–L427. [Google Scholar] [CrossRef] [PubMed]
- Whitsett, J.A.; Glasser, S.W. Regulation of surfactant protein gene transcription. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 1998, 1408, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Holzinger, D.; Gieldon, L.; Mysore, V.; Nippe, N.; Taxman, D.J.; Duncan, J.A.; Broglie, P.M.; Marketon, K.; Austermann, J.; Vogl, T.; et al. Staphylococcus aureus Panton-Valentine leukocidin induces an inflammatory response in human phagocytes via the NLRP3 inflammasome. J. Leukoc. Biol. 2012, 92, 1069–1081. [Google Scholar] [CrossRef] [Green Version]
- Loffler, B.; Niemann, S.; Ehrhardt, C.; Horn, D.; Lanckohr, C.; Lina, G.; Ludwig, S.; Peters, G. Pathogenesis of Staphylococcus aureus necrotizing pneumonia: The role of PVL and an influenza coinfection. Expert Rev. Anti Infect. Ther. 2013, 11, 1041–1051. [Google Scholar] [CrossRef]
- Niemann, S.; Ehrhardt, C.; Medina, E.; Warnking, K.; Tuchscherr, L.; Heitmann, V.; Ludwig, S.; Peters, G.; Loffler, B. Combined action of influenza virus and Staphylococcus aureus panton-valentine leukocidin provokes severe lung epithelium damage. J. Infect. Dis. 2012, 206, 1138–1148. [Google Scholar] [CrossRef]
- Seo, S.H.; Webster, R.G. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J. Virol. 2002, 76, 1071–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Gene Name | Sequence Forward | Sequence Reverse |
|---|---|---|
| human GAPDH | 5′- CTCTGCTCCTCCTGTTCGAC -3′ | 5′-CAATACGACCAAATCCGTTGAC -3′ |
| human TNF-α | 5′- GGAGAAGGGTGACCGACTCA -3′ | 5′- CTGCCCAGACTCGGCAA -3′ |
| human SP-A | 5′- GATGGGCAGTGGAATGACAGG -3′ | 5′- GGGAATGAAGTGGCTAAGGGTG -3′ |
| RPL37A | 5′- ATTGAAATCAGCCAGCACGC -3′ | 5′- AGGAACCACAGTGCCAGATCC -3′ |
| CD80 | 5′- TGGTGCTGGCTG GTCTTTC -3′ | 5′- CGTTGCCACTTCTTTCACTTCC -3′ |
| CD206 | 5′- TCGGGTTTATGGAGCAGGTG -3′ | 5′- TGAACGGGAATGCACAGGTT -3′ |
| murine GAPDH | 5′-CAACAGCAACTCCCACTCTTC-3′ | 5′-GGTCCAGGGTTTCTTACTCCTT-3′ |
| murine SP-A | 5′-GCAGAGATGGGAGAGATGGTATCAA -3′ | 5′-ATGGACCTCCATTAGCATGTGGGA-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schicke, E.; Cseresnyés, Z.; Rennert, K.; Vau, V.; Haupt, K.F.; Hornung, F.; Nietzsche, S.; Swiczak, F.; Schmidtke, M.; Glück, B.; et al. Staphylococcus aureus Lung Infection Results in Down-Regulation of Surfactant Protein-A Mainly Caused by Pro-Inflammatory Macrophages. Microorganisms 2020, 8, 577. https://doi.org/10.3390/microorganisms8040577
Schicke E, Cseresnyés Z, Rennert K, Vau V, Haupt KF, Hornung F, Nietzsche S, Swiczak F, Schmidtke M, Glück B, et al. Staphylococcus aureus Lung Infection Results in Down-Regulation of Surfactant Protein-A Mainly Caused by Pro-Inflammatory Macrophages. Microorganisms. 2020; 8(4):577. https://doi.org/10.3390/microorganisms8040577
Chicago/Turabian StyleSchicke, Elisabeth, Zoltán Cseresnyés, Knut Rennert, Vanessa Vau, Karoline Frieda Haupt, Franziska Hornung, Sandor Nietzsche, Fatina Swiczak, Michaela Schmidtke, Brigitte Glück, and et al. 2020. "Staphylococcus aureus Lung Infection Results in Down-Regulation of Surfactant Protein-A Mainly Caused by Pro-Inflammatory Macrophages" Microorganisms 8, no. 4: 577. https://doi.org/10.3390/microorganisms8040577
APA StyleSchicke, E., Cseresnyés, Z., Rennert, K., Vau, V., Haupt, K. F., Hornung, F., Nietzsche, S., Swiczak, F., Schmidtke, M., Glück, B., Koch, M., Schacke, M., Heller, R., Mosig, A. S., Figge, M. T., Ehrhardt, C., Löffler, B., & Deinhardt-Emmer, S. (2020). Staphylococcus aureus Lung Infection Results in Down-Regulation of Surfactant Protein-A Mainly Caused by Pro-Inflammatory Macrophages. Microorganisms, 8(4), 577. https://doi.org/10.3390/microorganisms8040577







