Effects of Halophyte Root Exudates and Their Components on Chemotaxis, Biofilm Formation and Colonization of the Halophilic Bacterium Halomonas Anticariensis FP35T
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Swimming Motility Assay
2.3. Identification of Chemosensory Systems in H. anticariensis FP35T
2.4. Cloning of CheA ORF in a Multicopy Plasmid
2.5. Extraction of Root Exudates from Salicornia Plants
2.6. Analysis of Salicornia Root Exudates Using UPL-HRMS Q-TOF
2.7. Chemotaxis Assays
2.7.1. Qualitative Capillary Chemotaxis Assay
2.7.2. Quantitative Capillary Assay
2.8. Biofilm Formation Assay
2.9. Salicornia Seed Bacterization and Germination Assay
2.10. Statistical Analyses
3. Results
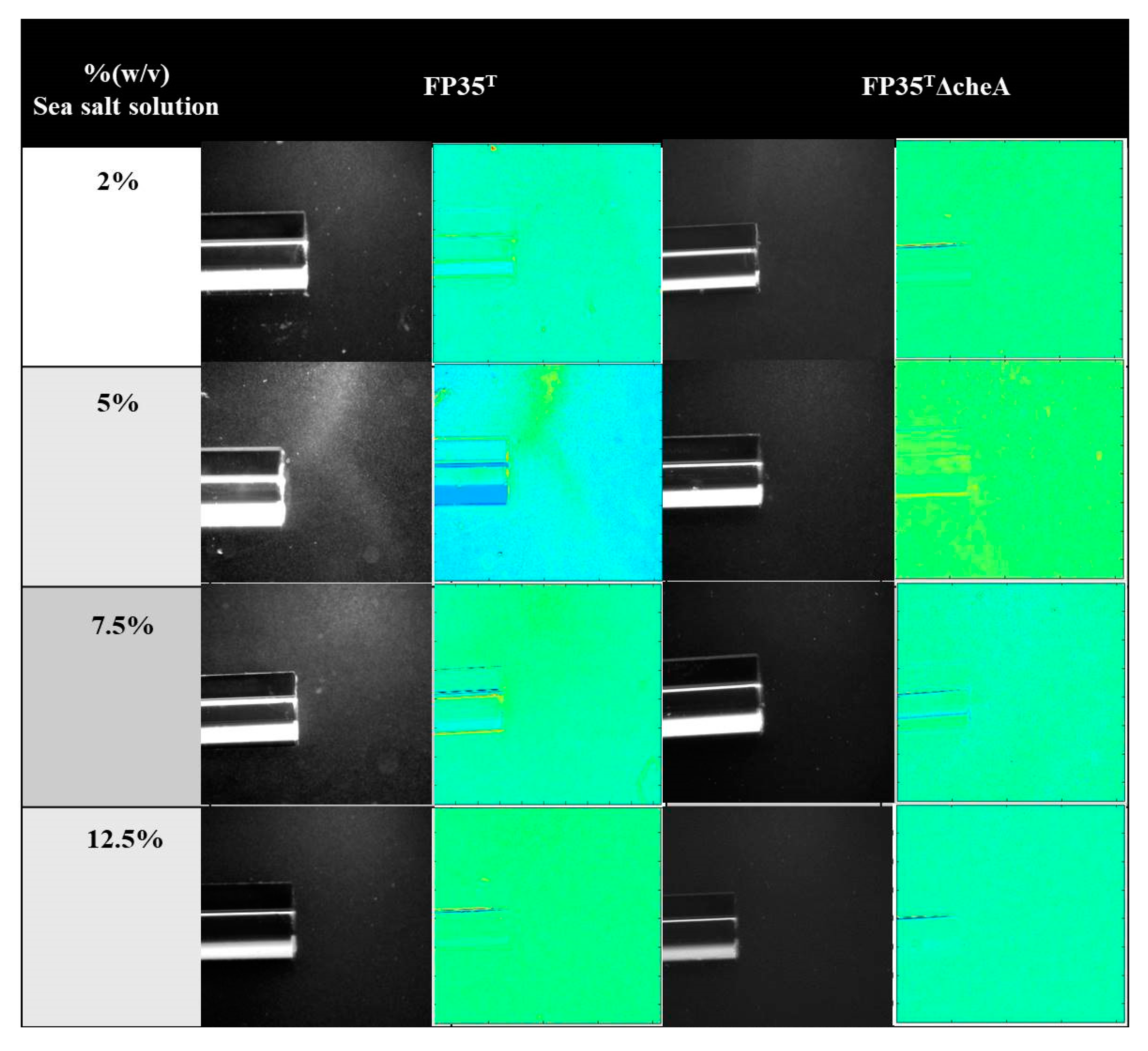
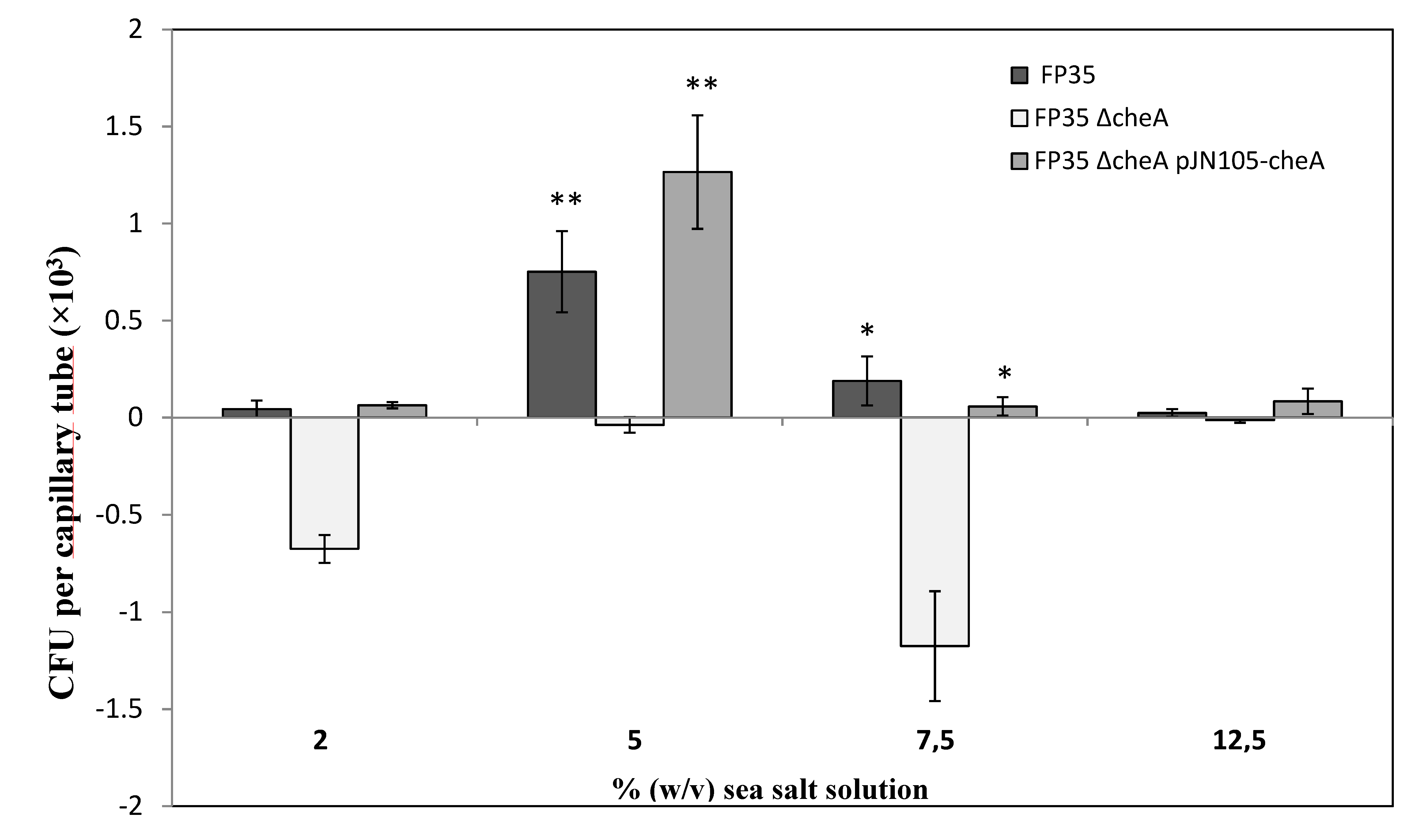
3.1. Assessment of the Chemotactic Response of H. anticariensis FP35T to Salicornia Root Exudates
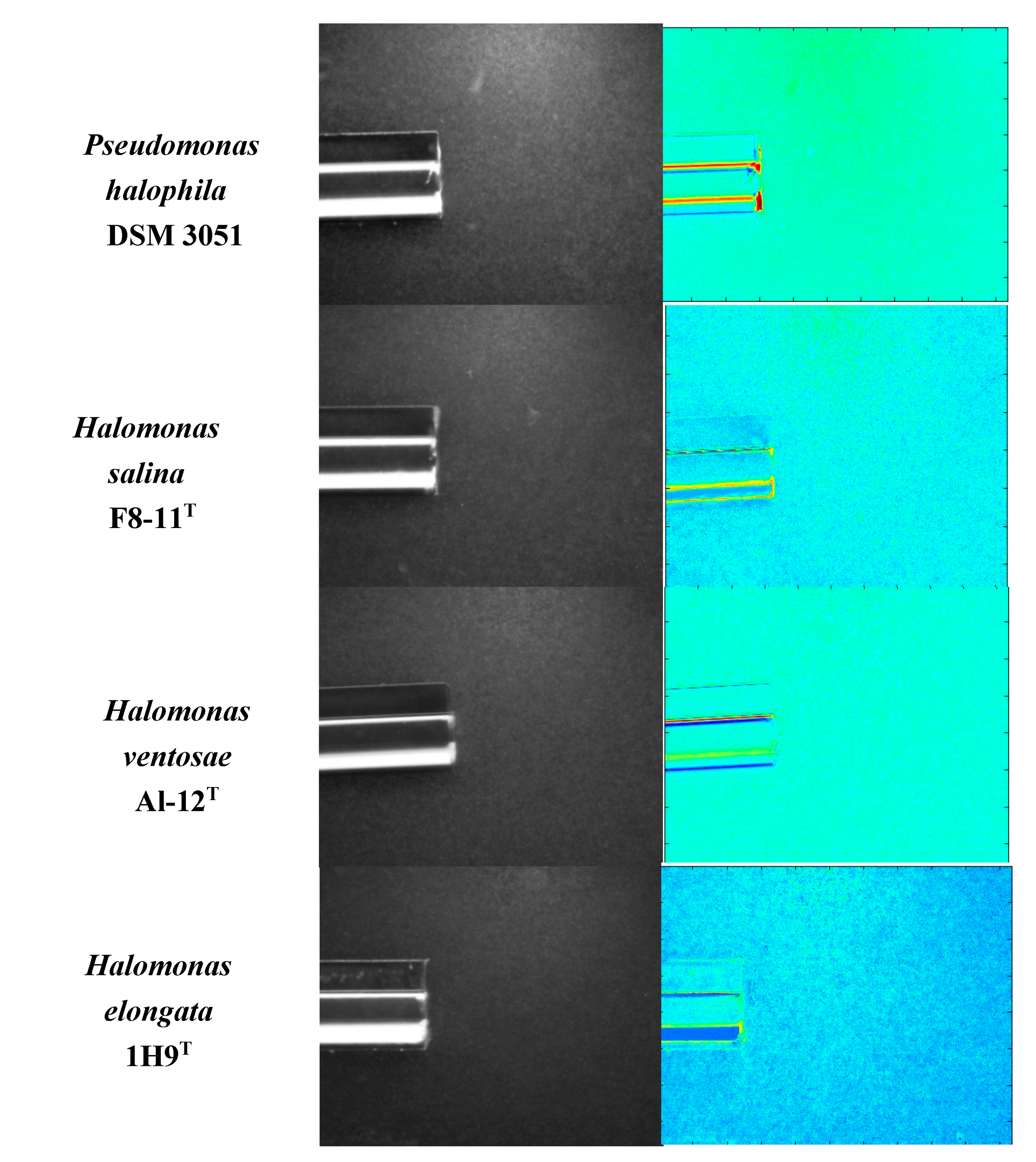
3.2. Chemoattraction to Salicornia Root Exudates in a Variety of Halophilic Bacteria
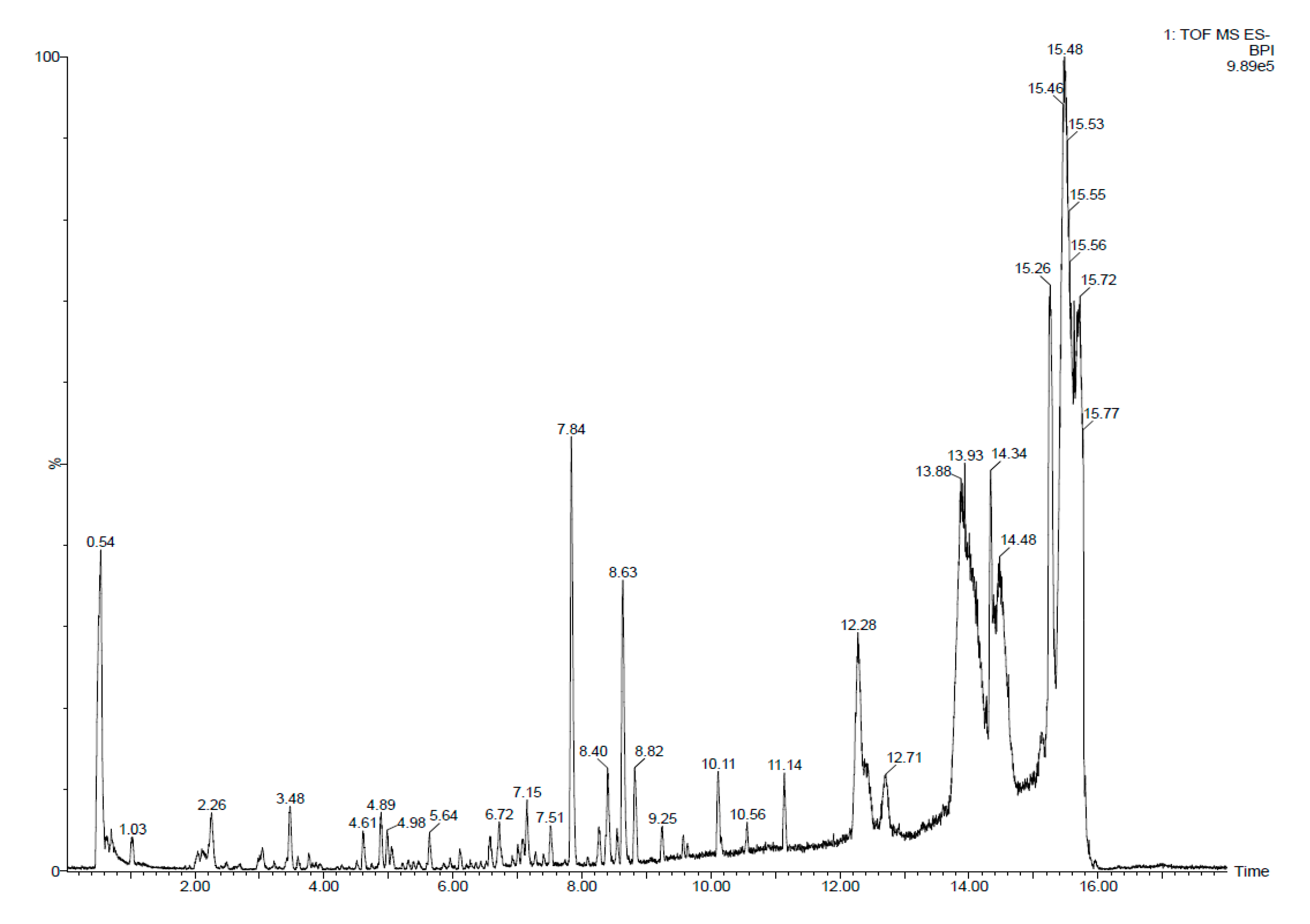
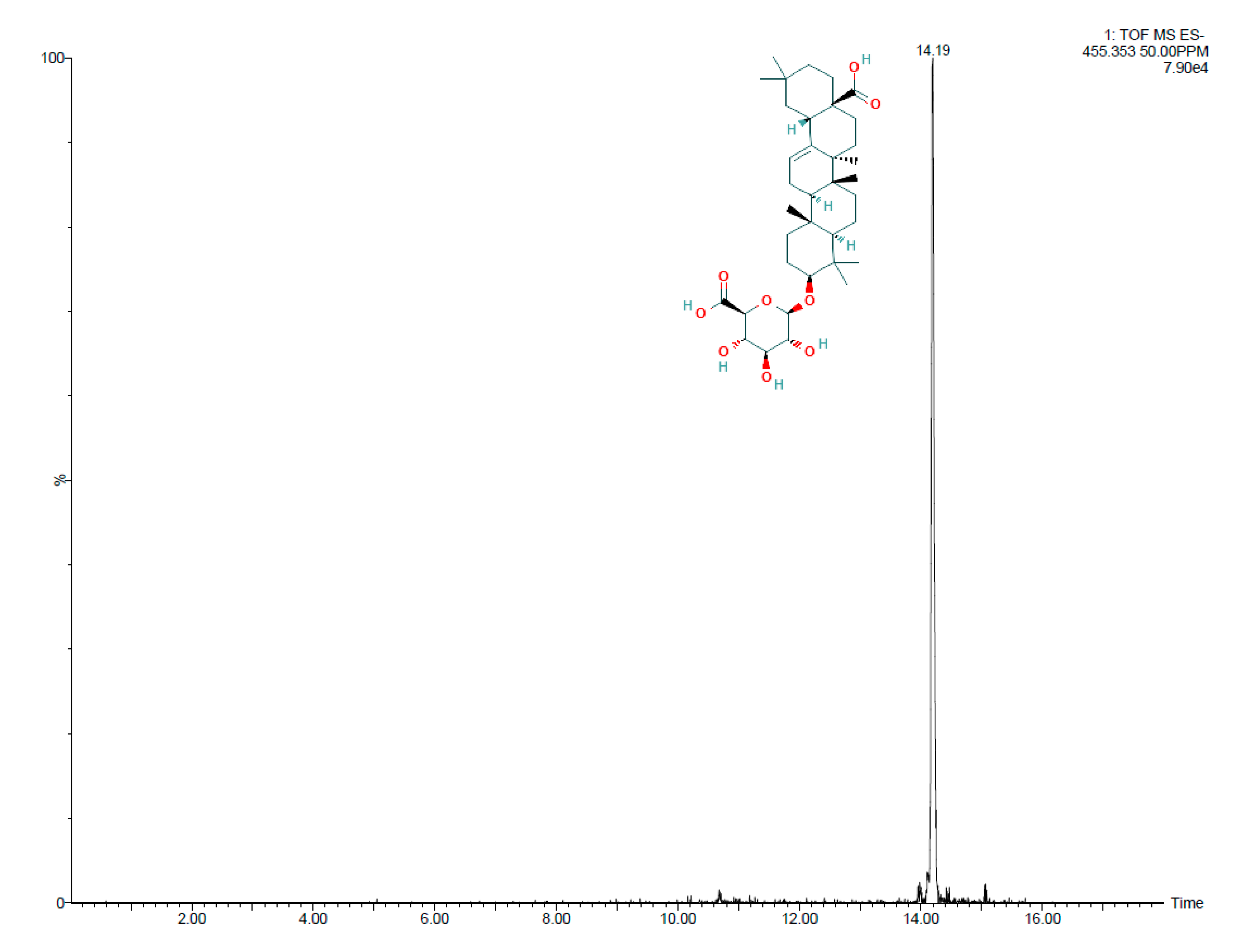
3.3. Identification of Salicornia Exudate Composition Using UPLC - HRMS Q-TOF
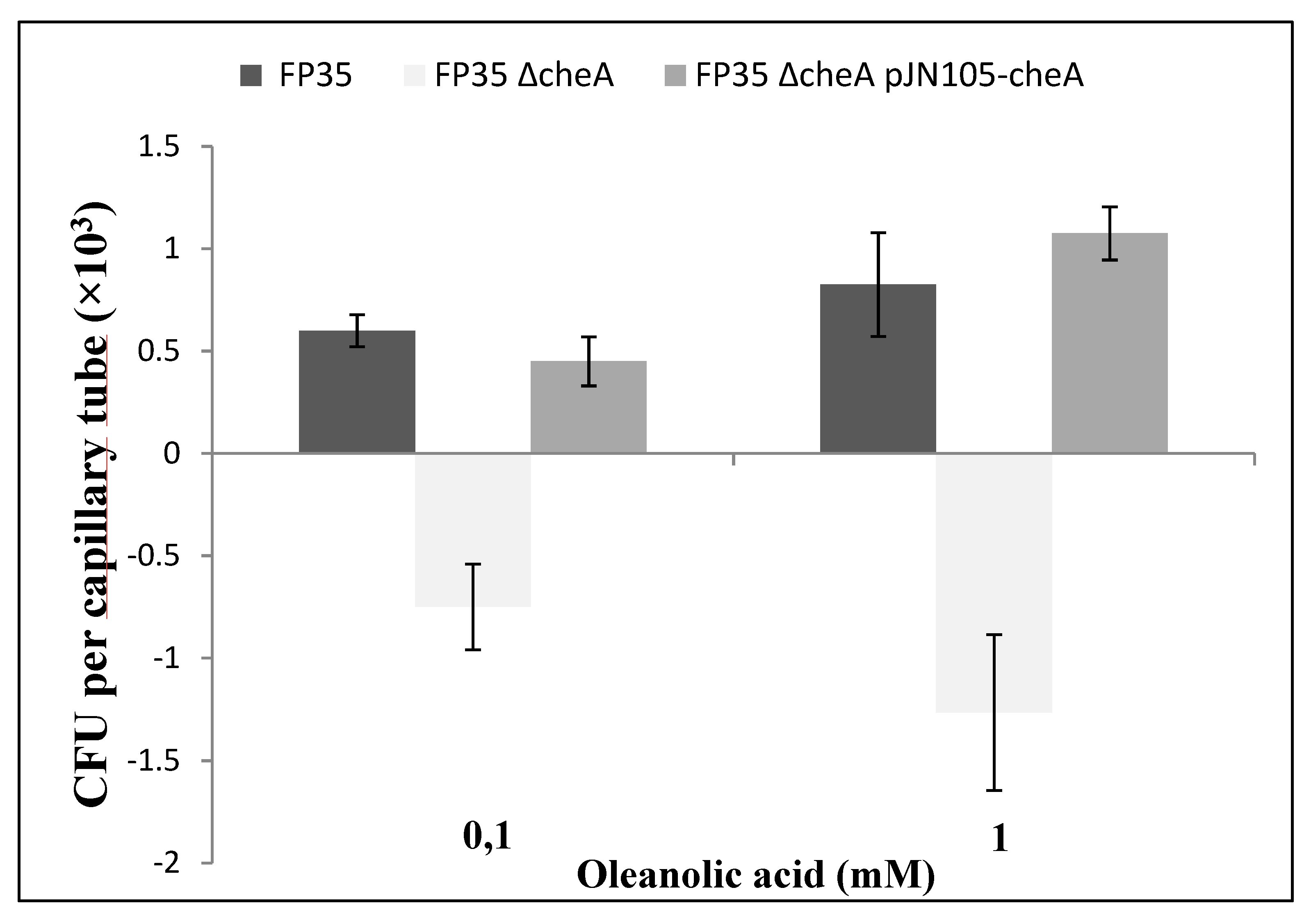
3.4. Assessment of Chemotactic Response of H. anticariensis FP35T to Oleanolic Acid
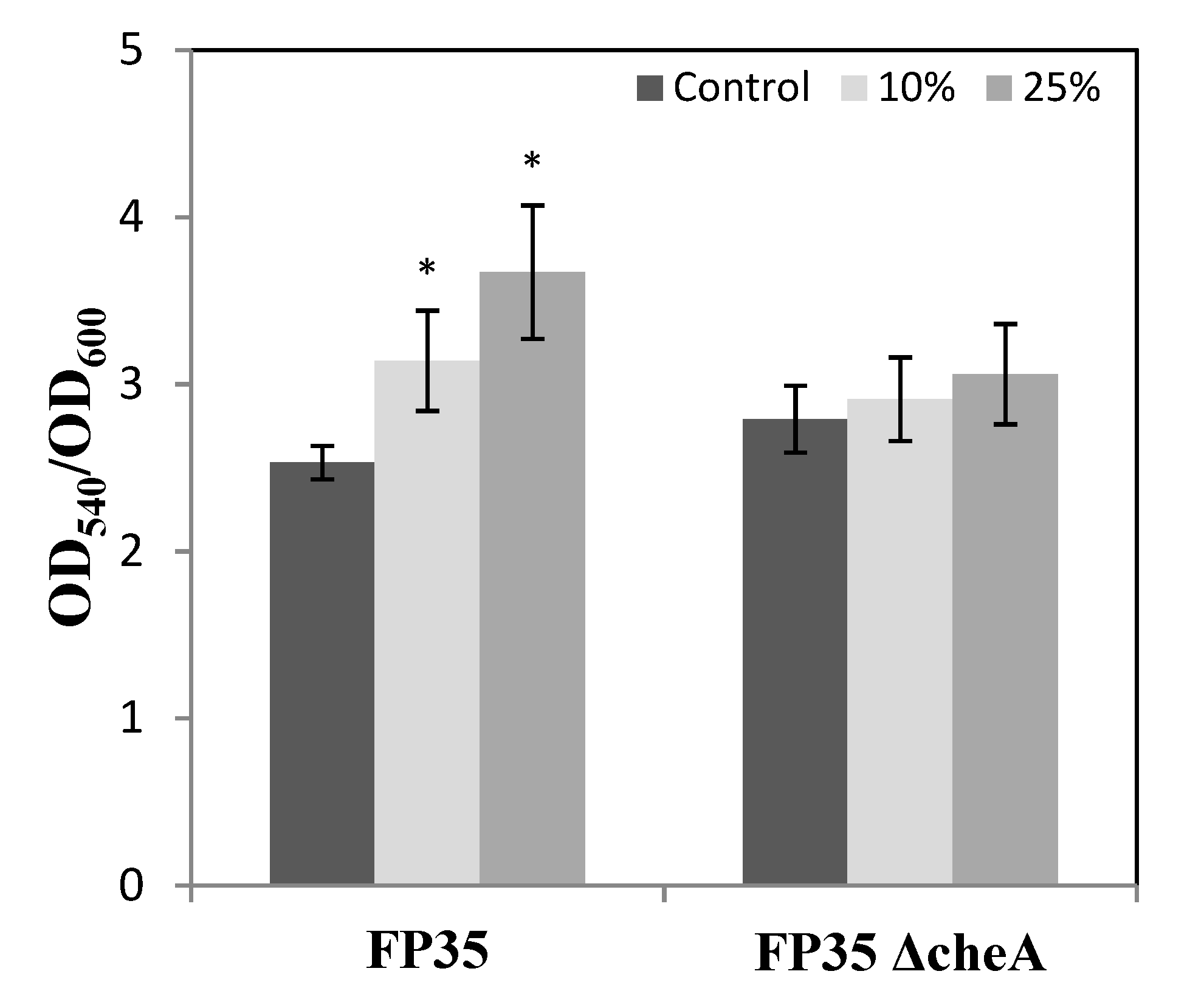
3.5. Evaluation of H. anticariensis FP35T Biofilm Formation in Response to Salicornia Root Exudates and Oleanolic Acid
3.6. Salicornia Plant Reactions to Inoculation with H. anticariensis strain FP35T and its Mutant Strain FP35ΔcheA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. PPB 2004, 42, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, K.C.; Venkatesan, K.; Balakrishnan, V.; Chellappan, K.P.; Balasubramanian, T. Restoration of saline land by halophytes for Indian soils. Soil Biol. Biochem. 2007, 39, 2661–2664. [Google Scholar] [CrossRef]
- Manousaki, E.; Kalogerakis, N. Halophytes—An emerging trend in phytoremediation. Int. J. Phytoremediat. 2011, 13, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Kadereit, G.; Ball, P.; Beer, S.; Mucina, L.; Sokoloff, D.; Teege, P.; Yaprak, A.E.; Freitag, H. A taxonomic nightmare comes true: Phylogeny and biogeography of glassworts (Salicornia L. Chenopodiaceae). Taxon 2007, 56, 1143–1170. [Google Scholar] [CrossRef]
- Ventura, Y.; Wuddineh, W.A.; Ephrath, Y.; Shpigel, M.; Sagi, M. Molybdenum as an essential element for improving total yield in seawater-grown Salicornia europaea (L.). Sci. Hortic. 2010, 126, 395–401. [Google Scholar] [CrossRef]
- Ventura, Y.; Sagi, M. Halophyte crop cultivation: The case for salicornia and sarcocornia. Environ. Exp. Bot. 2013, 92, 144–153. [Google Scholar] [CrossRef]
- Rhee, M.H.; Park, H.J.; Cho, J.Y. Salicornia herbacea: Botanical, chemical and pharmacological review of halophyte marsh plant. J. Med. Plants Res. 2009, 3, 548–555. [Google Scholar]
- Karthivashan, G.; Park, S.Y.; Kweon, M.H.; Kim, J.; Haque, M.E.; Cho, D.Y.; Kim, I.S.; Cho, E.A.; Ganesan, P.; Choi, D.K. Ameliorative potential of desalted Salicornia europaea L. extract in multifaceted Alzheimer’s-like scopolamine-induced amnesic mice model. Sci. Rep. 2018, 8, 7174. [Google Scholar] [CrossRef]
- Abdal, M.S. Salicornia production in Kuwait. World Appl. Sci. J. 2009, 6, 1033–1038. [Google Scholar]
- Moghaieb, R.E.A.; Saneoka, H.; Fujita, K. Effect of salinity on osmotic adjustment, glycinebetaine accumulation and the betaine aldehyde dehydrogenase gene expression in two halophytic plants, Salicornia europaea and Suaeda maritima. Plant Sci. 2004, 166, 1345–1349. [Google Scholar] [CrossRef]
- Glenn, E.P.; Pitelka, L.F.; Olsen, M.W. The use of halophytes to sequester carbon. Water Air Soil Pollut. 1992, 64, 251–263. [Google Scholar] [CrossRef]
- Ozawa, T.; Wu, J.; Fujii, S. Effect of inoculation with a strain of Pseudomonas pseudoalcaligenes isolated from the endorhizosphere of Salicornia europea on salt tolerance of the glasswort. Soil Sci. Plant Nutr. 2007, 53, 12–16. [Google Scholar] [CrossRef]
- Argandona, M.; Fernandez-Carazo, R.; Llamas, I.; Martinez-Checa, F.; Caba, J.M.; Quesada, E.; del Moral, A. The moderately halophilic bacterium Halomonas maura is a free-living diazotroph. FEMS Microbiol. Lett. 2005, 244, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Mapelli, F.; Marasco, R.; Rolli, E.; Barbato, M.; Cherif, H.; Guesmi, A.; Ouzari, I.; Daffonchio, D.; Borin, S. Potential for plant growth promotion of rhizobacteria associated with Salicornia growing in Tunisian hypersaline soils. BioMed Res. Int. 2013, 2013, 248078. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Canovas, M.J.; Bejar, V.; Martinez-Checa, F.; Quesada, E. Halomonas anticariensis sp. nov., from Fuente de Piedra, a saline-wetland wildfowl reserve in Malaga, southern Spain. Int. J. Syst. Evol. Microbiol. 2004, 54, 1329–1332. [Google Scholar] [CrossRef] [PubMed]
- Llamas, I.; Quesada, E.; Martinez-Canovas, M.J.; Gronquist, M.; Eberhard, A.; Gonzalez, J.E. Quorum sensing in halophilic bacteria: Detection of N-acyl-homoserine lactones in the exopolysaccharide-producing species of Halomonas. Extrem. Life Extrem. Cond. 2005, 9, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Mata, J.A.; Bejar, V.; Llamas, I.; Arias, S.; Bressollier, P.; Tallon, R.; Urdaci, M.C.; Quesada, E. Exopolysaccharides produced by the recently described halophilic bacteria Halomonas ventosae and Halomonas anticariensis. Res. Microbiol. 2006, 157, 827–835. [Google Scholar] [CrossRef]
- Sood, G. Chemotactic response of plant-growth-promoting bacteria towards roots of vesicular-arbuscular mycorrhizal tomato plants. FEMS Microbiol. Ecol. 2003, 45, 219–227. [Google Scholar] [CrossRef]
- Sampedro, I.; Parales, R.E.; Krell, T.; Hill, J.E. Pseudomonas chemotaxis. FEMS Microbiol. Rev. 2015, 39, 17–46. [Google Scholar]
- Flanary, P.L.; Allen, R.D.; Dons, L.; Kathariou, S. Insertional inactivation of the Listeria monocytogenes cheYA operon abolishes response to oxygen gradients and reduces the number of flagella. Can. J. Microbiol. 1999, 45, 646–652. [Google Scholar] [CrossRef]
- Foynes, S.; Dorrell, N.; Ward, S.J.; Stabler, R.A.; McColm, A.A.; Rycroft, A.N.; Wren, B.W. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 2000, 68, 2016–2023. [Google Scholar] [CrossRef] [PubMed]
- De Weert, S.; Vermeiren, H.; Mulders, I.H.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. MPMI 2002, 15, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Zhang, W.; Wang, D.; Mao, J.; Huang, Q.; Guo, S.; Shen, Q. Root Exudates from Grafted-Root Watermelon Showed a Certain Contribution in Inhibiting Fusarium oxysporum f. sp. niveum. PLoS ONE 2013, 8, e63383. [Google Scholar] [CrossRef] [PubMed]
- Rudrappa, T.; Czymmek, K.J.; Pare, P.W.; Bais, H.P. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef]
- Tena-Garitaonaindia, M.; Llamas, I.; Toral, L.; Sampedro, I. Chemotaxis of halophilic bacterium Halomonas anticariensis FP35 towards the environmental pollutants phenol and naphthalene. Sci. Total Environ. 2019, 669, 631–636. [Google Scholar] [CrossRef]
- Gasperotti, A.F.; Studdert, C.A.; Revale, S.; Herrera Seitz, M.K. Draft genome sequence of Halomonas sp. KHS3, a polyaromatic hydrocarbon-chemotactic strain. Genome Announc. 2015, 3, e00020-15. [Google Scholar] [CrossRef]
- Gasperotti, A.F.; Revuelta, M.V.; Studdert, C.A.; Herrera Seitz, M.K. Identification of two different chemosensory pathways in representatives of the genus Halomonas. BMC Genom. 2018, 19, 266. [Google Scholar]
- Li, S.; Zhang, N.; Zhang, Z.; Luo, J.; Shen, B.; Zhang, R.; Shen, Q. Antagonist Bacillus subtilis HJ5 controls Verticillium wilt of cotton by root colonization and biofilm formation. Biol. Fertil. Soils 2013, 49, 295–303. [Google Scholar] [CrossRef]
- Xu, Z.; Shao, J.; Li, B.; Yan, X.; Shen, Q.; Zhang, R. Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl. Environ. Microbiol. 2013, 79, 808–815. [Google Scholar] [CrossRef]
- Kasim, W.A.; Gaafar, R.M.; Abou-Ali, R.M.; Omar, M.N.; Hewait, H.M. Effect of biofilm forming plant growth promoting rhizobacteria on salinity tolerance in barley. Ann. Agric. Sci. 2016, 61, 217–227. [Google Scholar] [CrossRef]
- Llamas, I.; del Moral, A.; Martinez-Checa, F.; Arco, Y.; Arias, S.; Quesada, E. Halomonas maura is a physiologically versatile bacterium of both ecological and biotechnological interest. Antonie Van Leeuwenhoek 2006, 89, 395–403. [Google Scholar] [CrossRef]
- Qurashi, A.W.; Sabri, A.N. Bacterial exopolysaccharide and biofilm formation stimulate chikpea growth and soil aggregation under salt stress. Braz. J. Microbiol. 2012, 43, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Haynes, W.C.; Wicherham, L.J.; Hesseltine, C.W. Maintenance of cultures of industrially important microorganisms. Appl. Microbiol. 1955, 3, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Moraine, R.A.; Rogovin, P. Kinetics of polysaccharide B-1459 fermentation. Biotechnol. Bioeng. 1966, 8, 511–524. [Google Scholar] [CrossRef]
- Rodriguez-Valera, F.; Ruiz-Berraquero, F.; Ramos-Cormenzana, A. Characteristics of the heterotrophic bacterial populations in hypersaline environments of different salt concentrations. Microb. Ecol. 1981, 7, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, M.J.; Prado, B.; del Moral, A.; Rios, R.; Ramos-Cormenzana, A.; Campos, V. Numerical taxonomy of moderately halophilic gram-positive cocci isolated from the Salar de Atacama (Chile). Microbiologia 1991, 7, 35–41. [Google Scholar] [PubMed]
- Vreeland, R.H.; Litchfield, C.D.; Martin, E.L.; Elliot, E. Halomonas elongata, a New Genus and Species of Extremely Salt-Tolerant Bacteria. Int. J. Syst. Evol. Microbiol. 1980, 30, 485–495. [Google Scholar] [CrossRef]
- Fendrich, C. Halovibrio variabilis gen. nov. sp. nov., Pseudomonas halophila sp. nov. and a new halophilic aerobic coccoid Eubacterium from Great Salt Lake, Utah, USA. Syst. Appl. Microbiol 1988, 11, 36–43. [Google Scholar] [CrossRef]
- Martinez-Canovas, M.J.; Quesada, E.; Llamas, I.; Bejar, V. Halomonas ventosae sp. nov., a moderately halophilic, denitrifying, exopolysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 733–737. [Google Scholar] [CrossRef]
- Amjres, H.; Bejar, V.; Quesada, E.; Abrini, J.; Llamas, I. Halomonas rifensis sp. nov., an exopolysaccharide-producing, halophilic bacterium isolated from a solar saltern. Int. J. Syst. Evol. Microbiol. 2011, 61, 2600–2605. [Google Scholar] [CrossRef]
- Martinez-Checa, F.; Bejar, V.; Martinez-Canovas, M.J.; Llamas, I.; Quesada, E. Halomonas almeriensis sp. nov., a moderately halophilic, exopolysaccharide-producing bacterium from Cabo de Gata, Almería, south-east Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 2007–2011. [Google Scholar] [CrossRef] [PubMed]
- González-Domenech, C.M.; Martinez-Checa, F.; Quesada, E.; Bejar, V. Halomonas cerina sp. nov., a moderately halophilic, denitrifying, exopolysaccharide-producing bacterium. Int. J. Syst. Evol. Microbiol. 2008, 58, 803–809. [Google Scholar]
- Calvo, C.; Martínez-Checa, F.; Toledo, F.; Porcel, J.; Quesada, E. Characteristics of bioemulsifiers synthesised in crude oil media by Halomonas eurihalina and their effectiveness in the isolation of bacteria able to grow in the presence of hydrocarbons. Appl. Microbiol. Biotechnol. 2002, 60, 347–351. [Google Scholar] [PubMed]
- Llamas, I.; Bejar, V.; Martinez-Checa, F.; Martinez-Canovas, M.J.; Molina, I.; Quesada, E. Halomonas stenophila sp. nov., a halophilic bacterium that produces sulphate exopolysaccharides with biological activity. Int. J. Syst. Evol. Microbiol. 2011, 61, 2508–2514. [Google Scholar] [CrossRef] [PubMed]
- Luque, R.; Bejar, V.; Quesada, E.; Martinez-Checa, F.; Llamas, I. Halomonas ramblicola sp. nov., a moderately halophilic bacterium from Rambla Salada, a Mediterranean hypersaline rambla. Int. J. Syst. Evol. Microbiol. 2012, 62, 2903–2909. [Google Scholar] [CrossRef] [PubMed]
- García, T.; Mellado, E.; Ostos, J.C.; Ventosa, A. Halomonas organivorans sp. nov., a moderate halophile able to degrade aromatic compounds. Int. J. Syst. Evol. Microbiol. 2004, 54, 1723–1728. [Google Scholar]
- Ha, D.G.; Kuchma, S.L.; O’Toole, G. Plate-based assay for swimming motility in Pseudomonas aeruginosa. Methods Mol. Biol. 2014, 1149, 59–65. [Google Scholar]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Markowitz, V.M.; Chen, I.M.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P.; et al. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012, 40, D115–D122. [Google Scholar] [CrossRef]
- Newman, J.R.; Fuqua, C. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 1999, 227, 197–203. [Google Scholar] [CrossRef]
- Demarre, G.; Guerout, A.M.; Matsumoto-Mashimo, C.; Rowe-Magnus, D.A.; Marliere, P.; Mazel, D. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 2005, 156, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J.; Ruhmann, B.; Sieber, V.; Romero-Jimenez, L.; Sanjuan, J.; Perez-Mendoza, D. Screening of c-di-GMP-Regulated Exopolysaccharides in Host Interacting Bacteria. Methods Mol. Biol. 2018, 1734, 263–275. [Google Scholar] [PubMed]
- Zhao, F.J.; Hamon, R.E.; McLaughlin, M.J. Root exudates of the hyperaccumulator Thlaspi caerulescens do not enhance metal mobilization. New Phytol. 2001, 151, 613–620. [Google Scholar] [CrossRef]
- Parales, R.E.; Harwood, C.S. Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr. Opin. Microbiol. 2002, 5, 266–273. [Google Scholar] [CrossRef]
- Adler, J. Method for Measuring Chemotaxis and Use of Method to Determine Optimum Conditions for Chemotaxis by Escherichia coli. J. Gen. Microbiol. 1973, 74, 77–91. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. JoVE 2011, 47, 2437. [Google Scholar] [CrossRef]
- Jha, B.; Gontia, I.; Hartmann, A. The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant Soil 2012, 356, 265–277. [Google Scholar] [CrossRef]
- Abdul Baki, A.A.; Anderson, J.D. Vigour determination in soya bean by multiple criteria. Crop Sci. 1973, 13, 630–633. [Google Scholar] [CrossRef]
- Martins, S.J.; Medeiros, F.H.V.; Lakshmanan, V.; Bais, H.P. Impact of Seed Exudates on Growth and Biofilm Formation of Bacillus amyloliquefaciens ALB629 in Common Bean. Front. Microbiol. 2017, 8, 2631. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Ling, N.; Raza, W.; Ma, J.; Huang, Q.; Shen, Q. Identification and role of organic acids in watermelon root exudates for recruiting Paenibacillus polymyxa SQR-21 in the rhizosphere. Eur. J. Soil Biol. 2011, 47, 374–379. [Google Scholar] [CrossRef]
- Bais, H.P.; Park, S.W.; Weir, T.L.; Callaway, R.M.; Vivanco, J.M. How plants communicate using the underground information superhighway. Trends Plant Sci. 2004, 9, 26–32. [Google Scholar] [CrossRef]
- Marasco, R.; Mapelli, F.; Rolli, E.; Mosqueira, M.J.; Fusi, M.; Bariselli, P.; Reddy, M.; Cherif, A.; Tsiamis, G.; Borin, S.; et al. Salicornia strobilacea (Synonym of Halocnemum strobilaceum) grown under different tidal regimes selects rhizosphere bacteria capable of promoting plant growth. Front. Microbiol. 2016, 7, 1286. [Google Scholar] [CrossRef]
- Haas, D.; Defago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Tahrioui, A.; Quesada, E.; Llamas, I. Genetic and phenotypic analysis of the GacS/GacA system in the moderate halophile Halomonas anticariensis. Microbiology 2013, 159, 462–474. [Google Scholar] [CrossRef]
- D’Ippólito, S.; de Castro, R.E.; Herrera Seitz, M.K. Chemotactic response to gas oil of Halomonas spp. strains isolated from saline environments in Argentina. Rev. Argent. Microbiol. 2011, 43, 107–110. [Google Scholar]
- Parales, R.E.; Ditty, J.L.; Harwood, C.S. Toluene-degrading bacteria are chemotactic towards the environmental pollutants benzene, toluene, and trichloroethylene. Appl. Environ. Microbiol. 2000, 66, 4098–4104. [Google Scholar] [CrossRef]
- Vardar, G.; Barbieri, P.; Wood, T.K. Chemotaxis of Pseudomonas stutzeri OX1 and Burkholderia cepacia G4 toward chlorinated ethenes. Appl. Microbiol. Biotechnol. 2005, 66, 696–701. [Google Scholar] [CrossRef]
- Kato, J.; Shitashiro, M.; Yamamoto, M.; Kuroda, A.; Ikeda, T.; Takiguchi, N.; Ohtake, H. Chemotaxis by Pseudomonas aeruginosa toward 2,4-dichlorophenoxyacetate and volatile chlorinated aliphatic compounds. Abstr. Gen. Meet. Am. Soc. Microbiol. 2001, 101, 627. [Google Scholar]
- Shitashiro, M.; Kato, J.; Fukumura, T.; Kuroda, A.; Ikeda, T.; Takiguchi, N.; Ohtake, H. Evaluation of bacterial aerotaxis for its potential use in detecting the toxicity of chemicals to microorganisms. J. Biotechnol. 2003, 101, 11–18. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, D.; Liu, Y.; Li, S.; Shen, Q.; Zhang, R. Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil 2014, 374, 689–700. [Google Scholar] [CrossRef]
- Chen, Y.; Cao, S.; Chai, Y.; Clardy, J.; Kolter, R.; Guo, J.H.; Losick, R. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol. Microbiol. 2012, 85, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Petriacq, P.; Williams, A.; Cotton, A.; McFarlane, A.E.; Rolfe, S.A.; Ton, J. Metabolite profiling of non-sterile rhizosphere soil. Plant J. Cell Mol. Biol. 2017, 92, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.M.; Chakraborty, D.; Dey, S. Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 2010, 5, 359–368. [Google Scholar] [CrossRef]
- Pollier, J.; Goossens, A. Oleanolic acid. Phytochemistry 2012, 77, 10–15. [Google Scholar] [CrossRef]
- Wu, D.; Ding, W.; Zhang, Y.; Liu, X.; Yang, L. Oleanolic Acid Induces the Type III Secretion System of Ralstonia solanacearum. Front. Microbiol. 2015, 6, 1466. [Google Scholar] [CrossRef]
- Reyes-Darias, J.A.; Yang, Y.; Sourjik, V.; Krell, T. Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa. Mol. Microbiol. 2015, 96, 513–525. [Google Scholar] [CrossRef]
- Collins, K.D.; Andermann, T.M.; Draper, J.; Sanders, L.; Williams, S.M.; Araghi, C.; Ottemann, K.M. The Helicobacter pylori CZB cytoplasmic chemoreceptor TlpD forms an autonomous polar chemotaxis signalling complex that mediates a tactic response to oxidative stress. J. Bacteriol. 2016, 198, 1563–1575. [Google Scholar] [CrossRef]
- Tashiro, Y.; Inagaki1a, A.; Ono, K.; Inaba, T.; Yawata, Y.; Uchiyama, H.; Nomura, N. Low concentrations of ethanol stimulate biofilm and pellicle formation in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 2014, 78, 178–181. [Google Scholar] [CrossRef]
- Reyes-Darias, J.A.; Garcia, V.; Rico-Jimenez, M.; Corral-Lugo, A.; Lesouhaitier, O.; Juarez-Hernandez, D.; Yang, Y.; Bi, S.; Feuilloley, M.; Munoz-Rojas, J.; et al. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol. Microbiol. 2015, 97, 488–501. [Google Scholar] [CrossRef]
- Matilla, M.A.; Espinosa-Urgel, M.; Rodriguez-Herva, J.J.; Ramos, J.L.; Ramos-Gonzalez, M.I. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 2007, 8, R179. [Google Scholar] [CrossRef]
| Bacterial Inoculation | Shoot (cm) | % Increase | Root (cm) | % Increase | Germination (%) | Vigour Index | % Increase |
|---|---|---|---|---|---|---|---|
| Control | 0.159 ± 0.06a | 0.236 ± 0.11a | 29 | 12.52 | |||
| H. anticariensis FP35T | 0.179 ± 0.08ab | 12,5% | 0.679 ± 0.19b | 287% | 37 | 31.7 | 153% |
| H. anticariensis FP35 ΔcheA | 0.152 ± 0.05a | 0% | 0.548 ± 0.13b | 232% | 32 | 22.4 | 79% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampedro, I.; Pérez-Mendoza, D.; Toral, L.; Palacios, E.; Arriagada, C.; Llamas, I. Effects of Halophyte Root Exudates and Their Components on Chemotaxis, Biofilm Formation and Colonization of the Halophilic Bacterium Halomonas Anticariensis FP35T. Microorganisms 2020, 8, 575. https://doi.org/10.3390/microorganisms8040575
Sampedro I, Pérez-Mendoza D, Toral L, Palacios E, Arriagada C, Llamas I. Effects of Halophyte Root Exudates and Their Components on Chemotaxis, Biofilm Formation and Colonization of the Halophilic Bacterium Halomonas Anticariensis FP35T. Microorganisms. 2020; 8(4):575. https://doi.org/10.3390/microorganisms8040575
Chicago/Turabian StyleSampedro, Inmaculada, Daniel Pérez-Mendoza, Laura Toral, Esther Palacios, César Arriagada, and Inmaculada Llamas. 2020. "Effects of Halophyte Root Exudates and Their Components on Chemotaxis, Biofilm Formation and Colonization of the Halophilic Bacterium Halomonas Anticariensis FP35T" Microorganisms 8, no. 4: 575. https://doi.org/10.3390/microorganisms8040575
APA StyleSampedro, I., Pérez-Mendoza, D., Toral, L., Palacios, E., Arriagada, C., & Llamas, I. (2020). Effects of Halophyte Root Exudates and Their Components on Chemotaxis, Biofilm Formation and Colonization of the Halophilic Bacterium Halomonas Anticariensis FP35T. Microorganisms, 8(4), 575. https://doi.org/10.3390/microorganisms8040575







