Aestipascuomyces dupliciliberans gen. nov, sp. nov., the First Cultured Representative of the Uncultured SK4 Clade from Aoudad Sheep and Alpaca
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Isolation
2.3. Morphological Characterization
2.4. Substrate Utilization
2.5. Phylogenetic Analysis and Ecological Distribution
2.6. Data and Culture Accession
3. Results
3.1. Obtained Isolates
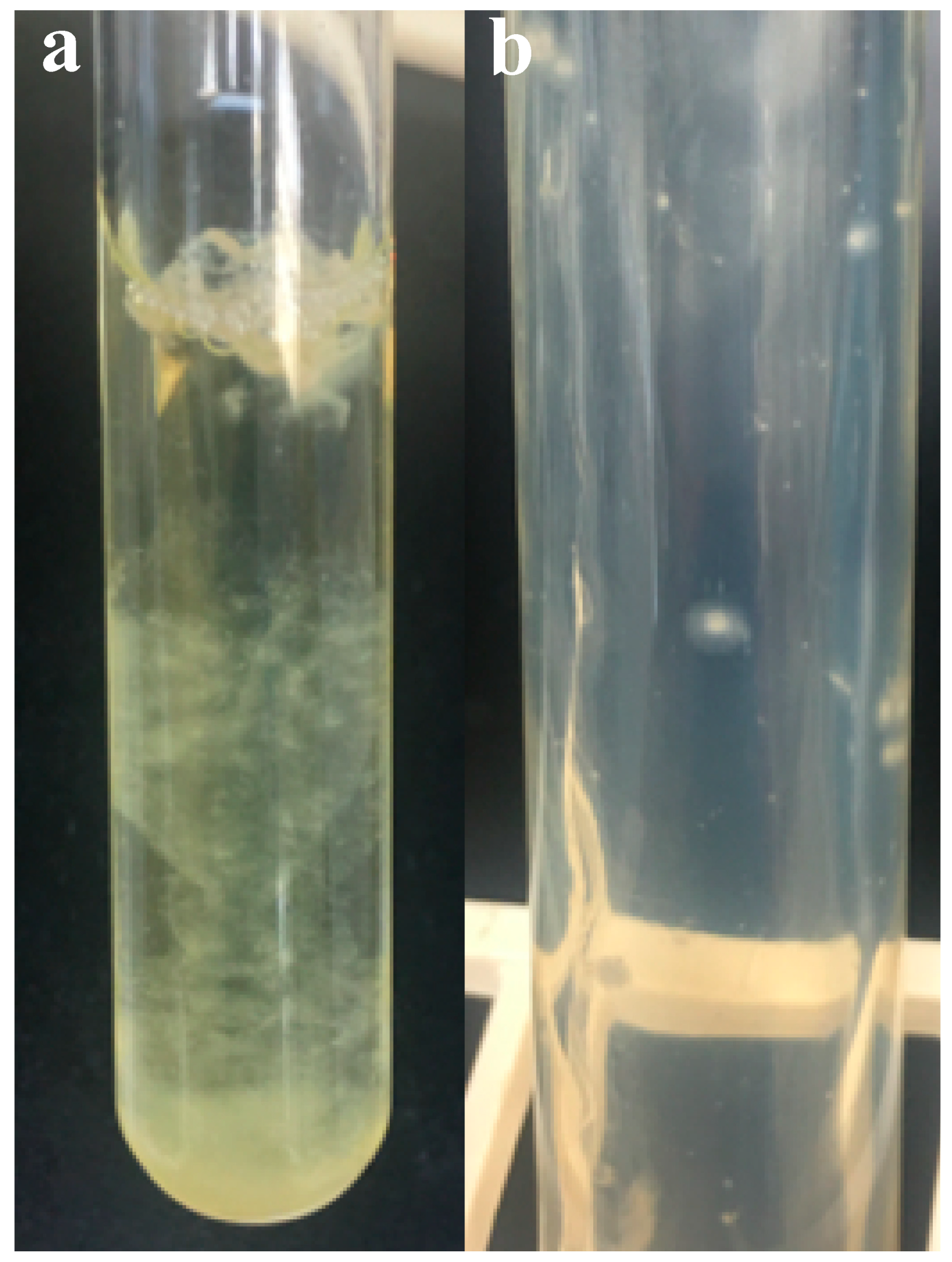
3.2. Colony Morphology and Liquid Growth Pattern
3.3. Microscopic Features
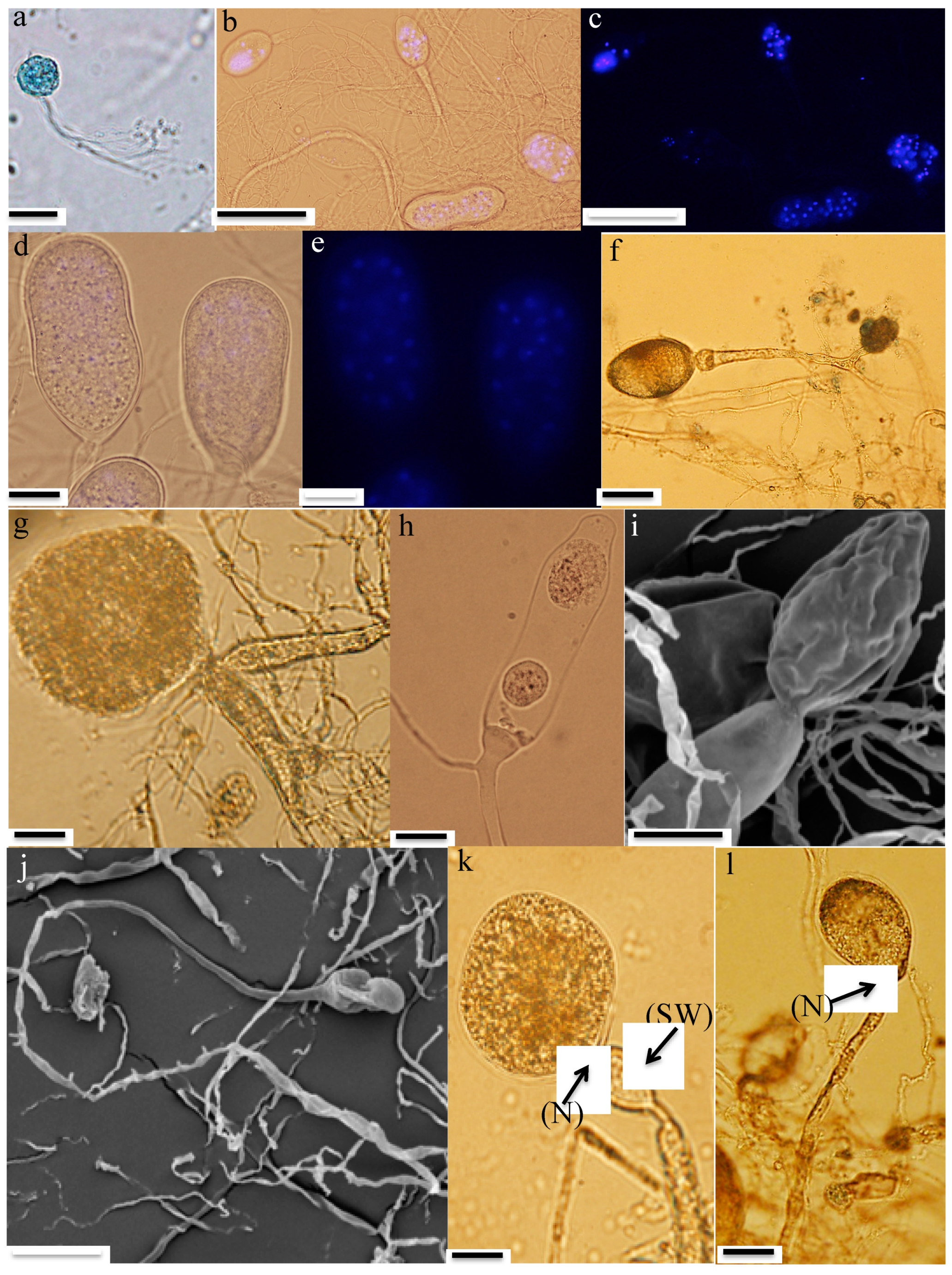
- Zoospores: Strain R4 produced globose zoospores with an average diameter of 9.3 ± 2.1 μm (standard deviation for 60 zoospores, range: 5–14 μm) (Figure 2a). All zoospores were polyflagellated, with 7–20 flagella and an average flagellum length of 28.1 ± 4.8 μm (average ± standard deviation from 60 zoospores, range: 19–36 μm). Strain A252 produced slightly larger spherical zoospores (10–20 μm, average 14 μm) with slightly longer flagella (35–49 μm, average 42 μm) (Figure 3a).
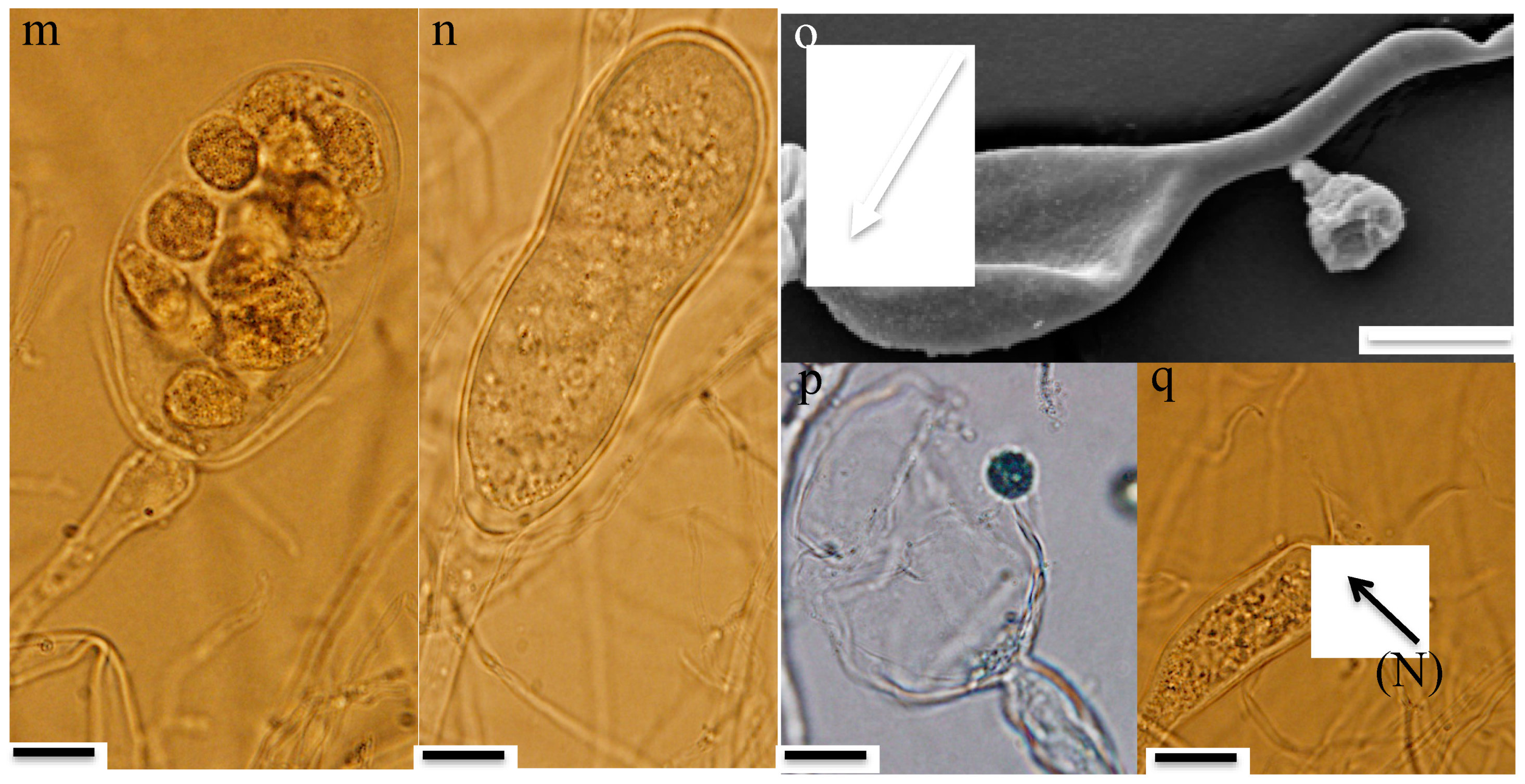
- Thalli and sporangia: Zoospore germination in strain R4 resulted in monocentric thalli with highly-branched anucleated rhizoids (Figure 2b–e). Strain R4 displayed both endogenous and exogenous thallus development. Endogenous thalli developed as a result of enlargement of zoospore cysts into sporangia with one (Figure 2f), or two adjacent (Figure 2g) rhizoidal systems. Endogenous sporangia displayed different shapes and sizes including ovoid (20–70 μm length (L) × 15–45 μm width (W) (Figure 2f), rhomboid (30–70 μm L × 40–85 μm W) (Figure 2g), and elongated (25–90 μm L × 15–40 μm W) (Figure 2h). No intercalary or pseudo-intercalary sporangia (sporangia present between two main rhizoidal systems) were observed. Exogenous sporangia were mainly developed at the end of unbranched sporangiophores that ranged in length between 10 and 300 μm (Figure 2i,j). Wide flattened sporangiophores (Figure 2i) and sporangiophores ending with sub-sporangial swellings (Figure 2k) were also frequently encountered. Mature exogenous sporangia ranged in size between 40 and 90 μm (L) and 15 and 35 μm (W), and they exhibited different morphologies including obpyriform (Figure 2i), ellipsoid (Figure 2j), globose (Figure 2k), ovoid (Figure 2m), and constricted ellipsoid (Figure 2n). Sporangial necks (the point between sporangia and rhizoids) were either tightly constricted (Figure 2f,k,q), or broad (Figure 2j,l,n,o). The neck opening, port, was either narrow (Figure 2k) or wide (Figure 2l). On the other hand, only endogenous sporangia were observed for strain A252, with one (Figure 3b) or, less frequently, two adjacent (Figure 3c) rhizoidal systems. Endogenous sporangia were mainly globose (diameter up to 145 µm) (Figure 3d), and ellipsoidal (up to 76 µm L × 48 µm W) (Figure 3e).
- Zoospore release: Zoospore release in strain R4 was achieved through two mechanisms, either from an apical pore (Figure 2o), as previously observed in Feramyces [14]), or through rupturing of the sporangial wall (Figure 2p), as commonly observed in Neocallimastix [16]. To our knowledge, the simultaneous utilization of both mechanisms by a single strain has not been previously reported in other AGF taxa. Sporangial walls either stayed intact (Figure 2o) or completely disintegrated after zoospore discharge (Figure 2q).
3.4. Substrate Utilization
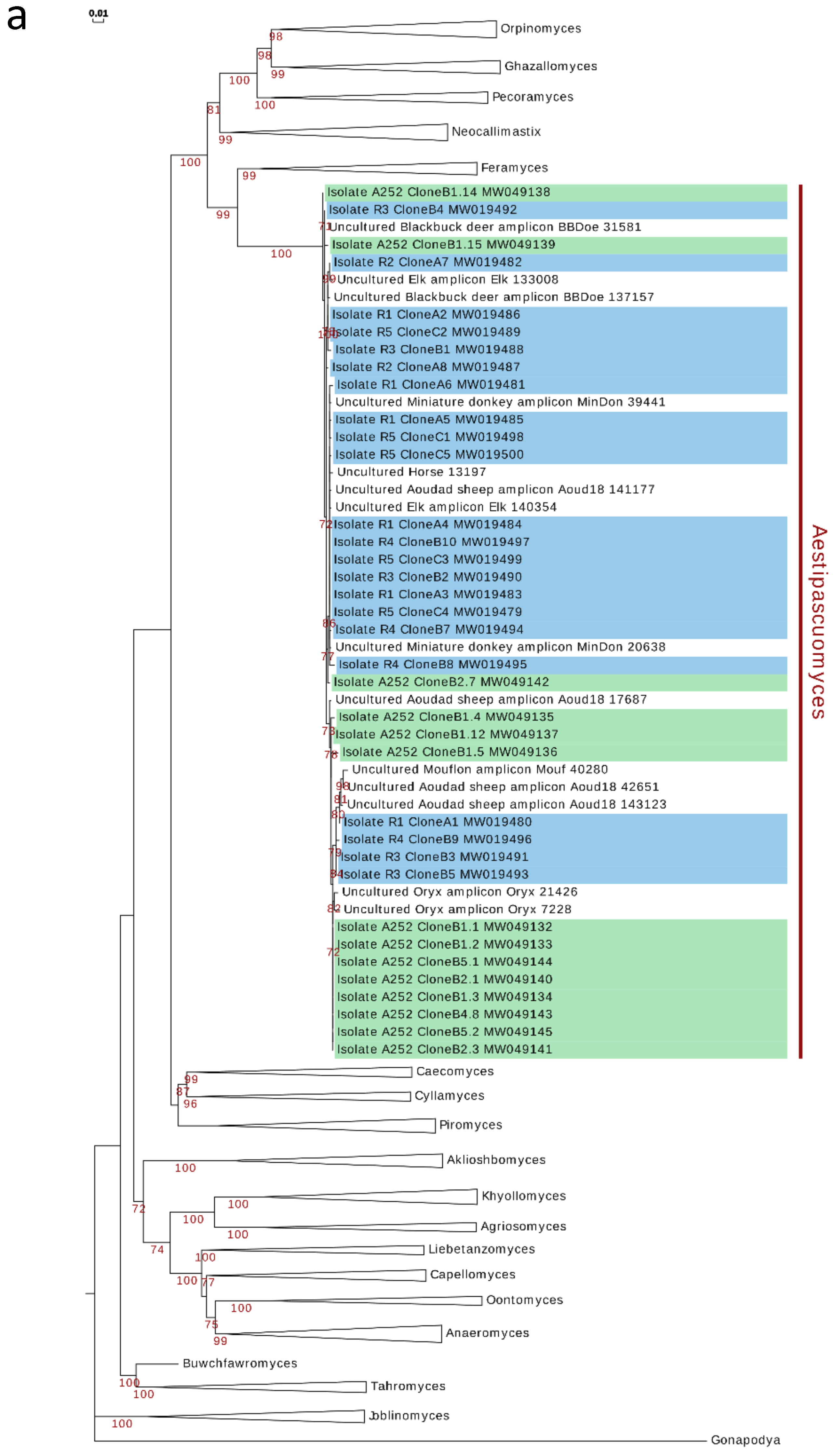
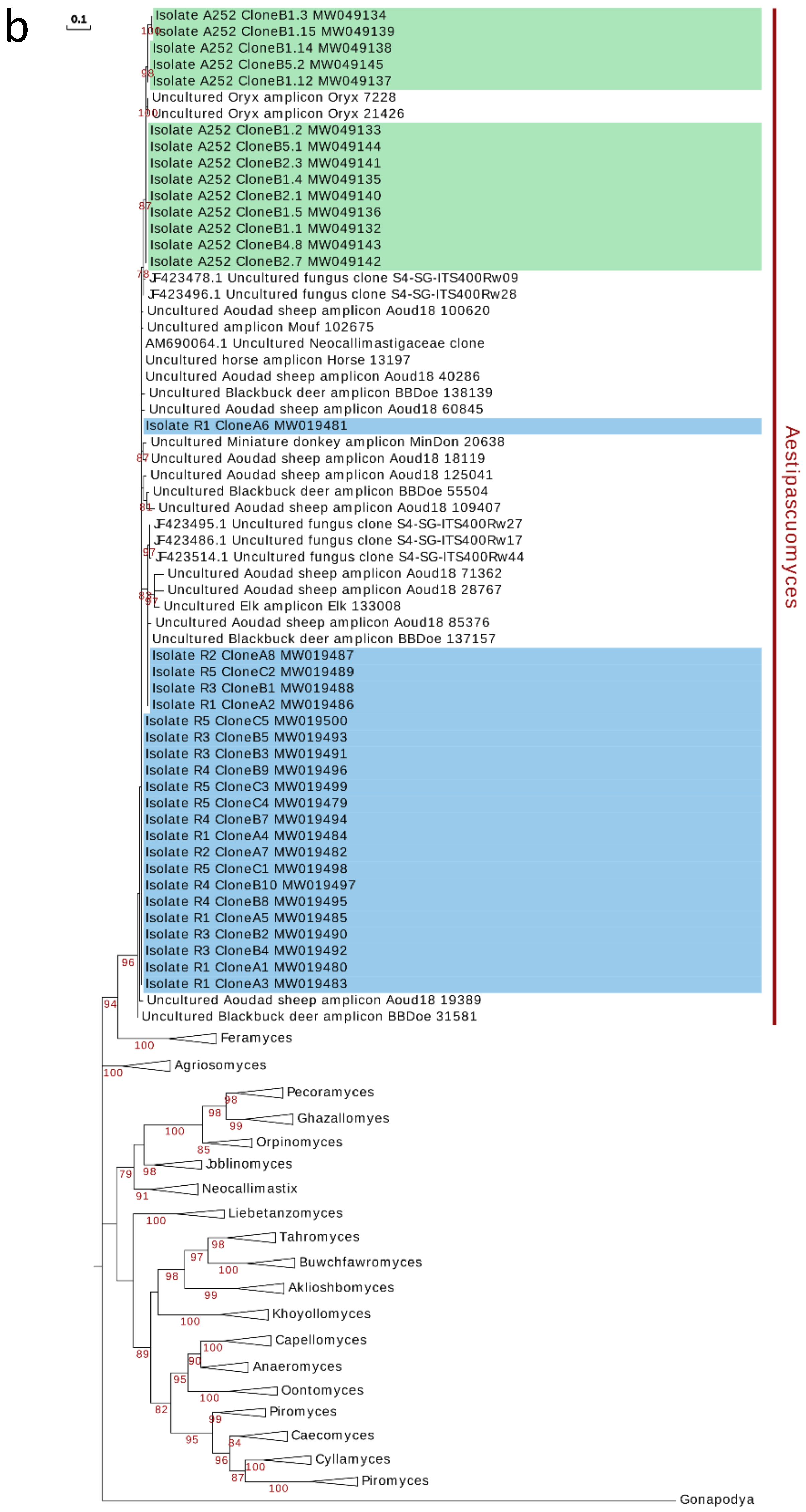
3.5. Phylogenetic Analysis and Ecological Distribution
4. Discussion
5. Taxonomy
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gruninger, R.J.; Puniyab, A.K.; Callaghanc, T.M.; Edwardsc, J.E.; Youssef, N.; Dagare, S.S.; Fliegerova, K.; Griffith, G.W.; Forster, R.; Tsang, A.; et al. Anaerobic Fungi (Phylum Neocallimastigomycota): Advances in understanding of their taxonomy, life cycle, ecology, role, and biotechnological potential. FEMS Microbiol. Ecol. 2014, 90, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Youssef, N.H.; Couger, M.B.; Struchtemeyer, C.G.; Liggenstoffer, A.S.; Prade, R.A.; Najar, F.Z.; Atiyeh, H.K.; Wilkins, M.R.; Elshahed, M.S. Genome of the anaerobic fungus Orpinomyces sp. C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 2013, 79, 4620–4634. [Google Scholar] [CrossRef] [Green Version]
- Hagen, L.H.; Brooke, C.G.; Shaw, C.A.; Norbeck, A.D.; Piao, H.; Arntzen, M.Ø.; Olson, H.M.; Copeland, A.; Isern, N.; Shukla, A.; et al. Proteome specialization of anaerobic fungi during ruminal degradation of recalcitrant plant fiber. ISME J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.E.; Forster, R.J.; Callaghan, T.M.; Dollhofer, V.; Dagar, S.S.; Cheng, Y.; Chang, J.; Kittelmann, S.; Fliegerova, K.; Puniya, A.K.; et al. PCR and omics based techniques to study the diversity, ecology and biology of anaerobic fungi: Insights, challenges and opportunities. Front. Microbiol. 2017, 8, 1657. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Zhou, M.; Ma, T.; Bi, S.; Wang, W.; Zhang, Y.; Huang, X.; Guan, L.L.; Long, R. Survey of rumen microbiota of domestic grazing yak during different growth stages revealed novel maturation patterns of four key microbial groups and their dynamic interactions. Anim. Microbiome 2020, 2, 23. [Google Scholar] [CrossRef]
- Haitjema, C.H.; Gilmore, S.P.; Henske, J.K.; Solomon, K.V.; Groot, R.d.; Kuo, A.; Mondo, S.J.; Salamov, A.A.; LaButti, K.; Zhao, Z.; et al. A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2017, 2, 17087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barr, D.J.S.; Kudo, H.; Jakober, K.D.; Cheng, K.-J. Morphology and development of rumen fungi: Neocallimastix sp., Piromyces communis, and Orpinomyces bovis gen. nov., sp. nov. Can. J. Bot. 1989, 67, 2815–2824. [Google Scholar] [CrossRef]
- Gold, J.J.; Heath, I.B.; Bauchop, T. Ultrastructural description of a new chytrid genus of caecum anaerobe, Caecomyces equi gen. nov., sp. nov., assigned to the Neocallimasticaceae. Biosystems 1988, 21, 403–415. [Google Scholar] [CrossRef]
- Breton, A.; Dusser, M.; Gaillard-Matinie, B.; Guillot, J.; Millet, L.; Prensier, G. Piromyces rhizinflata nov. sp., a strictly anaerobic fungus from faeces of the Saharian ass: A morphological, metabolic and ultrastructural study. FEMS Microbiol. Lett. 1991, 66, 1–8. [Google Scholar] [CrossRef]
- Heath, B.I.; Bauchop, T.; Skipp, R.A. Assignment of the rumen anaerobe Neocallimastix frontalis to the Spizellomycetales (Chytridiomycetes) on the basis of its polyflagellate zoospore ultrastructure. Can. J. Bot. 1983, 61, 295–307. [Google Scholar] [CrossRef]
- Dagar, S.S.; Kumar, S.; Griffith, G.W.; Edwards, J.E.; Callaghan, T.M.; Singh, R.; Nagpal, A.K.; Puniya, A.K. A new anaerobic fungus (Oontomyces anksri gen. nov., sp. nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol. 2015, 119, 731–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callaghan, T.M.; Podmirseg, S.M.; Hohlweck, D.; Edwards, J.E.; Puniya, A.K.; Dagar, S.S.; Griffith, G.W. Buwchfawromyces eastonii gen. nov., sp. nov.: A new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. MycoKeys 2015, 9, 11–28. [Google Scholar]
- Hanafy, R.A.; Elshahed, M.S.; Liggenstoffer, A.S.; Griffith, G.W.; Youssef, N.H. Pecoramyces ruminantium, gen. nov., sp. nov., an anaerobic gut fungus from the feces of cattle and sheep. Mycologia 2017, 109, 231–243. [Google Scholar] [CrossRef]
- Hanafy, R.A.; Elshahed, M.S.; Youssef, N.H. Feramyces austinii, gen. nov., sp. nov., an anaerobic gut fungus from rumen and fecal samples of wild Barbary sheep and fallow deer. Mycologia 2018, 110, 513–525. [Google Scholar] [CrossRef]
- Hanafy, R.A.; Lanjekar, V.B.; Dhakephalkar, P.K.; Callaghan, T.M.; Dagar, S.S.; Griffith, G.W.; Elshahed, M.S.; Youssef, N.H. Seven new Neocallimastigomycota genera from wild, zoo-housed, and domesticated herbivores greatly expand the taxonomic diversity of the phylum. Mycologia 2020. [Google Scholar] [CrossRef]
- Ho, Y.W.; Barr, D.J.S. Classification of anaerobic gut fungi from herbivores with emphasis on rumen fungi from malaysia. Mycologia 1995, 87, 655–677. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R.; et al. Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015, 75, 27–274. [Google Scholar] [CrossRef]
- Breton, A.; Bernalier, A.; Dusser, M.; Fonty, G.; Gaillard-Martinie, B.; Guitlot, J. Anaeromyces mucronatus nov. gen., nov. sp. A new strictly anaerobic rumen fungus with polycentric thallus. FEMS Microbiol. Lett. 1990, 70, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Ozkose, E.; Thomas, B.J.; Davies, D.R.; Griffith, G.W.; Theodorou, M.K. Cyllamyces aberensis gen.nov. sp.nov., a new anaerobic gut fungus with branched sporangiophores isolated from cattle. Can. J. Bot. 2001, 79, 666–673. [Google Scholar]
- Liggenstoffer, A.S.; Youssef, N.H.; Couger, M.B.; Elshahed, M.S. Phylogenetic diversity and community structure of anaerobic fungi (Phylum Neacallymastigales) in ruminant and non-ruminant herbivores. ISME J. 2010, 4, 1225–1235. [Google Scholar] [CrossRef]
- Kittelmann, S.; Naylor, G.E.; Koolaard, J.P.; Janssen, P.H. A proposed taxonomy of anaerobic fungi (class Neocallimastigomycetes) suitable for large-scale sequence-based community structure analysis. PLoS ONE 2012, 7, e36866. [Google Scholar] [CrossRef]
- Kittelmann, S.; Seedorf, H.; Walters, W.A.; Clemente, J.C.; Knight, R.; Gordon, J.I.; Janssen, P.H. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS ONE 2013, 8, e47879. [Google Scholar] [CrossRef]
- Koetschan, C.; Kittelmann, S.; Lu, J.; Al-Halbouni, D.; Jarvis, G.N.; Muller, T.; Wolf, M.; Janssen, P.H. Internal transcribed spacer 1 secondary structure analysis reveals a common core throughout the anaerobic fungi (Neocallimastigomycota). PLoS ONE 2014, 9, e91928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanafy, R.A.; Johnson, B.; Youssef, N.H.; Elshahed, M.S. Assessing anaerobic gut fungal diversity in herbivores using D1/D2 large ribosomal subunit sequencing and multi-year isolation. Environ. Microbiol. 2020, 22, 3883–3908. [Google Scholar] [CrossRef]
- Calkins, S.; Elledge, N.C.; Hanafy, R.A.; Elshahed, M.S.; Youssef, N.H. A fast and reliable procedure for spore collection from anaerobic fungi: Application for RNA uptake and long-term storage of isolates. J. Microbiol. Meth. 2016, 127, 206–213. [Google Scholar] [CrossRef] [Green Version]
- Hungate, R.E. A roll tube method for cultivation of strict anaerobes. Meth. Microbiol. 1969, 3, 117–132. [Google Scholar]
- Lowe, S.E.; Theodorou, M.K.; Trinci, A.P.; Hespell, R.B. Growth of anaerobic rumen fungi on defined and semi-defined media lacking rumen fluid. J. Gen. Microbiol. 1985, 131, 2225–2229. [Google Scholar] [CrossRef] [Green Version]
- Marvin-Sikkema, F.D.; Richardson, A.J.; Stewart, C.S.; Gottschal, J.C.; Prins, R.A. Influence of hydrogen-consuming bacteria on cellulose degradation by anaerobic fungi. Appl Environ. Microbiol 1990, 56, 3793–3797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Nicholson, M.J.; McSweeney, C.S.; Mackie, R.I.; Brookman, J.L.; Theodorou, M.K. Diversity of anaerobic gut fungal populations analysed using ribosomal ITS1 sequences in faeces of wild and domesticated herbivores. Anaerobe 2009, 16, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.S.; Bu, D.; Xu, J.; Hyde, K.D.; Yu, Z. A phylogenetic census of global diversity of gut anaerobic fungi and a new taxonomic framework. Fungal Divers. 2018, 89, 253–266. [Google Scholar] [CrossRef]
- Wang, Y.; Youssef, N.H.; Couger, M.B.; Hanafy, R.A.; Elshahed, M.S.; Stajich, J.E. Molecular dating of the emergence of anaerobic rumen fungi and the impact of laterally acquired genes. Msystems 2019, 4, e00247-19. [Google Scholar] [CrossRef] [Green Version]
- Lowe, S.E.; Griffith, G.G.; Milne, A.; Theodorou, M.K.; Trinci, A.P.J. The life cycle and growth kinetics of an anaerobic rumen fungus. Microbiology 1987, 133, 1815–1827. [Google Scholar] [CrossRef] [Green Version]
- Webb, J.; Theodorou, M.K. Neocallimastix hurleyensis sp.nov., an anaerobic fungus from the ovine rumen. Can. J. Bot. 1991, 69, 1220–1224. [Google Scholar] [CrossRef]
- Ahrendt, S.R.; Quandt, C.A.; Ciobanu, D.; Clum, A.; Salamov, A.; Andreopoulos, B.; Cheng, J.-F.; Woyke, T.; Pelin, A.; Henrissat, B.; et al. Leveraging single-cell genomics to expand the fungal tree of life. Nat. Microbiol. 2018, 3, 1417–1428. [Google Scholar] [CrossRef] [Green Version]
- Davis, W.J.; Amses, K.R.; James, E.S.; James, T.Y. A new 18S rRNA phylogeny of uncultured predacious fungi (Zoopagales). Mycologia 2019, 111, 291–298. [Google Scholar] [CrossRef] [PubMed]
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a fully resolved fungal tree of life. Ann. Rev. Microbiol 2020, 74, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef] [Green Version]
- Joshi, A.; Lanjekar, V.B.; Dhakephalkar, P.K.; Callaghan, T.M.; Griffith, G.W.; Dagar, S.S. Liebetanzomyces polymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. MycoKeys 2018, 40. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stabel, M.; Hanafy, R.A.; Schweitzer, T.; Greif, M.; Aliyu, H.; Flad, V.; Young, D.; Lebuhn, M.; Elshahed, M.S.; Ochsenreither, K.; et al. Aestipascuomyces dupliciliberans gen. nov, sp. nov., the First Cultured Representative of the Uncultured SK4 Clade from Aoudad Sheep and Alpaca. Microorganisms 2020, 8, 1734. https://doi.org/10.3390/microorganisms8111734
Stabel M, Hanafy RA, Schweitzer T, Greif M, Aliyu H, Flad V, Young D, Lebuhn M, Elshahed MS, Ochsenreither K, et al. Aestipascuomyces dupliciliberans gen. nov, sp. nov., the First Cultured Representative of the Uncultured SK4 Clade from Aoudad Sheep and Alpaca. Microorganisms. 2020; 8(11):1734. https://doi.org/10.3390/microorganisms8111734
Chicago/Turabian StyleStabel, Marcus, Radwa A. Hanafy, Tabea Schweitzer, Meike Greif, Habibu Aliyu, Veronika Flad, Diana Young, Michael Lebuhn, Mostafa S. Elshahed, Katrin Ochsenreither, and et al. 2020. "Aestipascuomyces dupliciliberans gen. nov, sp. nov., the First Cultured Representative of the Uncultured SK4 Clade from Aoudad Sheep and Alpaca" Microorganisms 8, no. 11: 1734. https://doi.org/10.3390/microorganisms8111734
APA StyleStabel, M., Hanafy, R. A., Schweitzer, T., Greif, M., Aliyu, H., Flad, V., Young, D., Lebuhn, M., Elshahed, M. S., Ochsenreither, K., & Youssef, N. H. (2020). Aestipascuomyces dupliciliberans gen. nov, sp. nov., the First Cultured Representative of the Uncultured SK4 Clade from Aoudad Sheep and Alpaca. Microorganisms, 8(11), 1734. https://doi.org/10.3390/microorganisms8111734





